Abstract
Bacteriophage λ integrase (Int) catalyzes the integration and excision of the phage λ chromosome into and out of the Esherichia coli host chromosome. The seven carboxy-terminal residues (C-terminal tail) of Int comprise a context-sensitive regulatory element that links catalytic function with protein multimerization and also coordinates Int functions within the multimeric recombinogenic complex. The experiments reported here show that the β5-strand of Int is not simply a placeholder for the C-terminal tail but rather exerts its own allosteric effects on Int function in response to the incoming tail. Using a mutant integrase in which the C-terminal tail has been deleted (W350ter), we demonstrate that the C-terminal tail is required for efficient and accurate resolution of Holliday junctions by tetrameric Int. Addition of a free heptameric peptide of the same sequence as the C-terminal tail partially reverses the W350ter defects by stimulating Holliday junction resolution. The peptide also stimulates the topoisomerase function of monomeric W350ter. Single residue alterations in the peptide sequence and a mutant of the β5 strand indicate that the observed stimulation arises from specific contacts with the β5 strand (residues 239–243). The peptide does not stimulate binding of W350ter to its cognate DNA sites and therefore appears to recapitulate the effects of the normal C-terminal tail intermolecular contacts in wild-type Int. Models for the allosteric stimulation of Int activity by β5 strand contacts are discussed.
Keywords: bacteriophage lambda, integrase, Holliday junction, site-specific recombination, peptide
Bacteriophage lambda integrase (Int) is a tyrosine site-specific recombinase that functions as a higher-order tetrameric complex to catalyze the integration and excision of the viral chromosome into and out of the Escherichia coli chromosome by means of a highly coordinated sequence of cleavage and ligation reactions at specific loci (att sites, see Figure 1(a)).1 Previous work has established that the seven carboxy-terminal residues (C-terminal tail) of Int are integral to regulatory mechanisms that link catalytic function with protein multi-merization and also coordinate Int functions within the multimeric recombinogenic complex.2–6 Here we extend these results by demonstrating that the β5 strand of Int is not simply a placeholder for the multi-positional C-terminal tail but rather exerts its own allosteric effects on Int function in response to the incoming tail.
Figure 1.
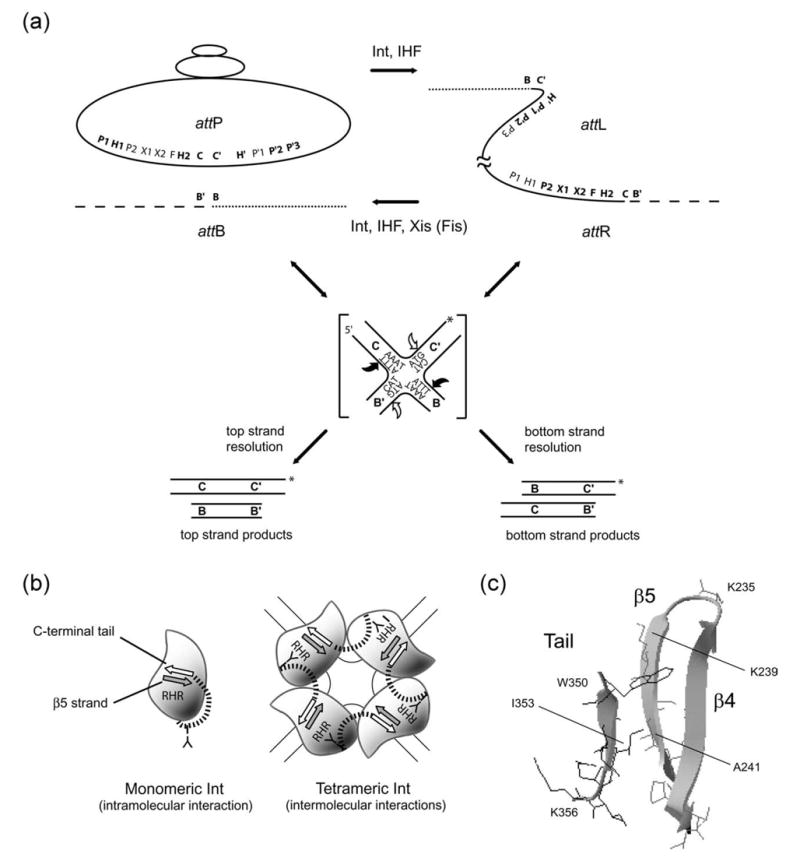
Schematic diagram of the λ site-specific recombination reactions and the alternate positions of the carboxy-terminal tail of Int. (a) Integrative recombination between attP and attB generates an integrated prophage bounded by the product sites, attL and attR, which are also partners in the excisive recombination that regenerates attP and attB. The indicated proteins required for each reaction use overlapping ensembles of DNA sites, indicated in bold for each reaction: core-type sites (C, C′, B, and B′), bound by the catalytic domain of Int, and the arm-type sites (P1, P2, P′1, P′2, and P′ 3), bound by the amino-terminal domain of Int, are separated by sites for the accessory DNA bending proteins IHF (H1, H2, and H′), Xis (X1 and X2), and Fis (F). A mimic of the Holliday junction recombination intermediate (indicated in brackets) was fabricated by annealing four synthetic DNA strands containing only the Int core-sites; it was used to assay the features of Int-dependent Holliday junction resolution. The Holliday junction used in these experiments has been described previously14; it is analogous to the intermediate formed during integrative recombination and has branch lengths of 24 (C), 23 (C′), 15 (B), and 16 (B′) bp. A pair of coordinated Int cleavages (black arrows) at the top strand core-type sites (C and B) generates a pair of product duplexes (COC′ and BOB′) that can be electrophoretically separated from the products (BOC′ and COB′) that are generated by cleavages (white arrows) at the bottom strand core-type sites (C′ and B′). A unique 32P-label (asterisk) on the C’OC strand of the Holliday junction is transferred to one of the products in each of the two resolution reactions. (b) Schematic cartoon of intra versus inter-molecular docking of the bi-positional C-terminal tail within the catalytic domain(s) of Int (the N-domains of Int are not depicted). In monomeric Int, intramolecular contacts between the C-terminal tail (white arrow) and strand β5 (gray arrow) pull the Tyr342 nucleophile (Y) away from the active site RHR triad. Intermolecular contacts, which form upon tetramerization (depicted here on a Holliday junction), position Tyr342 proximal to the active site triad of the donating Int protomer. (c) A structural view of the interacting surfaces between the carboxy-terminal tail and β5 (adapted from Aihara et al.4). Residues referred to in the text are identified.
Int is a member of the tyrosine recombinase family, whose hallmark is an ordered pair of transesterification reactions that first generate and then resolve a Holliday junction recombination intermediate (Figure 1(a)).7–9 The transesterification reactions are mediated by transient covalent 3′ phospho-tyrosine bonds that are formed at each of the four “core-type” Int binding sites. Within the tyrosine family of recombinases, Int belongs to the subgroup of heterobivalent DNA binding proteins that bind and bridge two distinct and distant DNA sequences. Int possesses a small amino-terminal domain (residues 1–70) that binds with high affinity to “arm-type” DNA sites that are distant from lower affinity core-type sites where the carboxy-terminal domain (residues 75–365) binds and executes DNA strand cleavage and ligation. Int-mediated bridging between the arm and core-type sites is assisted by DNA bends induced by accessory proteins (IHF, Xis, and Fis) bound to sites located between the two types of Int binding sequences. Among the ensemble of five arm-type Int binding sites and six sites for the accessory DNA-bending proteins, two overlapping subsets of sites are used to assemble the higher-order “intasomes” that will execute either integrative or excisive recombination (Figure 1(a)).
A deletion of the carboxy-terminal residues 350–356 of Int (W350ter) was first isolated in a selection for dominant negative mutants.10,11 It was independently re-isolated and functionally characterized by the Gardner and Gumport laboratory in a selection for Int mutants defective in different steps in recombination.6 They demonstrated that although W350ter was defective in catalyzing recombination, it displayed enhanced topoisomerase activity relative to wild-type Int.
More recent work has greatly elaborated on the complex role of the C-terminal tail.2,3,5 In monomeric Int, the C-terminal tail docks against the β5 strand (residues 239–243). As a result of these intramolecular contacts, the Tyr342 nucleophile is held approximately 20 Å away from the active site triad residues R212, H308, and R311,12 resulting in the repression of monomeric activity (Figure 1(b)). Consistent with the predictions of this structure, deletion of the C-terminal tail alleviates this repression and enhances the monomeric activity of Int.2,5,6 In contrast, in an Int tetramer, which likely comprises the functional core of the recombinogenic complex, the C-terminal tail contacts the β5 strand of a neighboring Int to form a cyclic arrangement of linkages.13 As a result of these intermolecular contacts (Figure 1(c)), the Tyr342 nucleophile is positioned near the active site (Figure 1(b)).4,13 When Int functions as a multimer, such as during cleavage of a full att site, the C-terminal tail enhances activity and, as a result, W350ter is deficient in these functions.5 These data indicate that, when present, the C-terminal tail acts as a damper of monomeric Int activity, while it acts to stimulate multimeric Int function. The C-terminal tail thus affords an Int regulatory mechanism that links catalytic function with protein multimerization, ensuring that the required chromosomal cleavages occur only in the context of the multi-meric recombinogenic complex.
In contrast to the large amount of information available regarding the C-terminal tail, very little is known about the role and function of the β5 strand. We have therefore designed experiments to test whether the β5 strand exerts any influence upon Int function as a consequence of its contact with the C-terminal tail. We first show that the C-terminal tail is required for efficient and accurate resolution of Holliday junctions into canonical top and bottom strand products. We then demonstrate that the β5 strand is more than a passive docking site for the C-terminal tail of a neighboring Int. Rather, β5 responds to the incoming tail with an allosteric stimulation of Int activity and fidelity.
C-terminal tail is required for efficient and coordinated Holliday junction resolution
To assay the ability of W350ter to resolve Holliday junction recombination intermediates we used as a substrate the previously characterized synthetic Holliday junction, referred to as CM7.14 As a result of the four different arm lengths on this Holliday junction, the resolution products resulting from cleaving either a pair of top or bottom strands differ from each other, and are readily distinguished by gel electrophoresis (Figure 1(a) lower).14,15 The reactions were carried out in the presence of oligonucleotides encoding the P′1 and P′2 arm-type sites under conditions that optimize the fidelity of resolution, i.e. the least amount of aberrant resolution products.14
We observe that W350ter is severely deficient in the ability to resolve synthetic Holliday junctions and is also greatly diminished in the accuracy with which it carries out resolution (Figure 2(a)). Over a range of protein concentrations, the reduced accuracy of W350ter is reflected by the higher ratio of aberrant products that migrate faster than the normal resolution products that predominate with wild-type Int (Figure 2(b)). We believe that these aberrant hairpin products arise from uncoordinated DNA cleavages at “non-partner” Int binding sites, either during or after resolution.14,16 These findings extend previous results2,3,5,6 indicating that while the C-terminal tail is not required for DNA cleavage by monomeric Int, it plays a critical role in the function and proper coordination of multimeric Int.
Figure 2.
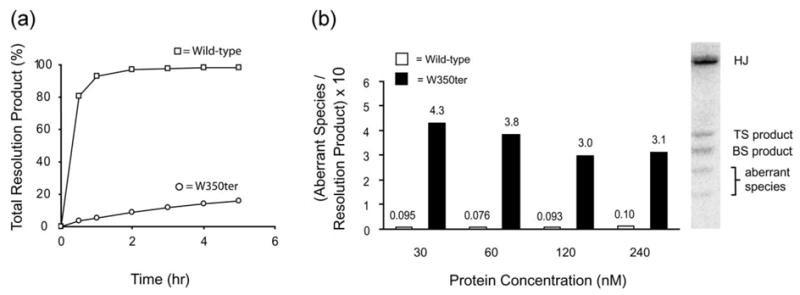
The C-terminal tail is required for efficient and accurate Holliday junction resolution. (a) Amount of total Holliday junction resolution product (top and bottom strand cleavages) by wild-type Int or W350ter. (b) Ratio of aberrant species to total Holliday junction resolution product at indicated protein concentrations of wild-type Int (open bars) and W350ter (filled bars). The aberrant species are likely single arm hairpins generated by uncoordinated Int cleavages, either during or after resolution.14,16 All Holliday junction resolution reactions ((a) and (b) and Figure 3(a)) were incubated at 20 °C and contain 120 nM integrase in assay volumes ranging from 10–100 μl with 50 mM Tris–HCl (pH 7.5), 50 mM NaCl, 0.5 mg/ml bovine serum albumin, 5 mM dithiothreitol, 10 nM 32P-labeled Holliday junction substrate, and 240 nM P′ 1,2 arm oligonucleotides. Reactions were quenched with a SDS gel loading solution and separated on 7% (w/v) polyacrylamide Tris–glycine (pH 8.0), 0.1% (w/v) SDS gels. Gels were subsequently dried and analyzed with the Fuji BAS-2500 phosphorimager system. Wild-type and mutant Int proteins were prepared as described.23 Int mutations were created via site-directed PCR mutagenesis with a Quikchange kit (Stratagene). Protein concentrations were determined by comparing the intensities of Coomasie-stained Int bands on an SDS-PAGE gel with those of a reference Int preparation quantified by amino acid composition (WM Keck Facility, Yale University). Preparation and sequences of synthetic Holliday junctions and P′ 1,2 oligonucleotides have been described.14
Intermolecular contacts between β5 and the C-terminal tail stimulate Holliday junction resolution
To isolate the known effects of the C-terminal tail in positioning the Tyr342 nucleophile from any potential consequences of its interaction with the β5 strand, we attempted to compensate for the missing tail in W350ter by adding to reactions a heptameric peptide of the same sequence as the C-terminal tail. We reasoned that by providing the C-terminal tail sequence in solution as a free peptide (n-WDKIEIK-c), it would be unable to affect the position of the Tyr342 nucleophile but could still bind and interact with the β5 strand.
In assays containing W350ter, 32P-labeled Holliday junction substrate, and P′1,2 oligonucleotides, the addition of 2 mg/ml tail peptide stimulates the rate of Holliday junction resolution (Figure 3(a)). The observed stimulation is peptide concentration-dependent (Figure 3(b)). The observed stimulation is also sequence-specific, as addition of 2 mg/ml heptameric peptide in which the wild-type sequence is scrambled (n-DIWKKIE-c) fails to stimulate W350ter resolution activity (Figure 3(a)). We additionally assayed the addition of 2 mg/ml of two tail peptides containing the point mutations W350G (n-GDKIEIK-c) and I353M (n-WDKMEIK-c), which we predicted from previous studies would disrupt specific contacts between the C-terminal tail and the β5 strand (Figure 1(c)).2–4 Both of these mutant peptides fail to stimulate W350ter resolution activity (Figure 3(a)).
Figure 3.
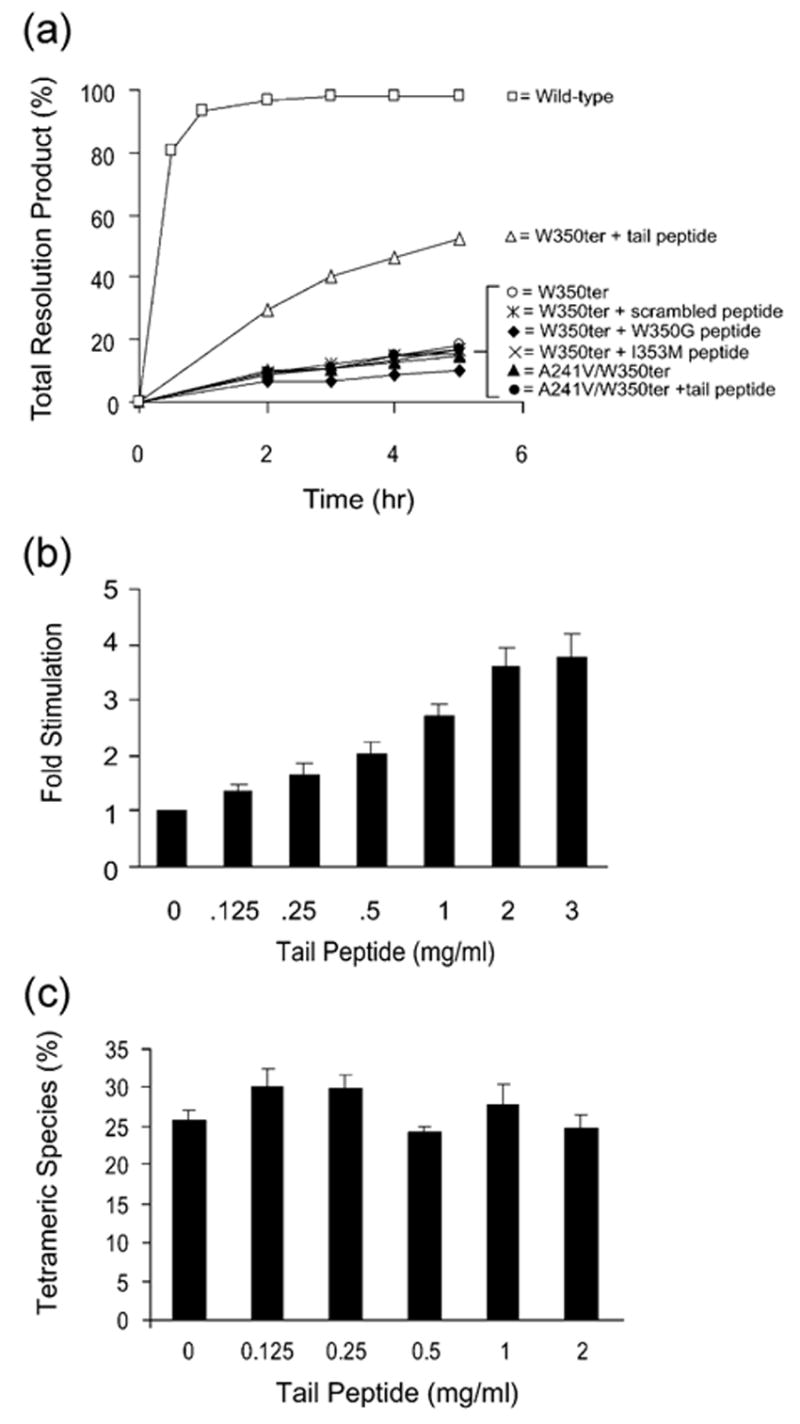
W350ter Holliday junction resolution activity is specifically stimulated by the heptameric C-terminal tail peptide. (a) Holliday junction resolution by the indicated wild-type or mutant Int proteins in the absence or presence of the indicated heptameric peptide (2 mg/ml). (b) Relative fold stimulation of W350ter Holliday junction resolution by tail peptide in varying concentrations in comparison to reactions without tail peptide incubated at 20 °C for four hours. (c) Gel mobility shift assay of 32P-labeled Holliday junction incubated with W350ter at 20 °C for ten minutes in the absence or presence of heptameric C-terminal tail peptide. Reaction conditions were the same as the resolution assays (Figures 2(a) and (b) and 3(a)) except for the presence of EDTA (1 mM) and glycerol (5%). Binding reactions were analyzed by electrophoresis at 25 °C on 7% polyacrylamide Tris–borate–EDTA (pH 8.1) gels. Quantification with a Fuji BAS-2500 phosphorimager was used to determine the percent of tetrameric Int–Holliday junction complex; any small amounts of resolution product (less than 3%) were considered to have been part of the Int–Holliday junction complex. The concentration of W350ter was determined by titration to be well within the linear range for tetramer formation and the amount of tetrameric complex bound at ten minutes is below half-maximal (data not shown). All peptides were custom synthesized and HPLC purified to 95% purity by AnaSpec, Inc. and stored lyophilized at −20 °C. Stock solutions of peptide were dissolved in sterile, de-ionized water to a concentration of 4 mg/ml and stored frozen at either −20 °C or in liquid nitrogen.
As an additional control for peptide specificity, we assayed the A241V mutation in the background of the W350ter mutation. A241 is a residue in β5 that has been functionally implicated in interactions with the C-terminal tail (Figure 1(c)),2,4 and we predicted that this mutation may disrupt the tail–β5 strand contacts. As predicted, the double mutant A241V-W350ter is not stimulated by the addition of 2 mg/ml of wild-type heptameric tail-peptide (Figure 3(a)). Mixing of the of tail peptide with wild-type Int (either prior to or after the addition of Holliday junction substrate) has no stimulatory or inhibitory effect on the resolution activity of wild-type Int (data not shown), suggesting, as expected, that the tail peptide is unable to out-compete the covalently attached tail of wild-type Int. Of note, the heptameric tail peptide increases the fidelity (i.e. resolution products/aberrant species) of W350ter by virtue of stimulating the formation of resolution products, while the amount of aberrant species appears to remain constant in both the presence and absence of peptide (data not shown). Taken together, these data indicate that the heptameric C-terminal tail peptide added in solution is indeed stimulating Holliday junction resolution by W350ter via direct interaction with the β5 strand.
The C-terminal tail is not thought to play a significant role in DNA binding because W350ter and wild-type Int bind core-type sites similarly.2 If the heptameric peptide is indeed closely mimicking the interactions of the C-terminal tail, it should not affect binding to core-type Int sites. This was tested in an electrophoretic gel mobility shift assay for the formation of tetrameric complexes between W350ter and 32P-labeled Holliday junction (Figure 1(a)). The presence of increasing concentrations of peptide in the binding mixtures does not increase the amount of tetrameric W350ter–Holliday junction complex relative to the absence of peptide (Figure 3(c)), indicating that the observed peptide stimulation of W350ter resolution activity is not due to a significant enhancement of its core-type site binding affinity. The differing effects of the tail peptide, i.e. the increase of W350ter resolution activity versus no enhancement of Holliday junction binding, are consistent with the suggestion that the peptide serves as a valid model for the canonical interactions between the C-terminal tail and the β5 strand.
Peptide stimulation of topoisomerase activity
All of the experiments described thus far, which indicate that the β5 strand stimulates Int function in response to contacts by the heptameric tail peptide, were carried out in the context of a tetrameric complex (Holliday junction resolution). We next sought to examine the tail peptide’s effect on monomeric Int acting as a topoisomerase. It was previously shown that W350ter has higher topoisomerase activity than wild-type Int.2,6 We hypothesized that this was due to the fact that the Tyr342 nucleophile in W350ter was not pulled out of the catalytic pocket by a C-terminal tail making intramolecular contacts with β5, as it does in wild-type Int.2,12 If this is correct, the data reported above strongly imply that the previously observed topoisomerase activity of W350ter reflects the net effect of two opposing consequences of deleting the C-terminal tail: (1) enhancement of activity, due to removal of the C-terminal tail influence on Tyr342; and (2) reduction of activity, due to loss of the contacts with β5. If such a competition exists, W350ter should have an even greater topoisomerase activity in the presence of the heptameric tail peptide.
In order to assay Int topoisomerase activity we used gel electrophoresis to visualize the conversion of supercoiled plasmid DNA to species with a lower linking number (slower mobility) and leading ultimately to fully relaxed DNA.17 Consistent with the data and interpretations presented above, we find that addition of 1 mg/ml of heptameric tail peptide further stimulates the topoisomerase activity of W350ter, which, as reported earlier, is already greater than wild-type Int (Figure 4). In agreement with the results of the resolution assays, the addition of 1 mg/ml of control peptides W350G and I353M to the W350ter reactions, as well as addition of 1 mg/ml of tail peptide to the reactions with double mutant A241V/W350ter, does not significantly stimulate topoisomerase activity (Figure 4).
Figure 4.
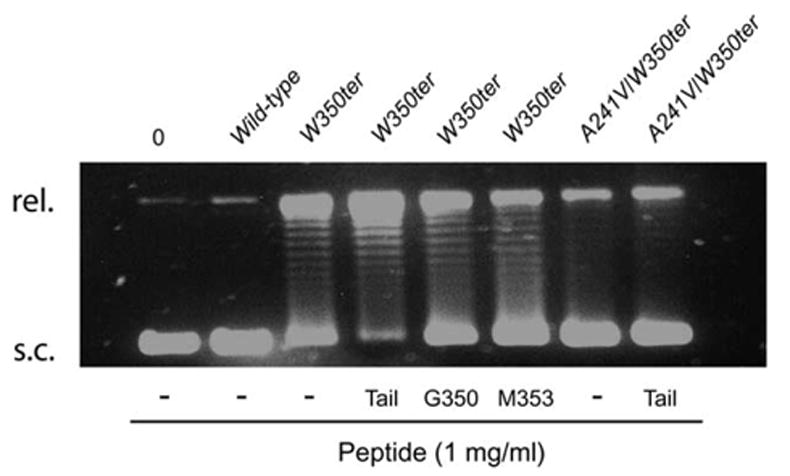
Tail peptide effects on topoisomerase activity. Topoisomerase assay of wild-type Int, W350ter, and A241V-W350ter in the absence or presence of the heptameric C-terminal tail peptide (Tail), and W350G and I353M peptides. Activity was assayed by the conversion of 0.4 μg of pBR327 supercoiled DNA (s.c.) to a relaxed circle (rel.). Reactions were incubated at 25 °C for 15 minutes in the same buffer as the Holliday junction resolutions (Figure 2), except for the presence of EDTA (1 mM), and contained 120 nM of protein. Reactions were quenched with gel loading buffer containing 1% SDS, electrophoresed on 1% (w/v) agarose Tris–acetate–EDTA (pH 8.2) gels, and visualized by staining with ethidium bromide.
Implications for intermolecular stimulation by the C-terminal tail in recombination
The results reported here show that the C-terminal tail of λ Int is required for efficient and accurate resolution of synthetic Holliday junctions (Figures 2(a) and (b) and 3(a)). Preliminary analysis shows that E349ter has a phenotype similar to W350ter and can also be stimulated by heptameric peptide (data not shown). Of greatest interest is the finding that the W350ter defect is partially reversed by addition of the C-terminal heptameric peptide in solution (Figure 3(a)).
Peptides carrying point mutations directed to specific contacts on β5 failed to stimulate resolution, and a mutation within β5 believed to disrupt interactions with the C-terminal tail was not stimulated by the wild-type peptide (Figure 3(a)). Together these results strongly indicate that contacts between the C-terminal tail and the β5 strand4,12,13 are responsible for mediating the stimulatory effects of the heptameric tail peptide.
It is not surprising that the peptide is unable to restore the resolution activity of W350ter back to the levels of wild-type Int, where the local tail concentration is much higher (peptide concentrations above 3 mg/ml are inhibitory). Additionally, the C-terminal tail peptide and/or the Tyr342 nucleophile may not be oriented precisely as they are in the wild-type Int, since the physical linkage provided by the cyclic intermolecular interactions of the C-terminal tail is not recapitulated by the free tail peptide. Indeed, the fact that the peptide is as effective as it is in stimulating W350ter, along with previous biochemical results on monomeric Int functions, indicates that the Tyr342 nucleophile can be positioned in the active site without the assistance of the C-terminal tail.
The coordination of sequential pairs of DNA cleavage, exchange, and ligation is an essential element in the pathway of all tyrosine recombinases. A common motif for achieving some of this coordination involves the intermolecular contact between a “donated” carboxy-terminal element and an “acceptor” target on a neighboring protomer. For the Cre recombinase of phage P1, crystallographic evidence suggests that the C-terminal helix M-N linker region contacts strand β3 and could modulate the positioning of the β2-β3 loop, which contains K201 (analogous to λ K235), within the active site of active Cre protomers.9 In the XerC/D recombinases of E. coli, C-terminal contacts between XerC and XerD are likewise implicated in the control and coordination of cleavage activity,18,19 underscoring the commonality of this regulatory motif throughout the members of the tyrosine recombinase family.
Crystallographic data have been indispensable in offering insight into to the various functional “lifestyles” of the C-terminal tail. The crystal structure of the catalytic domain of λ Int at 1.9 Å offers a clear view of the intramolecular contacts between the C-terminal tail and strand β5 that likely occur in monomeric Int in solution.12 In this structure, the C-terminal tail is secured to β5 by a series of hydrogen bonds and hydrophobic interactions involving residues W350 and I353, among others. These intramolecular contacts would appear to render Int inactive, since they position the Tyr342 nucleophile far from the active site. Although NMR studies indicate this region is fairly mobile in the absence of DNA,20 biochemical studies are consistent with the inferences from the crystal structure.2,3,5,6,12
The co-crystal structure of C75 Int and att-site DNA at 3 Å clearly models a rearrangement and extension of the C-terminal tail to form intermolecular contacts with β5 of a neighboring protomer in a similar fashion as the intramolecular contacts seen in monomeric Int, with the exception of an apparently unique salt bridge that forms between E349 and K239.4 More importantly, this structure shows how the intermolecular C-terminal tail contacts could permit the placement of Tyr342 in the active site, which, by virtue of the design of the suicide att-site substrate DNA, is covalently linked to the scissile phosphate. Recent crystal structures of Int tetramers in complex with Holliday junctions depict the cyclic nature of these intermolecular contacts between the C-terminal tail and β5.13 Interestingly, the structures suggest that these contacts differ in the active and inactive subunits. This results in a structural asymmetry that may locally alter the position of Tyr342 in the active site and thus provide a mechanistic basis for coordination of the cleavage activity of partner Ints.13
The Int crystal structures are congruent with and support the biochemical and genetic studies on the role of the C-terminal tail (see also above). Because of the limits of resolution and the static nature of the crystal structures however, they yield little insight on the potential mechanistic role of the β5 strand in response to contacts from a neighboring C-terminal tail. Based upon the results presented here, we conclude that the β5 strand responds to the presence of a donated C-terminal tail by allosterically stimulating DNA cleavage and ligation. The molecular basis of this stimulation is not known, though it is plausible that contact between the C-terminal tail and β5 may act to alter the position of the β4-β5 loop, which contains K235 (see Figure 1(c)), an essential and conserved catalytic residue in Int (S. N. Düby, unpublished results), Cre, and XerD.9 Indeed, the positioning of this particular lysine and its homologues in other recombinases has been speculated as a common means of active site regulation within the tyrosine recombinase family.9 K235 is believed to be directly involved in the chemistry of cleavage and ligation, possibly as a general acid in a manner similar to the analogous residue (K167) in vaccinia topoisomerase IB,7,21 and structural data indicate that K235 is poised within the active site in a position favorable for proton-donation or transition-state intermediate stabilization.4 Additional studies have indicated that Thr236, another β4-β5 loop residue, is also required for catalytic function, pointing to another potential avenue for the β5 strand to exert an influence on catalysis.6,22
Another possible mechanism for the β5-mediated allosteric stimulation is the promotion of new or enhanced protein–protein interactions within the Int tetrameric complex. Evidence for or against this class of mechanisms might come from crystal structures of tetrameric Int–Holliday junction complexes containing W350ter Int and/or genetic and biochemical experiments designed to address this specific question.
The present experiments clearly demonstrate that an Int protomer is stimulated by specific intermolecular contacts with the β5 strand. We do not know, however, if this stimulation reflects a basal activation of all four Int protomers by the tail, or if it reflects the differential stimulation that distinguishes active from inactive partner Ints, i.e. whether some other structural feature of the recombinogenic complex modulates the β5 stimulations described here.
Acknowledgments
We thank Tapan Biswas and Hideki Aihara of the Ellenberger laboratory, Troy Shirangi, and members of the Landy Laboratory for helpful discussions and comments on the manuscript, Christine Lank for technical assistance and Joan Boyles for help with preparation of the manuscript. This work was supported by NIH grants GM33928 and GM2723 to A.L.
Abbreviations used
- HJ
Holliday junction
- TS
top strand
- BS
bottom strand
- s.c.
supercoiled
- rel.
relaxed
Footnotes
Edited by J. Karn
References
- 1.Campbell A. Episomes. Advan Genet. 1962;11:104–145. [Google Scholar]
- 2.Tekle M, Warren DJ, Biswas T, Ellenberger T, Landy A, Nunes-Duby SE. Attenuating functions of the C terminus of lambda integrase. J Mol Biol. 2002;324:649–665. doi: 10.1016/s0022-2836(02)01108-7. [DOI] [PubMed] [Google Scholar]
- 3.Kazmierczak RA, Swalla BM, Burgin AB, Gumport RI, Gardner JF. Regulation of site-specific recombination by the C-terminus of lambda integrase. Nucl Acids Res. 2002;30:5193–5204. doi: 10.1093/nar/gkf652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aihara H, Kwon HJ, Nunes-Duby SE, Landy A, Ellenberger T. A conformational switch controls the DNA cleavage activity of lambda integrase. Mol Cell. 2003;12:187–198. doi: 10.1016/s1097-2765(03)00268-5. [DOI] [PubMed] [Google Scholar]
- 5.Lee SY, Aihara H, Ellenberger T, Landy A. Two structural features of lambda integrase that are critical for DNA cleavage by multimers but not by monomers. Proc Natl Acad Sci USA. 2004;101:2770–2775. doi: 10.1073/pnas.0400135101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han YW, Gumport RI, Gardner JF. Mapping the functional domains of bacteriophage lambda integrase protein. J Mol Biol. 1994;235:908–925. doi: 10.1006/jmbi.1994.1048. [DOI] [PubMed] [Google Scholar]
- 7.Azaro M, Landy A. Lambda integrase and the lambda Int family. In: Craig NL, Craigie R, Gellert M, Lambovitz A, editors. Mobile DNA II. ASM Press; Washington DC: 2002. pp. 118–148. [Google Scholar]
- 8.Nash H. Site-specific recombination: integration, excision, resolution, and inversion of defined DNA segments. In: Neidhardt FC, Curtiss RI, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press; Washington, DC: 1996. pp. 2363–2376. [Google Scholar]
- 9.Van Duyne GD. A structural view of tyrosine recombinase site-specific recombination. In: Craig NL, Craigie R, Gellert M, Lambovitz A, editors. Mobile DNA II. ASM Press; Washington DC: 2002. pp. 93–113. [Google Scholar]
- 10.Enquist LW, Weisberg RA. A genetic analysis of the att-int-xis region of coliphage lambda. J Mol Biol. 1977;111:97–120. doi: 10.1016/s0022-2836(77)80117-4. [DOI] [PubMed] [Google Scholar]
- 11.Bear SE, Clemens JB, Enquist LW, Zagursky RJ. Mutational analysis of the lambda int gene: DNA sequence of dominant mutations. J Bacteriol. 1987;169:5880–5883. doi: 10.1128/jb.169.12.5880-5883.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwon HJ, Tirumalai R, Landy A, Ellenberger T. Flexibility in DNA recombination: structure of the lambda integrase catalytic core. Science. 1997;276:126–131. doi: 10.1126/science.276.5309.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Biswas T, Aihara H, Radman-Livaja M, Filman D, Landy A, Ellenberger T. A structural basis for allosteric control of DNA recombination by lambda integrase. Nature. 2005;435:1059–1066. doi: 10.1038/nature03657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radman-Livaja M, Shaw C, Azaro M, Biswas T, Ellenberger T, Landy A. Arm sequences contribute to the architecture and catalytic function of a lambda integrase-Holliday junction complex. Mol Cell. 2003;11:783–794. doi: 10.1016/s1097-2765(03)00111-4. [DOI] [PubMed] [Google Scholar]
- 15.Hsu PL, Landy A. Resolution of synthetic att-site Holliday structures by the integrase protein of bacteriophage lambda. Nature. 1984;311:721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franz B, Landy A. Interactions between lambda Int molecules bound to sites in the region of strand exchange are required for efficient Holliday junction resolution. J Mol Biol. 1990;215:523–535. doi: 10.1016/s0022-2836(05)80165-2. [DOI] [PubMed] [Google Scholar]
- 17.Keller W, Wendel I. Stepwise relaxation of supercoiled SV40 DNA. Cold Spring Harbor Symp Quant Biol. 1975;39:199–208. doi: 10.1101/sqb.1974.039.01.026. [DOI] [PubMed] [Google Scholar]
- 18.Hallet B, Arciszewska LK, Sherratt DJ. Reciprocal control of catalysis by the tyrosine recombinases XerC and XerD: an enzymatic switch in site-specific recombination. Mol Cell. 1999;4:949–959. doi: 10.1016/s1097-2765(00)80224-5. [DOI] [PubMed] [Google Scholar]
- 19.Ferreira H, Butler-Cole B, Burgin A, Baker R, Sherratt DJ, Arciszewska LK. Functional analysis of the C-terminal domains of the site-specific recombinases XerC and XerD. J Mol Biol. 2003;330:15–27. doi: 10.1016/s0022-2836(03)00558-8. [DOI] [PubMed] [Google Scholar]
- 20.Subramaniam S, Tewari AK, Nunes-Duby SE, Foster MP. Dynamics and DNA substrate recognition by the catalytic domain of lambda integrase. J Mol Biol. 2003;329:423–439. doi: 10.1016/s0022-2836(03)00469-8. [DOI] [PubMed] [Google Scholar]
- 21.Krogh BO, Shuman S. Proton relay mechanism of general acid catalysis by DNA topoisomerase IB. J Biol Chem. 2002;277:5711–5714. doi: 10.1074/jbc.C100681200. [DOI] [PubMed] [Google Scholar]
- 22.Bankhead T, Segall AM. Characterization of a mutation of bacteriophage lambda integrase. Putative role in core binding and strand exchange for a conserved residue. J Biol Chem. 2000;275:36949–36956. doi: 10.1074/jbc.M004679200. [DOI] [PubMed] [Google Scholar]
- 23.Sarkar D, Radman-Livaja M, Landy A. The small DNA binding domain of lambda integrase is a context-sensitive modulator of recombinase functions. EMBO J. 2001;20:1203–1212. doi: 10.1093/emboj/20.5.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]


