Abstract
The HIV envelope glycoprotein (Env) is composed of two non-covalently associated subunits: gp120 and gp41. Panning of phage-displayed antibody libraries against Env-based antigens has resulted mostly in selection of anti-gp120 antibodies. Native gp41 in the absence of gp120 is unstable. The use of gp41 fragments as antigens has resulted in selection of antibodies with relatively modest neutralizing activity. To enhance selection of antibodies specific for gp41 in the context of the whole Env we have developed a methodology termed competitive antigen panning (CAP). By using CAP we identified a novel gp41-specific human monoclonal antibody (hmAb), m48, from an immune library derived from long-term nonprogressors with high titers of broadly cross-reactive neutralizing antibodies (bcnAbs). M48 was only selected by CAP but not by the conventional preincubation methodology. It neutralized a panel of HIV-1 primary isolates from different clades in assays based on spreading infection in peripheral blood mononuclear cells (PBMCs) with potency higher than that of the well characterized broadly cross-reactive HIV-1-neutralizing antibodies IgG1 4E10 and Fab Z13. These results may have implications for selection of novel gp41-specific bcnAbs, and for the development of HIV-1 inhibitors and vaccine immunogens.
Keywords: HIV, Antibody, Phage display, gp41, Inhibitors, Vaccines
1. Introduction
HIV uses various strategies to escape immune responses including rapid generation of mutants that outpace the development of neutralizing antibodies (Wei et al., 2003; Richman et al., 2003; Garber et al., 2004) and hiding important for replication conserved structures of its envelope glycoprotein (Env) by variable loops and extensive glycosylation, transient exposure, occlusion within the oligomer, and conformational masking (Poignard et al., 2001; Burton, 2002; Johnson et al., 2002; Wei et al., 2003). As a result, elicitation of cross-reactive neutralizing antibodies in vivo is rareand usually occurs after relatively long periods of maturation (Burton et al., 1997) (Zolla-Pazner, 2004a). Only several Env-specific hmAbs have been identified (Zolla-Pazner, 2004a) that exhibited neutralizing activity to primary isolates from different clades including the anti-gp120 human monoclonal antibodies b12 (Burton et al., 1994; Roben et al., 1994), 2G12 (Trkola et al., 1996; Sanders et al., 2002; Scanlan et al., 2002), m14 (Zhang et al., 2004b), m18 (Bouma et al., 2003), F105 ( ?), 447-52D (Gorny et al., 1992) and Fab X5 (Moulard et al., 2002), and the anti-gp41 antibodies 2F5 (Muster et al., 1993), 4E10 (Stiegler et al., 2001; Zwick et al., 2001) and Fab Z13 (Zwick et al., 2001). Identification and characterization of novel cross-reactive HIV-1-neutralizing human monoclonal antibodies may provide insights into the closely guarded conserved structures that still could serve as epitopes for neutralizing antibodies, and has implications for development of vaccines as well as for understanding mechanisms of HIV entry and evasion of immune responses, and for design of entry inhibitors.
Many HIV-1-neutralizing human monoclonal antibodies (nhmAbs) have been identified by immortalization of B lymphocytes from HIV-infected patients eitherby EBV transformation (Gorny et al., 1989; Robinson et al., 1990) or by cell fusion (Grunow et al., 1988; Buchacher et al., 1994) (hybridomas obtained by fusion of EBV transformants with heteromyeloma cells have also been extensively used (Posner et al., 1987; Gorny et al., 1991; Posner et al., 1991)] followed by screening of their supernatants for antigen-specific antibodies. Selection of HIV-1-neutralizing antibodies from phage-displayed human antibody libraries has also been used by panning against one antigen (Burton et al., 1991) or several antigens sequentially (Zhang et al., 2003); it is a powerfull and versatile approach that allows modifications of the panning process for enhanced selection of antibodies with desirable properties (Zhang and Dimitrov, 2006, in press).
The first identified human Fabs against the gp41 subunit of the HIV-1 envelope glycoprotein isolated by panning of phage-displayed antibody libraries have not neutralized HIV-1 laboratory strains at biologically significant concentrations (Binley et al., 1996). Later a gp41-specific bcnAb, Fab Z13, was selected by panning against the MN peptide 2031 containing the sequence ELDKWA that corresponds to the 2F5 core epitope, and against whole HIV-1MN virions (Zwick et al., 2001). However, Fab Z13 is considered to exhibit relatively modest inhibitory activity compared to the other two known gp41-specific bcnAbs, 2F5 and 4E10. Although 2F5 and 4E10 also bind to peptides derived from the gp41 membrane-proximal external region (MPER), efforts to use such peptides as vaccine immunogens or as antigens for screening of phage libraries were not successful in elicitation or selection of 2F5 or 4E10-like antibodies. Recently, two reports have described selection of anti-gp41 antibodies by using gp41 fragments for panning of phage antibody libraries but the selected antibodies exhibited relatively modest neutralizing activity (Louis et al., 2005; Miller et al., 2005). These results are consistent with the findings that the use of gp41 fragments that contain exposed antibody-accessible surfaces and resemble some fusion intermediates or post-fusion structure as vaccine immunogens leads to elicitation of weakly neutralizing antibodies or antibodies lacking broadly neutralizing activity against primary isolates (Zolla-Pazner, 2004b).
We have hypothesized that screening of immune phage libraries against purified Env ectodomains, gp140s, which contain both gp120 and truncated gp41 (lacking transmembrane domains and cytoplasmic tails) could lead to selection of anti-gp41 antibodies that exhibit neutralizing activity. However, we have previously found that panning of immune human antibody phage libraries against gp140, results mostly in selection of anti-gp120 antibodies (Zhang et al., 2003; Zhang et al., 2004b). Isolated gp41 cannot be used as a screening antigen because it is unstable and aggregates, and its conformation even if stabilized in the absence of gp120 could be different from that in the native Env state. To enhance the selection of gp41-specific antibodies we developed an approach termed competitive antigen panning (CAP), which is based on the idea to outcompete gp120-specific antibodies with an excess of gp120.. By using the CAP methodology we selected a novel hmAb, m48, specific for gp41 from an immune human antibody phage library derived from long-term nonprogressors with high titers of bcnAbs. M48 was only selected by CAP and not by the conventional pre-incubation methodology. It neutralized several HIV-1 primary isolates from different clades with potency better than that of 4E10 and Fab Z13 in PBMC-based assays. Unlike 2F5, 4E10 and Z13, which bind to denatured gp140s and linear peptides derived from the membrane-proximal external region (MPER), m48 does not bind to denatured gp140s and thirty-five linear peptides derived from extracellular portion of gp41, indicating a conformational nature of its epitope. The newly identified anti-gp41 antibody may have potential as an HIV-1 inhibitor in combination with other known anti-gp120 and anti-gp41 neutralizing antibodies and its epitope may have potential as vaccine immunogen capable of eliciting the same or similar antibodies in vivo. The CAP methodology may also have implications for selection of other antibodies to domains of whole proteins that are unstable in isolation.
2. Materials and Methods
2.1 Virus, gp120, gp140, peptides and antibodies
HIV-1 isolates were obtained from the NIH AIDS Research and Reference Reagent Program (ARRRP). Thirty-five gp41-derived peptides including N36, C34, DP178 and 2031 were also obtained from the ARRRP. Recombinant gp12089.6 gp120CM243, gp14089.6 and gp140CM243 were produced by using recombinant vaccinia viruses (the viruses containing the gene encoding gp12089.6 and gp14089.6 were a gift from R. Doms, University of Pennsylvania, Philadelphia, PA) with a combination of lentil lectin affinity chromatography and size exclusion chromatography. The human monoclonal antibodies 2F5, 4E10, Z13 and m14 were produced in our laboratories. The plasmid encoding Z13 was provided by M. Zwick and D. Burton (The Scripps Research Institute). The following antibodies were purchased: HRP conjugated monoclonal mouse anti-M13 antibody (Pharmacia, Uppsala, Sweden), HRP conjugated polyclonal anti-human IgG F(ab’)2 antibodies (Jackson ImmunoResearch, Westgrove, PA), and HRP conjugated streptavidin (Zymed Laboratories Inc., San Francisco, CA).
2.2 Competitive Antigen Panning (CAP) and screening
The phage library was constructed using pComb3H phagemid vector and 30 cc of bone marrow obtained from three long-term nonprogressors (A, H and K) (Dreyer et al., 1999) whose sera exhibited the broadest (against six primary isolates (Montefiori et al., 1996)) and most potent (against JR-FL at 1:40 dilution) HIV-1 neutralization activity among 37 HIV-infected individuals (provided by T. Evans, University of California, Davis) (Zhang et al., 2003). Competitive antigen panning (CAP) was performed as follow. Phage (5×1012 cfu/ml) in a total volume of 1 ml were pre-adsorbed on streptavidin-M280-Dynabeads in PBS for 1 h at room temperature. The phage bound to beads were removed using a magnetic separator (Dynal). The phage library (supernatant) was transferred to a new eppendorf tube and incubated with 50 nM biotinylated HIV-1 envelope glycoprotein gp140CM243 and 250 nM non-biotinylated gp120CM243 (5-fold more on molar level than biotinylated gp140CM243) for 2 h at room temperature on a head-to-tail rotator. The panning against Env gp14089.6 was done in parallel the same way as panning against gp140CM243. In a control panning, the phage library was depleted with 250 nM biotinylated gp120CM243 or gp12089.6 prior to incubation with 50 nM biotinylated gp140CM243 or gp14089.6. Phage bound to biotinylated gp140CM243 or gp14089.6 were separated from the phage library using streptavidin-M280-Dynabeads and a magnetic separator. After washing 20 times with 1 ml of PBS containing 0.1 % Tween-20 and another 20 times with 1 ml of PBS, bound phage were eluted from the beads using 1 ml of 100 mM Triethanolamine followed by neutralization with 0.5 ml of 1M, pH7.5 Tris-HCl. For the 2nd round of panning, 10 nM (2 nM for the 3rd round) of biotinylated gp140CM243, gp14089.6 and 5-fold excess of non-biotinylated gp120CM243, gp12089.6 were used as antigens. The control phage libraries were also panned 2nd and 3rd times as described above, but with decreased amount of biotinylated gp140CM243 or gp14089.6 antigens (10 nM for the 2nd round and 2 nM for the 3rd round) after depletion with five-fold more biotinylated gp120CM243 or gp12089.6. After 3rd round of panning, 96 individual clones from each panned library were screened for binding to gp140/120CM243 and gp140/12089.6 by phage ELISA as described previously (Zhang et al., 2004b). Briefly, 96-well Nunc-Immunok Maxisorpk surface plates (Nalge Nunc International) were coated overnight at 4ºC with 100 μl of gp120 or gp140 (1 μg/ml in sodium bicarbonate buffer, pH 8.3). The microwells were blocked in 100 μl of 4% non-fat dry milk in PBS for 1 h at 37ºC. After four washes with washing buffer (0.05% Tween-20 in PBS), wells were incubated with 100 μl of monoclonal phage supernatant for 2 h at 37ºC. Bound phage were detected by using horse radish peroxidase (HRP) labeled anti-M13 monoclonal antibody (Pharmacia) with incubation for 1 h at 37ºC and revealed by adding ready-to-use ABTS substrate (Pharmacia). Color development was performed at room temperature for 15 min and monitored at 405 nm.
2.3 Preparation of soluble Fab and binding assays
Soluble Fab m48 was produced as described (Barbas et al., 2001). Protein G columns were used for purification. ELISA binding assays with soluble Fabs and recombinant HIV-1 gp140s or gp41-derived peptides were performed by directly coating gp140s at 1 μg/ml or peptides at 4 μg/ml on 96-well plates followed by addition of three-fold serially diluted soluble anti-gp41 Fabs. When denatured gp140s were used, purified gp140s were diluted in 1% sodium dodecyl sulfate-50 mM dithiothreitol to 10μg/ml and / or boiled for 5 min, and then diluted 1:10 in PBS and coated on 96-well plates. Competition ELISAs of free gp12089.6 with immobilized gp14089.6 for binding to anti-gp41 antibodies were performed by coating 1 μg/ml gp14089.6 on 96-well plates followed by simultaneous addition of ten-fold serially diluted free gp12089.6 and soluble anti-gp41 Fabs at constant concentrations corresponding to 70% maximum binding. Bound Fabs were detected using HRP conjugated to anti-human IgG, F(ab’)2 and ABTS substrate.
2.4 HIV-1 neutralization assays
Two PBMC-based assays using infectious viruses were performed to test potency of the selected monoclonal antibody. In the PBMC-P24 assay, infectious replication-competent virus (500 TCID50 in 50 μl) was incubated with the antibody (4x concentration in 50 μl complete RPMI 1640 medium) for 30 min at 37°C in a well of a 96 well plate followed by addition of PHA-activated PBMC (5 x 104 in 100 μl of complete RPMI 1640 medium); the antibody concentration in the final volume of 200 μl is referred as the test concentration. After overnight incubation at 37°C 150 μl of the culture supernatant was removed and 150 μl of fresh culture medium was added without adding antibody and virus; on day four 100 μl of the culture supernatant was exchanged with 100 μl of fresh culture medium. Seven days after initiation of the infection duplicate samples were harvested for measurement of p24 by ELISA as described previously (Zhao et al., 2002; Jiang et al., 2004). The percentage (%) inhibition of the p24 production was calculated by using the formula [1-(E-N)/(P-N)]x100, where P is the ELISA optical density (OD) value of the positive control (50 μl virus + 50 μl medium + 100 μl PBMCs), N - OD value of the negative control (50 μl virus + 150 μl medium), and E - the OD value of the experimental result (50 μl test sample + 50 μl virus + 100 μl PBMCs). The PBMCs used for this assay were isolated from the blood of healthy donors at the New York Blood Center by standard density gradient centrifugation using Histopaque-1077 (Sigma). The cells were plated in 75 cm2 plastic flasks and incubated at 37 °C for 2 hrs. The non-adherent cells were collected and resuspended at 5 x 106 in 10 ml RPMI-1640 medium containing 10% FBS, 5 μg/ml PHA and 100 U/ml IL-2 (Sigma), followed by incubation at 37 °C for 3 days. The PHA-stimulated cells were then used for the assay.
In the PBMC-RT assay infectious replication-competent virus (100 TCID50 in 50 μl) was incubated with the antibody (2x concentration in 100 μl complete RPMI with Interleukin-2 (IL-2)) for 30 min at 37°C in a well of a 96 well plate followed by addition of phytohemagglutinin (PHA)-activated PBMC (5 x 104 in 50 μl of complete RPMI 1640 medium with IL-2) pooled from two donors four days after separation; the antibody concentration in the final volume of 200 μl is referred as the test concentration. The antibody and virus were not washed but remained in the culture, and seven days later at the infection peak triplicate samples were harvested for measurement of the reverse transcriptase (RT) activity. Percentage neutralization was calculated by using the formula [1-(E-N)/(P-N)]x100, where P is the RT activity of the positive control (mock culture without antibody - 50 μl virus + 100 μl medium + 50 μl PBMCs), N - the negative control (50 μl virus + 100 μl medium containing 0.1 μM AZT + 50 μl PBMCs), and E - the experimental result (100 μl containing antibody + 50 μl virus + 50 μl PBMCs). AZT was used as a positive control for inhibition with 100% inhibition at concentration equal to 0.1 μM, where the measured cell toxicity was undetectable (IC50 = 0.002 μM, TC50 > 1 μM), and negative control for lack of residual virus and for background subtraction.
2. 5 Statistical analysis
To compare neutralization activity of different antibodies we used Student's paired two-tailed t-test. Calculated levels of significance are presented as p values; differences with p equal to or lower than 0.05 were considered significantly different. The mean values of neutralizing activities and standard errors (SD/sqrt n, where n is the number of replicates typically three) were also calculated. The medians for all isolates were calculated by using all triplicate values and not directly from the means for each isolate.
3. Results
3.1 Selection of anti-gp41 phage-displayed Fabs by CAP
We have previously used recombinant soluble Env ectodomains (gp140s) as antigens for panning of phage libraries, and selected anti-gp120 but not anti-gp41 antibodies (Zhang et al., 2003; Zhang et al., 2004a; Zhang et al., 2004b). This is likely due to the higher concentration of high-affinity anti-gp120 antibodies in our immune antibody library compared to the concentration of high-affinity anti-gp41 antibodies. One of the strategies to increase the likelihood of selecting anti-gp41 antibodies would be to pre-incubate the library with gp120 to eliminate most of the anti-gp120 antibodies. However, such a strategy could also result in non-specific retention of anti-gp41 antibodies that would be lost in the subsequent panning against gp140. We have hypothesized that a better strategy would be to incubate simultaneously gp140 with excess of gp120 and then selectively extract the gp140 in complex with presumably anti-gp41 antibodies (Fig. 1). Under these equilibrium conditions most of the high-affinity anti-gp120 antibodies will be bound to gp120 and those anti-gp41 antibodies that bind non-specifically to gp120 or else will bind to gp41 on gp140.
Fig. 1. Schematic representation of the competitive antigen panning (CAP) methodology.

CAP allows enhanced selection of antibodies to subunits of whole proteins. For example, gp41 is difficult to isolate in native state. To enhance selection of such antibodies against gp41 the library is panned against the whole protein, in this case soluble HIV Env, gp120-gp41 (gp140), in the presence of excess gp120, which binds most of the gp120-specific antibodies. Then isolation of the tagged whole protein with bound antibodies will lead to enrichment of gp41-specific antibodies.
To test our CAP strategy we biotinylated recombinant gp140 from two different isolates (89.6 and CM243 denoted as gp14089.6 and gp140CM243, respectively) and mixed each of them with 5-fold molar excess of the corresponding non-biotinylated recombinant gp120. This mixture was used for panning of an immune antibody phage library. After three rounds of panning, screening of individual phage clones was performed with phage ELISA by using gp14089.6 and gp140CM243, and the corresponding gp120s. The CAP resulted in significant number of phage-displayed antibodies that bound to gp140 but did not bind to gp120 (gp41 binders) (Fig. 2). To assess the efficiency of the CAP methodology, depletion of the antibody library with gp12089.6 and gp120CM243 was performed in separate experiments prior to panning against gp14089.6 and gp140CM243. In this case the number of gp41 binders was smaller compared to the number of clones selected by CAP (Fig. 2). We have not identified any antibodies specific for gp41 from the 89.6 isolate when the antibody library was depleted with gp12089.6 prior to panning against gp14089.6.
Fig. 2. Efficient selection of gp41-specific phage-displayed antibodies by using CAP against gp140s from different isolates.

Total of 384 clones (96 for each panned library) were screened by phage ELISA for binding to homologous gp14089.6, CM243 or gp12089.6, CM243 after the third round of panning against gp140s from 89.6 or CM243 by using CAP or gp12089.6, CM243 pre-depletion. Those clones that did not bind gp120 but did bind gp140 were designated as gp41 binders. Gp120 pre-depletion denotes depletion of gp120 binders before panning against gp140s without using CAP.
Sequencing of the DNA from clones encoding for gp41-specific antibodies that exhibited significant binding as measured by phage ELISA (optical density (OD) at 405 nm > 1.0) showed that most of them were either with identical sequences or differed by few amino acid residues in the framework. One clone, m48, was selected for further characterization. M48 was extensively enriched in the libraries panned against gp14089.6 using CAP, but not selected by pre-depletion of the library with gp12089.6, which indicates that m48 would have been lost if the panning was performed by traditional pre-depletion strategy. This result suggests that CAP is an efficient methodology for selection of gp41-specific antibodies by using the entire Env ectodomain (gp140).
3.2 Specificity of the anti-gp41 antibody against soluble gp140s
We expressed the newly identified antibody as soluble Fab and confirmed its gp41 specificity by a competition ELISA using free gp120 (Fig. 3). Binding of m48 to gp14089.6 was not affected by the presence of free gp12089.6 to any significant degree except at high concentration (1μM) of gp12089.6, which explains why m48 was not selected from the panned libraries pre-depleted with gp12089.6. The anti-gp120 antibody m14 and the anti-gp41 antibodies 2F5 and Z13 were used as controls. The binding of m14 to coated gp14089.6 decreased significantly while 2F5 binding did not change with an increase in the gp120 concentration. The Z13 binding decreased at the highest concentration (1μM) of gp12089.6 similarly to m48.
Fig. 3. Competition of free gp12089.6 with immobilized gp14089.6 for binding to m48.

One μg/ml of gp14089.6 was coated on 96-well plates. Ten-fold serially diluted free gp12089.6 was added to the wells and soluble anti-gp41 antibodies at constant concentrations corresponding to 70% maximum binding were simultaneously added to the wells. Bound antibodies were detected using HRP conjugated to anti-human IgG, F(ab’)2 and ABTS substrate. The binding of antibodies in the absence of free gp120 was taken as 100%. The relative binding was calculated by using the formula: (optical density at 405 nm in the presence of free gp120 / optical density at 405 nm in the absence of free gp120) × 100.
3.3 Neutralizing activity of m48 against primary HIV-1 isolates from different clades
To begin to evaluate the possibility for broad neutralizing activity of m48 we tested its activity in two PBMC assays as described in the Materials and Methods. Both of them are based on spreading virus infection but one (PBMC-p24) measures p24 and the other (PBMC-RT) reverse transcriptase (RT) activity.
In the PBMC-p24 assay Fab m48 neutralized four or two out of ten isolates with an IC50 or IC90 lower than 40 μg/ml, respectively. Fab Z13, which was used for comparison, neutralized, one out of ten isolates with an IC50 lower than 40 μg/ml and none with an IC90 lower than 40 μg/ml (Table 1). Fab m48 neutralized isolates from clades B, C, D and F. Fab Z13 neutralized only the clade B isolate. The isolates from clade A, clade G and group O isolates were not neutralized by both antibodies tested. These data and their statistical analysis showed that Fab m48 is significantly more potent than Fab Z13 for this panel of primary isolates as tested by the PBMC-based assay.
Table 1. Inhibitory activity of anti-gp41 mAbs against a panel of primary HIV-1 isolates measured by the PBMC-24 assay.
The PBMC-p24 assay is based on inhibition of spreading infections of PBMCs by primary isolates and measurement of p24 seven days post infection (see Materials and Methods for details). The highest concentration of antibody tested was 40 μg/ml. If the inhibitory activity was <50% or 90 % at this concentration, the IC50 or IC90 were recorded as >40 μg/ml.
| HIV-1 isolate | Clade | Coreceptor specificity | IC50 (μg/ml) (Mean ± SD)
|
|
| Fab m48 | Fab Z13 | |||
|
| ||||
| 92RW008 | A | R5 | >40 | >40 |
| 94UG103 | A | X4R5 | >40 | >40 |
| 92US657 | B | R5 | 19±0.3 | 20±1.7 |
| 93IN101 | C | R5 | 9.0±0.4 | >40 |
| 92UG001 | D | R5 | 14±0.2 | >40 |
| 93THA051 | E | X4R5 | >40 | >40 |
| 93THA009 | E | R5 | >40 | >40 |
| 93BR020 | F | X4R5 | 22±4.8 | >40 |
| RU570 | G | R5 | >40 | >40 |
| BCF02 | group O | R5 | >40 | >40 |
|
| ||||
| HIV-1 isolate | Clade | Coreceptor specificity | IC90 (μg/ml) (Mean ± SD)
|
|
| Fab m48 | Fab Z13 | |||
|
| ||||
| 92RW008 | A | R5 | >40 | >40 |
| 94UG103 | A | X4R5 | >40 | >40 |
| 92US657 | B | R5 | 32±0.5 | >40 |
| 93IN101 | C | R5 | >40 | >40 |
| 92UG001 | D | R5 | 27±1.7 | >40 |
| 93THA051 | E | X4R5 | >40 | >40 |
| 93THA009 | E | R5 | >40 | >40 |
| 93BR020 | F | X4R5 | >40 | >40 |
| RU570 | G | R5 | >40 | >40 |
| BCF02 | group O | R5 | >40 | >40 |
To confirm and further extend these findings to another panel of primary isolates from different clades we used a second PBMC assay based on measurement of RT. To potentially increase the Fab potency, and confer biological effector functions and long half-life in vivo, as well as to better mimic the in vivo neutralization, Fab 48 was converted to full antibody in an IgG1 format, and the PBMC-RT assay was used to evaluate its inhibitory activity against a range of primary isolates from different clades (Table 2). IgG1 4E10 and Fab Z13 were used as controls. M48 neutralized (about 90% median neutralization at 50 μg/ml) most of the primary isolates except the clade A isolate UG029. The clade D isolate UG001 was neutralized only by Fab m48 (Table 2). The comparison between the Fab and IgG1 formats of the same antibody, m48, showed that on average the Fab appears more potent than the IgG1. Significant difference in their neutralizing potency was observed for the isolates UG001 (clade D) and BCF 03 (clade O). Fab m48 completely neutralized these two isolates while the IgG1 m48 neutralizing activity was less than 50% at 50 μg/ml. IgG1 m48 was, however, more potent than Fab m48 for other isolates, e.g. the clade C isolates BR025 and ZA003 (Table 2).
Table 2. Inhibitory activity of anti-gp41 antibodies (IgG1s and Fabs) against a panel of HIV-1 primary isolates from different clades measured by the PBMC-RT assay.
The PBMC-RT assay is based on inhibition of spreading infections of PBMCs by primary isolates and measurement of reverse transcriptase (RT) seven days post infection. The percentage inhibition of RT activity (mean ± standard error) in the culture supernatant of HIV-1-infected PBMCs at day seven is presented as a measure of the antibody inhibitory activity (see Materials and Methods for details). The medians for all isolates were calculated by using all triplicate values and not directly from the means for each isolate. The antibody concentration was 50 μg/ml.
| HIV-1 isolate | clade | IgG1 m48 | IgG1 4E10 | Fab m48 | Fab Z13 |
|---|---|---|---|---|---|
| 92UG029 | A | 20±14 | 0+14 | 43+11 | 1.0+17 |
| 92HT594 | B | 65±21 | 31+8.6 | 100+0.1 | 55+23 |
| SHIV 89.6p | B | 83±12 | 25+13 | 97+1.0 | 96+1.2 |
| 92BR025 | C | 68+1.6 | 58+3.2 | 43+16 | 63+3.1 |
| 97ZA003 | C | 94±1.3 | 49+12 | 72+7.7 | 75+7.9 |
| 93IN101 | C | 99±0.1 | 99+0.1 | 100+0.1 | 99+0.1 |
| 93MW959 | C | 97±0.6 | 53+10 | 100+0.0 | 92+1.5 |
| 92UG001 | D | 38±6.4 | 29+0.7 | 99+0.2 | 49+10 |
| 93TH073 | E | 96±0.1 | 68+2.6 | 99+0.0 | 93+1.1 |
| 93BR029 | F | 76±16 | 62+12 | 94+3.3 | 83+11 |
| G3 | G | 93±0.6 | 57+16 | 92+0.4 | 85+3.6 |
| BCF03 | O | 44±16 | 34+7.5 | 99+0.6 | 42+21 |
|
| |||||
| Median | 87 | 51 | 98 | 79 | |
Our statistical analysis of these data demonstrated that in the PBMC-RT assay (Table 2) IgG1 m48 was significantly (p < 0.01) more potent than IgG1 4E10. Fab m48 was also significantly (p < 0.01) more potent than Fab Z13 (p < 0.01). Although IgG1 m48 was on average also more potent than Fab Z13, the difference was not statistically significant (p=0.48). The clade D isolate 92UG001 was tested in both PBMC-based assays (Table 1 and 2) and exhibited similar antibody neutralization pattern for Fab m48 and Fab Z13 indicating good correlation between the two assays. These results suggest that the newly identified anti-gp41 antibody exhibit broad neutralizing activity against two panels of primary isolates from different clades with a potency on average higher than the potency of Fab Z13 and IgG1 4E10 in two PBMC-based assays.
3.4 Initial characterization of the m48 epitope
The epitopes of the three known broadly neutralizing antibodies, 2F5, 4E10 and Z13, are localized in the membrane-proximal external region (MPER) of gp41 and include stretches of known sequences. They do bind to peptides containing these sequences and denatured gp140. To find out whether m48 binds to the same region and to begin to characterize its epitope, we measured m48 binding to peptides derived from different regions of gp41. Thirty-four peptides derived from gp41 including six peptides from the MPER were used in an ELISA assay to test m48 binding. M48 did not bound specifically to any significant degree to the peptides tested including N36, C34, DP178 and peptide 2031 (LLELDKWASLWNWFDITNWLW) (Fig. 4 and data not shown) indicating that this antibody recognizes a conformational epitope. This was confirmed by the observation that m48 bound to non-denatured but not to denatured gp140 (Fig. 4). To find which denaturing factor (boiling or reducing) affect its binding, we prepared two types of denatured gp140s: by boiling only and by adding a reducing reagent only. Boiling of gp140 did not significantly affect the binding of m48 to gp140, but addition of reducing reagent (5 mM DTT) abolished its binding (Fig. 5), indicating that disulfide bond in gp41 are important for the structural integrity of the epitope. In contrast, the control antibodies 2F5 and Z13 still bound to denatured (boiled or reduced) gp140 without significant change in affinity. These results indicate that the new anti-gp41 antibody recognizes a conformational epitope on gp41, which is different from those of the known broadly HIV-1-neutralizing anti-gp41 antibodies 2F5, 4E10 and Z13.
Fig. 4. Binding of anti-gp41 Fab m48 to selected peptides derived from gp41.
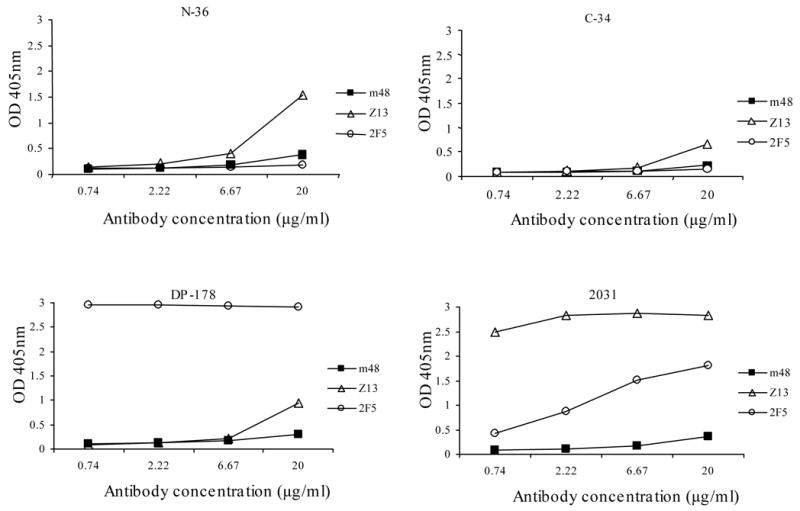
Gp41-derived peptides N36, C34, DP178 and 2031 at 4 μg/ml were coated on 96-well plates by incubation at 4°C overnight. The plates were blocked and three-fold serially diluted anti-gp41 antibodies Fab m48, Z13 and IgG 2F5 with starting concentration of 20 μg/ml were added to the wells. Bound antibodies were detected using HRP conjugated to anti-human IgG, F(ab’)2 and ABTS substrate.
Fig. 5. Binding of anti-gp41 Fabs to non-denatured (A) and denatured gp14089.6 (B, C).

Recombinant non-denatured gp14089.6 or denatured gp14089.6 at 1 μg/ml (prepared as described in Materials and Methods) was coated on 96-well plates. The plates were blocked using 3% BSA in PBS, and three-fold serially diluted anti-gp41 antibodies with starting concentration of 10 μg/ml were added to the wells. Bound antibodies were detected using HRP-conjugated anti-human IgG, F(ab’)2 and ABTS as substrate. Optical density at 405 nm was measured after color development at room temperature for 30 minutes.
4. Discussion
The major results of this study are the development of a methodology specifically designed for the enhanced selection of antibodies to gp41 presented in the context of gp140 and the identification of a novel cross-reactive HIV-1 nhmAb by. To our knowledge this is the first report describing a CAP methodology successfully used for selection of antibodies against recombinant proteins. In contrast to the previously characterized broadly neutralizing anti-gp41 hmAbs 2F5, 4E10 and Fab Z13, the newly identified antibody described here does not bind peptides or Envs that lack conformational integrity, and on average. it exhibits higher neutralizing activity than Z13 and 4E10 against 20 primary isolates from different clades in PBMC-based assays.
The CAP methodology can be also applied to selection of antibodies against epitopes that are genetically (e.g. mutants) or biologically modified (e.g. phosphoration, sulfation, etc.).. CAP would be superior to traditional pre-depletion method in terms of avoiding potential loss of target-specific antibodies. Our findings not only provide a novel methodology for identification of high-affinity hmAbs targeting protein domains in the context of whole proteins but also illustrate the use of this methodology by selection of a novel gp41-specific bcnAb which could have potential as a therapeutic and aid in the design of AIDS vaccines based on the structure of its epitope.
Acknowledgments
We wish to thank Michael Zwick and Dennis Burton for providing DNA encoding Z13. We also thank Robert Blumenthal, Peter Kwong and Hana Golding for helpful discussions. This project was supported by the NIH Intramural AIDS Targeted Antiviral Program (IATAP), the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and the Gates Foundation to DSD, DHHS NO1-CO-12400 to MYZ, and NIH AI48380 to CCB.
Abbreviations
- Env
envelope glycoprotein
- CAP
competitive antigen panning
- hmAb
human monoclonal antibody
- bcnAb
broadly cross-reactive neutralizing antibody
- PBMCs
peripheral blood mononuclear cells
- nhmAb
neutralizing human monoclonal antibody
- HRP
horse radish peroxidase
- RT
reverse transcriptase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barbas CF, Burton DR, Scott JK, Silverman GJ. Phage Display: A Laboratory Mannual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2001. [Google Scholar]
- Binley JM, Ditzel HJ, Barbas CF3, Sullivan N, Sodroski J, Parren PW, Burton DR. Human antibody responses to HIV type 1 glycoprotein 41 cloned in phage display libraries suggest three major epitopes are recognized and give evidence for conserved antibody motifs in antigen binding. AIDS Res Hum Retroviruses. 1996;12:911–924. doi: 10.1089/aid.1996.12.911. [DOI] [PubMed] [Google Scholar]
- Binley JM, Wrin T, Korber B, Zwick MB, Wang M, Chappey C, Stiegler G, Kunert R, Zolla-Pazner S, Katinger H, Petropoulos CJ, Burton DR. Comprehensive cross-clade neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J Virol. 2004;78:13232–13252. doi: 10.1128/JVI.78.23.13232-13252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouma P, Leavitt M, Zhang PF, Sidorov IA, Dimitrov DS, Quinnan GV., Jr Multiple interactions across the surface of the gp120 core structure determine the global neutralization resistance phenotype of human immunodeficiency virus type 1. J Virol. 2003;77:8061–8071. doi: 10.1128/JVI.77.14.8061-8071.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A, Katinger H. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS ResHumRetroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- Burton DR. Antibodies, viruses and vaccines. NatRevImmunol. 2002;2:706–713. doi: 10.1038/nri891. [DOI] [PubMed] [Google Scholar]
- Burton DR, Barbas CF, Persson MA, Koenig S, Chanock RM, Lerner RA. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. ProcNatlAcadSciUSA. 1991;88:10134–10137. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Montefiori DC. The antibody response in HIV-1 infection. AIDS. 1997;11(Suppl A):S87–S98. [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL, et al. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Dimitrov DS. Virus entry: molecular mechanisms and biomedical applications. NatRevMicrobiol. 2004;2:109–122. doi: 10.1038/nrmicro817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer K, Kallas EG, Planelles V, Montefiori D, McDermott MP, Hasan MS, Evans TG. Primary isolate neutralization by HIV type 1-infected patient sera in the era of highly active antiretroviral therapy. AIDS ResHumRetroviruses. 1999;15:1563–1571. doi: 10.1089/088922299309856. [DOI] [PubMed] [Google Scholar]
- Garber DA, Silvestri G, Feinberg MB. Prospects for an AIDS vaccine: three big questions, no easy answers. Lancet InfectDis. 2004;4:397–413. doi: 10.1016/S1473-3099(04)01056-4. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Conley AJ, Karwowska S, Buchbinder A, Xu JY, Emini EA, Koenig S, Zolla-Pazner S. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. JVirol. 1992;66:7538–7542. doi: 10.1128/jvi.66.12.7538-7542.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Gianakakos V, Sharpe S, Zolla-Pazner S. Generation of human monoclonal antibodies to human immunodeficiency virus. ProcNatlAcadSciUSA. 1989;86:1624–1628. doi: 10.1073/pnas.86.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Xu JY, Gianakakos V, Karwowska S, Williams C, Sheppard HW, Hanson CV, Zolla-Pazner S. Production of site-selected neutralizing human monoclonal antibodies against the third variable domain of the human immunodeficiency virus type 1 envelope glycoprotein. ProcNatlAcadSciUSA. 1991;88:3238–3242. doi: 10.1073/pnas.88.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunow R, Jahn S, Porstmann T, Kiessig SS, Steinkellner H, Steindl F, Mattanovich D, Gurtler L, Deinhardt F, Katinger H. The high efficiency, human B cell immortalizing heteromyeloma CB-F7. Production of human monoclonal antibodies to human immunodeficiency virus. J Immunol Methods. 1988;106:257–265. doi: 10.1016/0022-1759(88)90206-2. [DOI] [PubMed] [Google Scholar]
- Jiang S, Lu H, Liu S, Zhao Q, He Y, Debnath AK. N-substituted pyrrole derivatives as novel human immunodeficiency virus type 1 entry inhibitors that interfere with the gp41 six-helix bundle formation and block virus fusion. AntimicrobAgents Chemother. 2004;48:4349–4359. doi: 10.1128/AAC.48.11.4349-4359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Desrosiers RC. Viral persistance: HIV's strategies of immune system evasion. AnnuRevMed. 2002;53:499–518. doi: 10.1146/annurev.med.53.082901.104053. [DOI] [PubMed] [Google Scholar]
- Louis JM, Bewley CA, Gustchina E, Aniana A, Clore GM. Characterization and HIV-1 fusion inhibitory properties of monoclonal Fabs obtained from a human non-immune phage library selected against diverse epitopes of the ectodomain of HIV-1 gp41. J Mol Biol. 2005;353:945–951. doi: 10.1016/j.jmb.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Miller MD, Geleziunas R, Bianchi E, Lennard S, Hrin R, Zhang H, Lu M, An Z, Ingallinella P, Finotto M, Mattu M, Finnefrock AC, Bramhill D, Cook J, Eckert DM, Hampton R, Patel M, Jarantow S, Joyce J, Ciliberto G, Cortese R, Lu P, Strohl W, Schleif W, McElhaugh M, Lane S, Lloyd C, Lowe D, Osbourn J, Vaughan T, Emini E, Barbato G, Kim PS, Hazuda DJ, Shiver JW, Pessi A. A human monoclonal antibody neutralizes diverse HIV-1 isolates by binding a critical gp41 epitope. Proc NatlAcad Sci USA. 2005;102:14759–14764. doi: 10.1073/pnas.0506927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC, Pantaleo G, Fink LM, Zhou JT, Zhou JY, Bilska M, Miralles GD, Fauci AS. Neutralizing and infection-enhancing antibody responses to human immunodeficiency virus type 1 in long-term nonprogressors. JInfectDis. 1996;173:60–67. doi: 10.1093/infdis/173.1.60. [DOI] [PubMed] [Google Scholar]
- Moulard M, Phogat SK, Shu Y, Labrijn AF, Xiao XD, Binley JM, Zhang MY, Sidorov IA, Broder CC, Robinson J, Parren PWHI, Burton DR, Dimitrov DS. Broadly cross-reactive HIV-1-neutralizing human monoclonal Fab selected for binding to gp120-CD4-CCR5 complexes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6913–6918. doi: 10.1073/pnas.102562599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. JVirol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poignard P, Saphire EO, Parren PW, Burton DR. Gp120: biologic aspects of structural features. AnnuRevImmunol. 2001;19:253–274. doi: 10.1146/annurev.immunol.19.1.253. [DOI] [PubMed] [Google Scholar]
- Posner MR, Elboim H, Santos D. The construction and use of a human-mouse myeloma analogue suitable for the routine production of hybridomas secreting human monoclonal antibodies. Hybridoma. 1987;6:611–625. doi: 10.1089/hyb.1987.6.611. [DOI] [PubMed] [Google Scholar]
- Posner MR, Hideshima T, Cannon T, Mukherjee M, Mayer KH, Byrn RA. An IgG human monoclonal antibody that reacts with HIV-1/GP120, inhibits virus binding to cells, and neutralizes infection. JImmunol. 1991;146:4325–4332. [PubMed] [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. ProcNatlAcadSciUSA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roben P, Moore JP, Thali M, Sodroski J, Barbas CF3, Burton DR. Recognition properties of a panel of human recombinant Fab fragments to the CD4 binding site of gp120 that show differing abilities to neutralize human immunodeficiency virus type 1. JVirol. 1994;68:4821–4828. doi: 10.1128/jvi.68.8.4821-4828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JE, Holton D, Pacheco-Morell S, Liu J, McMurdo H. Identification of conserved and variant epitopes of human immunodeficiency virus type 1 (HIV-1) gp120 by human monoclonal antibodies produced by EBV-transformed cell lines. AIDS ResHumRetroviruses. 1990;6:567–579. doi: 10.1089/aid.1990.6.567. [DOI] [PubMed] [Google Scholar]
- Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. JVirol. 2002;76 :7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan CN, Pantophlet R, Wormald MR, Ollmann SE, Stanfield R, Wilson IA, Katinger H, Dwek RA, Rudd PM, Burton DR. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1-->2 mannose residues on the outer face of gp120. JVirol. 2002;76:7306–7321. doi: 10.1128/JVI.76.14.7306-7321.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, Katinger H. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS ResHumRetroviruses. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. JVirol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Zhang MY, Shu Y, Phogat S, Xiao X, Cham F, Bouma P, Choudhary A, Feng YR, Sanz I, Rybak S, Broder CC, Quinnan GV, Evans T, Dimitrov DS. Broadly cross-reactive HIV neutralizing human monoclonal antibody Fab selected by sequential antigen panning of a phage display library. J ImmunolMethods. 2003;283:17–25. doi: 10.1016/j.jim.2003.07.003. [DOI] [PubMed] [Google Scholar]
- Zhang MY, Shu Y, Sidorov I, Dimitrov DS. Identification of a novel CD4i human monoclonal antibody Fab that neutralizes HIV-1 primary isolates from different clades. Antiviral Res. 2004a;61:161–164. doi: 10.1016/j.antiviral.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Zhang MY, Xiao X, Sidorov IA, Choudhry V, Cham F, Zhang PF, Bouma P, Zwick M, Choudhary A, Montefiori DC, Broder CC, Burton DR, Quinnan GV, Jr, Dimitrov DS. Identification and characterization of a new cross-reactive human immunodeficiency virus type 1-neutralizing human monoclonal antibody. J Virol. 2004b;78:9233–9242. doi: 10.1128/JVI.78.17.9233-9242.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Ernst JT, Hamilton AD, Debnath AK, Jiang S. XTT formazan widely used to detect cell viability inhibits HIV type 1 infection in vitro by targeting gp41. AIDS ResHumRetroviruses. 2002;18:989–997. doi: 10.1089/08892220260235353. [DOI] [PubMed] [Google Scholar]
- Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. NatRevImmunol. 2004b;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S. Identifying epitopes of HIV-1 that induce protective antibodies. NatRevImmunol. 2004a;4:199–210. doi: 10.1038/nri1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, Parren PW. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


