Abstract
Medulloblastoma represents greater than 25% of childhood intracranial neoplasms and is considered a highly malignant tumor. This tumor, which arises predominantly in the cerebellar vermis, preferentially affects children between the ages of 5 and 15. Although the etiology of medulloblastomas in humans remains unknown, results from several experiments have indicated that the human neurotropic JC virus (JCV) is able to induce cerebellar neoplasms in rodents that exhibit a phenotype similar to that of human medulloblastomas. JCV is a polyomavirus that is widespread in the human population, with infection occurring most frequently in early childhood. In this study, we have examined the possible association of JCV with human medulloblastomas. By using PCR techniques we demonstrate that 11 of 23 samples of tumor tissue contain DNA sequences corresponding to three different regions of the JCV genome. More importantly, we demonstrate the presence of DNA sequences encoding the N- and C-terminal regions of the JCV oncogenic protein, T antigen, in 11 of 23 samples and the production of T antigen in the nuclei of 4 samples of tumor tissue. These observations provide evidence for a possible association of JCV with human medulloblastomas.
Medulloblastoma, a malignant invasive tumor of the cerebellum, represents one of the most common neoplasms of the nervous system in children with an annual incidence of approximately 1 per 200,000 (1). Nearly 70% of medulloblastomas occur in children under 16 years of age (2) and are rarely seen in patients older than 50 years of age. Although both sexes are affected, there is a slight predominance of male patients (65% male). Histologically, medulloblastomas are composed of densely cellular tumors with frequent apoptotic cells and mitotic figures. Infrequently, vascular proliferation and hemorrhage are observed. Although a small percentage of these embryonal neuroblastic tumors are related to genetically defined heritable syndromes associated with a predisposition toward tumorigenesis, the majority of medulloblastomas are sporadic and their etiology remains unknown. Previous results from molecular and cytogenetic studies have pointed to the possible involvement of chromosomes 1, 17, and, to a lesser degree, 6, 9, 10, 11, and 16 in the development of medulloblastoma (3, 4). Loss of heterozygosity in portions of chromosome 17 (17p) has been reported in 30–45% of medulloblastoma (5). The presence of the gene responsible for the production of the tumor suppressor protein, p53, on chromosome 17 led to the early speculation that this protein may play a key role in the development of medulloblastoma. However, more recent studies have shown that only a small percentage of these tumors (between 5 and 10%) contain mutations in the p53 gene (6, 7) and that in the majority of medulloblastoma, the p53 genome remains intact after the observed chromosome 17 deletion. Other studies have revealed elevated expression of the paired box (PAX) gene family of transcription factors in medulloblastoma (8). The importance of these observations in the development of medulloblastomas remains elusive.
JC virus (JCV) is a human neurotropic polyomavirus infecting more than 80% of the human population early in life (9). The virus originally was isolated from the brain of a patient with the central nervous system (CNS) demyelinating disease, progressive multifocal leukoencephalopathy (PML) (10). This fatal CNS disease usually is associated with lymphoproliferative disorders, immunosuppressive conditions, and, recently, AIDS (11). It is believed that the demyelination observed in PML is caused by the cytolytic destruction of oligodendrocytes, the myelin-producing cells of the CNS. Similar to other polyomaviruses, production of the viral early protein, T antigen, is essential for the subsequent events of the virus lytic cycle, which include viral DNA replication and viral late gene transcription (12). The T antigen of JCV exhibits greater than 70% homology with the related, well characterized protein from the primate polyomavirus, SV40. Similar to SV40 T antigen, the JCV early protein can form complexes with several cellular regulatory proteins such as p53 and pRb, two growth regulators that play an important role in tumorigenesis (13–16). Studies from our laboratory have shown the association of JCV T antigen with p53 and pRb in tumors of CNS origin that developed in transgenic animals as well as deregulation of their target proteins, which are involved in the control of cell growth and differentiation (17).
Recently, we have described the development of a transgenic animal model by using the early region of the genome of the human ubiquitous JCV, which contains the viral early promoter expressing the early gene encoding the oncogenic protein, JCV T antigen (18). Histological examination of the CNS revealed no features of hypomyelination, an established pathology that is seen upon expression of JCV T antigen in brain (19). Unexpectedly, cerebellar tumors that closely parallel human medulloblastomas in location, histological appearance, and expression of differentiation markers were found in the affected animals. Although the oncogenic potential of JCV has been demonstrated previously in several experimental animals (20, 21), the transgenic animal points to the possible direct involvement of the JCV early protein in the development of cerebellar medulloblastomas. In this study we have used PCR techniques and have provided evidence for the presence of the JCV genome sequence in large numbers of human medulloblastomas. Immunohistochemical evaluation of these tumor samples revealed expression of the viral oncogenic protein, T antigen, in tumor cells.
MATERIALS AND METHODS
Clinical Samples.
A total of 23 human medulloblastoma samples were obtained from the archives of the following institutions: Rhode Island Hospital, Providence, RI (7 samples); University of Virginia Hospitals, Charlottesville, VA (6 samples); St. Christopher’s Hospital for Children, Philadelphia, PA (9 samples); and Hahnemann University Hospital, Philadelphia, PA (1 sample). Formalin-fixed, paraffin-embedded surgical resections were histologically graded according to the World Health Organization classification.
Histological and Immunohistochemical Analysis.
Paraffin-embedded samples were sectioned at 4-μm thickness and stained with hematoxylin and eosin for routine histological analysis. Immunohistochemistry was performed by using the avidin-biotin-peroxidase complex system according to the manufacturer’s instructions (Vectastain Elite ABC Peroxidase Kit; Vector Laboratories). Briefly, sections were deparaffinized in xylene and rehydrated and then heated in 0.01 M sodium citrate buffer (pH 6.0) to 95°C under vacuum for 40 min and allowed to cool for 20 min at room temperature for nonenzymatic antigen retrieval. The slides then were rinsed and incubated in MeOH/3% H2O2 for 20 min to quench endogenous peroxidase. Sections then were washed in PBS and blocked in PBS/0.1% BSA containing 5% normal horse serum for 2 h at room temperature. Slides then were incubated overnight at room temperature with antibodies specific for viral proteins or cellular markers. Antibodies utilized to detect viral proteins included a mAb specific for SV40 T antigen, which cross-reacts with JCV T antigen (1:100 dilution, pAb416; Oncogene Science), and rabbit antisera raised against the JCV capsid protein, VP1 (1:1,000 dilution, a kind gift of Eugene Major, National Institute of Neurological Disorders and Stroke, National Institutes of Health). Paraffin-embedded tumor tissue from Syrian hamsters intracerebrally inoculated with JC virus and brain tissue from patients with PML were used as positive controls for detection of T antigen and capsid VP-1, respectively. Cellular markers were detected with mAbs specific for glial fibrillary acidic protein (1:100 dilution, clone 6F2; Dako); synaptophysin (1:500 dilution, clone SY38; Boehringer Mannheim), and class III β-tubulin (1:500 dilution, clone TUJ1, a kind gift of Anthony Frankfurter, Department of Biology, University of Virginia). Next, biotinylated horse anti-mouse or anti-rabbit IgG and avidin-biotin peroxidase complex steps were performed according to the manufacturer’s instructions (Vector Laboratories). Finally, the sections were developed with diaminobenzidine substrate, lightly counterstained with hematoxylin, and coverslipped with Permount.
DNA Extraction from Paraffin Sections.
DNA was prepared from 100 μm of paraffin-embedded tissue by using the QIAamp Tissue Kit (Qiagen) according to the manufacturer’s instructions. Several precautions were taken to avoid cross-contamination of the samples including use of a microtome dedicated for human PCR sample analysis housed in a location separate from the main laboratory. The microtome and surrounding benchtop were sterilized, and block and knife holders were washed and autoclaved immediately before use. A fresh, disposable blade was used for each specimen, sections were handled with single-use items such as cotton-tipped applicators, and the microtome was cleaned with xylene and ethanol between samples. In addition, sections of negative control tissue were cut on the microtome between each specimen. Extractions were performed in a clean, dust-free area with sterile and disposable materials.
PCR Amplification.
PCR amplification was performed on extracted DNA by using three individual sets of primers: PEP1 and PEP2 (nucleotides 4255–4274 and 4408–4427, respectively), which amplify sequences in the N-terminal region encoding the Rb pocket-binding domain of JCV T antigen, TC1 and TC2 (nucleotides 2578–2600 and 2776–2797, respectively), which amplify a region within the C-terminal coding region of JCV T antigen, and VP2 and VP3 (nucleotides 1828–1848 and 2019–2039, respectively), which amplify a portion of the VP1 capsid gene sequence. The nucleotide sequences of the primers and amplified fragments are shown in Fig. 3. Amplification was carried out on 500 ng of template DNA with AmpliTaq DNA Polymerase (Perkin–Elmer) in a total volume of 50 μl. Hot-start PCR using AmpliWax PCR Gem 100 (Perkin–Elmer) according to the manufacturer’s instructions was performed in the presence of 2.5 mM MgCl2 and 0.5 μM of each primer (Oligos, Etc., Guilford, CT). A Perkin–Elmer Gene Amp 9600 PCR System was used exclusively for this study with denaturation at 95°C for 90 sec, followed by 45 cycles of denaturation at 95°C for 15 sec, annealing for 30 sec, and extension at 72°C for 30 sec. The annealing steps were performed at temperatures of 55°C for the PEP primers, 52°C for the TC primers, and 54°C for the VP primers. As a termination step, the extension time of the last cycle was increased to 7 min. Samples amplified in the absence of template DNA served as a negative control, whereas inclusion of serial dilutions of the plasmid, pBJC, containing the JCV genome as template served as a positive control for each procedure.
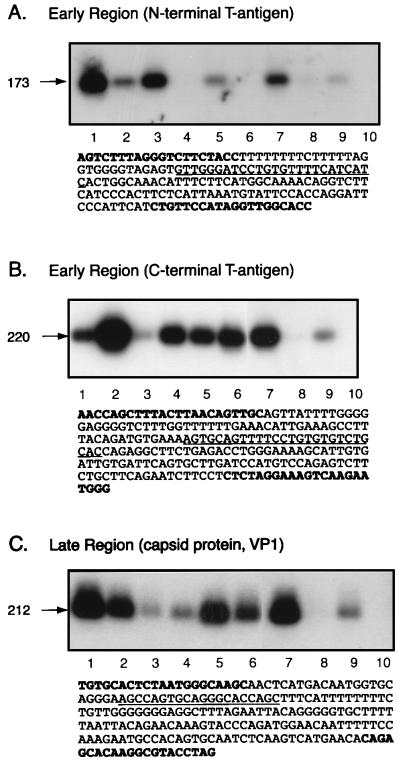
Figure 3.
Detection of JCV DNA sequences in human medulloblastomas. DNA from nine clinical samples was analyzed for the presence of JCV sequences by PCR using primers derived from the N terminus of T antigen (A), the C terminus of T antigen (B), and the late region of the viral genome (C). PCR products were analyzed by Southern blot analysis by using specific DNA probes as shown in Fig. 2. The nucleotide composition of the amplified DNA for each region is illustrated. Nucleotides depicting the position of PCR primers are shown in bold, and underlined nucleotides show the region detected by probes used for Southern blotting. Lane 10 represents a negative control (A, B, and C).
Southern Blot Hybridization.
Southern blot analysis was performed by using 15 μl of each PCR reaction separated by 2% agarose gel electrophoresis. Gels were depurinated in 0.2 M HCl, denatured in 1.5 M NaCl/0.5 M NaOH, and then neutralized in 1.5 M NaCl/0.5 M Tris⋅HCl, pH 7.4, for 15 min each. Amplified fragments then were transferred from the gel to a nylon membrane (Hybond-N; Amersham) in 20× SSC by using a semidry transfer apparatus (Turboblot; Schleicher & Schuell). The blots then were prehybridized in 0.5% SDS/10 mM EDTA/6× SSC/5× Denhardt’s solution/0.1 μg/μl salmon sperm DNA for 4 h followed by hybridization in the same solution containing 5 × 105 cpm/ml [γ-32P]ATP end-labeled oligonucleotide probe overnight at 65°C. Blots were washed twice in 2× SSC/0.1% SDS at 55°C for 5 min each and then three to five times in 0.1× SSC/0.1% SDS at 55°C for 3 min per wash. The membranes then were subjected to autoradiography overnight at −70°C. Probes used for Southern blotting included JCV probe (nucleotides 4303–4327) to detect fragments amplified with PEP primers, TC probe (nucleotides 2663–2688) for sequences amplified with TC primers, and VP probe (nucleotides 1872–1891) for those amplified with VP primers. Nucleotide sequences of the probes are shown in Fig. 3.
Subcloning and Sequencing of PCR-Amplified Fragments.
Amplified DNA fragments initially identified by Southern blot hybridization were excised from preparatory agarose gels stained with ethidium bromide, and DNA was purified by using the QIAquick PCR Purification Kit according to the manufacturer’s instructions (Qiagen). After extraction, the DNA was subcloned via the pBlue T7 Perfectly Blunt Cloning Kit by using the supplier’s protocol (Novagen), and clones were sequenced by automated fluorescent DNA cycle sequencing using the Applied Biosystems Prism 377 DNA Sequencer-XL.
RESULTS
Several earlier reports have described cases of patients with PML and concomitant CNS tumors including astrocytoma, oligodendroglioma, and CNS lymphoma (22). As well, two cases have described the occurrence of CNS tumors associated with JCV in the absence of immunosuppression, including an oligoastrocytoma and a childhood xanthoastrocytoma (23, 24). The development of medulloblastomas in transgenic mice harboring the JCV genome prompted us to initiate studies on pediatric medulloblastomas to evaluate the association of JCV with these tumors.
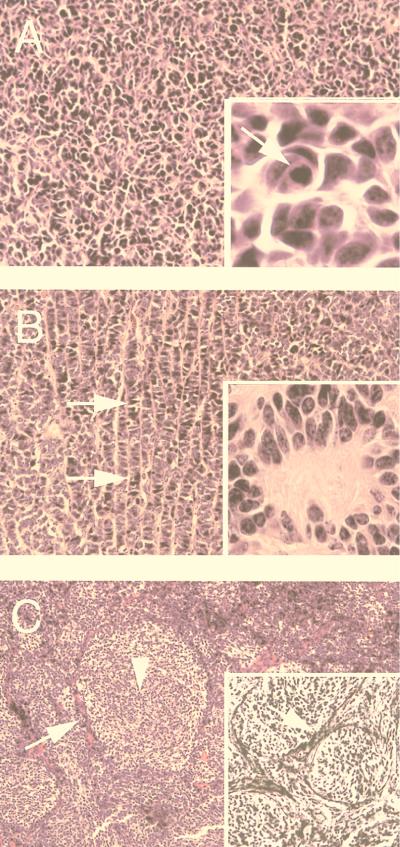
Toward this end, a collection of medulloblastomas (23 specimens) was assembled from various institutions in the United States, and, after histological assessment, they were examined for the presence of JCV DNA sequences by PCR and production of JCV early and late proteins by immunohistochemistry. Medulloblastoma samples were histologically classified according to the criteria of the World Health Organization, examples of which are shown in Fig. 1. Nine specimens exhibited the classic pattern with dense cellularity (Fig. 1A), four samples represented those with neuroblastic differentiation featuring rosettes and indian filing (Fig. 1B), and 10 cases featured the desmoplastic variant demonstrating a nodular pattern with mantles of poorly differentiated cells surrounding islands of neoplastic neuritogenesis (pale islands) (Fig. 1C). All three variants demonstrated a high nuclear-to-cytoplasmic ratio, frequent mitotic figures, and apoptotic bodies.
Figure 1.
Histological classification of medulloblastoma samples. Tumor specimens were classified as classic with a high degree of cellularity but no secondary architectural features (A), neuroblastic indicated by the presence of indian filing (see arrows in B) and rosettes (B Inset), or desmoplastic exhibiting pale islands (C). (C) Arrow and arrowhead indicate dark, poorly differentiated cells in the mantle zone surrounding a pale island and the pale island itself, respectively. (A Inset) The presence of apoptotic bodies. (C Inset) The reticulin impregnation characteristic of the compact, mantle-like zone of desmoplastic medulloblastoma. In all variants, a high nuclear-to-cytoplasmic ratio was observed [hematoxylin and eosin, original magnification, ×200; ×100 (C and C Inset); ×1,000 (all other Insets)].
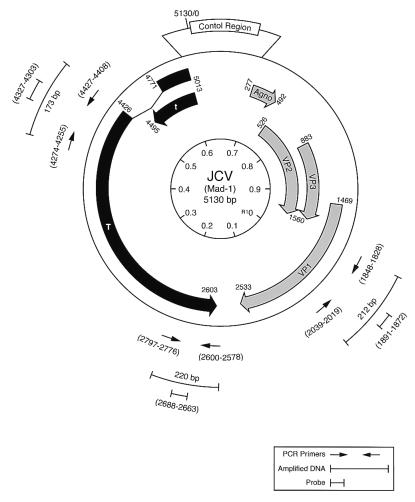
For detection of JCV gene sequences in the tumor samples, total DNA was extracted from paraffin-embedded tissues and evaluated by the PCR technique. Three pairs of primers were used in this study to amplify the N- and C-terminal regions of JCV T antigen between nucleotides 4255–4427 and 2578–2797, respectively, and the region corresponding to the viral capsid protein, VP1, between nucleotides 1828 and 2039. Fig. 2 schematizes the JCV genomic structure, depicting the positions of the primers, amplified sequences, and the DNA probes that were utilized for detection of the amplified DNA by Southern blot analysis. As a control, DNA from five samples of normal tissue from patients ages between 1 and 10 years old (not associated with tumor or PML) were prepared and examined for the presence of the JCV sequences. All PCR products were resolved by agarose gel electrophoresis and analyzed by Southern blot hybridization by using JCV-specific probes derived from the internal regions of the amplified fragments as specified in Fig. 2. Sequence analysis of amplified DNA fragments from seven samples were carried out after subcloning of the PCR products followed by automated DNA sequencing.
Figure 2.
Structural organization of the JCV genome and the position of the oligonucleotide primers and probes used for PCR amplification and Southern blot analysis. The numbers within the inner circle represent the map positions, with 0.0 being the EcoRI site (25). Thick arrows depict coding regions for the viral early protein T antigen (solid) and late proteins (shaded). The position of the viral control region between the initiation sites for early and late proteins is shown. The locations of the PCR primers are shown by thin arrows outside of the circle. The sizes of the amplified DNAs and the DNA probes used for Southern blot analysis for the detection of PCR products are depicted.
Fig. 3 illustrates representative Southern blot analysis of the PCR products from nine tumor samples and the sequence composition of the amplified DNA. As is evident, six and eight of nine samples shown in Fig. 3 (A and B, respectively) contain DNA fragments corresponding to the N- and C-terminal regions of T antigen. Further, eight of nine samples contain the DNA sequence corresponding to the late region of JCV (Fig. 3C). DNA from normal tissue showed no detectable signals corresponding to the JCV sequences (data not shown). DNA amplification and Southern blot analysis were repeated multiple times with different preparations of primers, probes, and DNA to verify the reproducibility of the data. Only those samples in which PCR amplification was repeatedly detected were considered as positive specimens for JCV sequences. Table 1 summarizes the characterization of the tumor samples based on their pathological diagnoses, anatomical location, immunocytochemical analysis, and the presence of JCV sequences. As shown in this table, 20 samples (87%) are positive for the N-terminal region of JCV T antigen, 13 samples (56.5%) contain sequences corresponding to the JCV T antigen C-terminal region, and 20 samples (87%) possess sequences of the VP1 region. Furthermore, these data indicate that 11 samples (48%) of the tumor tissue contain the DNA sequences of the JCV genome that correspond to all three amplified regions, including the N and C termini of T antigen and the viral capsid, VP1. The presence of SV40 DNA sequences also was evaluated by using a set of specific primers that recognize the C terminus of SV40 early genome as described previously (26). The preliminary observations indicated that 5 of 23 samples (sample numbers 2, 6, 10, 16, and 22 in Table 1) contain SV40 DNA sequences in addition to JCV. This observation is in agreement with recent reports on the detection of SV40 sequences in 29% of pediatric medulloblastomas (27).
Table 1.
Clinical, immunohistochemical, and DNA analysis of human medulloblastomas
| No. | Sex | Age, years | Diagnosis | Location | PCR amplification
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Immunohistochemistry
|
T-Ag (N-term) | T-Ag (C-term) | VP-1 | ||||||||
| T-Ag | Class III | Syn | GFAP | ||||||||
| 1 | M | 5 | Classic | Vermis | N/A | + | + | − | + | − | + |
| 2 | M | 7 | Desmoplastic | Right hemisphere | N/A | + | + | − | + | + | + |
| 3 | M | 6 | Desmoplastic | Left hemisphere | N/A | + | + | − | + | − | + |
| 4 | M | 4 | Classic | Vermis | N/A | N/A | N/A | N/A | + | + | + |
| 5 | M | 5 | Desmoplastic | Left hemisphere | N/A | + | + | − | + | − | + |
| 6 | F | 11 | Classic | Midline/vermis | N/A | + | + | − | + | − | + |
| 7 | M | 2 | Neuroblastic | Vermis | N/A | + | + | + | + | + | + |
| 8 | M | 1.5 | Desmoplastic | Left hemisphere | − | + | − | − | + | − | + |
| 9 | F | 4 | Desmoplastic | Left hemisphere | − | + | − | − | + | + | + |
| 10 | M | 15 | Desmoplastic | Left hemisphere | + | + | + | − | + | − | − |
| 11 | F | 42 | Desmoplastic | Midline/vermis | − | + | − | − | + | − | − |
| 12 | M | 7 | Classic | Midline/vermis | − | + | + | − | + | + | + |
| 13 | F | 18 | Neuroblastic | Midline/vermis | − | + | − | − | + | − | + |
| 14 | F | 8 | Desmoplastic | Posterior fossa mass | − | + | − | − | + | − | + |
| 15 | M | 7 | Classic | Midline/vermis | + | + | + | − | + | + | + |
| 16 | M | 9 | Classic | Midline/vermis | − | + | + | − | + | + | + |
| 17 | F | 3 | Desmoplastic | Midline/vermis | + | + | + | − | + | + | + |
| 18 | F | 9 | Desmoplastic | Posterior fossa mass | − | + | + | − | − | + | + |
| 19 | M | 5 | Neuroblastic | Posterior fossa mass | + | + | + | − | + | + | + |
| 20 | F | 2 | Classic | Posterior fossa mass | − | + | + | − | − | + | + |
| 21 | M | Newborn | Classic | Midline/vermis | − | + | + | − | + | + | + |
| 22 | M | 12 | Classic | Left hemisphere | − | + | − | − | − | − | − |
| 23 | M | 5 | Neuroblastic | Posterior fossa mass | − | + | + | + | + | + | + |
Diagnosis of the tumors is based on the World Health Organization criteria for medulloblastomas as detailed in Materials and Methods. The anatomical location of the tumors is shown. The age at the time of tumor resection and gender (F, female; M, male) of each patient is shown. N/A indicates that samples were not available or were not in appropriate condition for immunohistochemical analysis. T-Ag, T antigen; Class III, class III β-tubulin; Syn, synaptophysin; GFAP, glial fibrillary acidic protein.
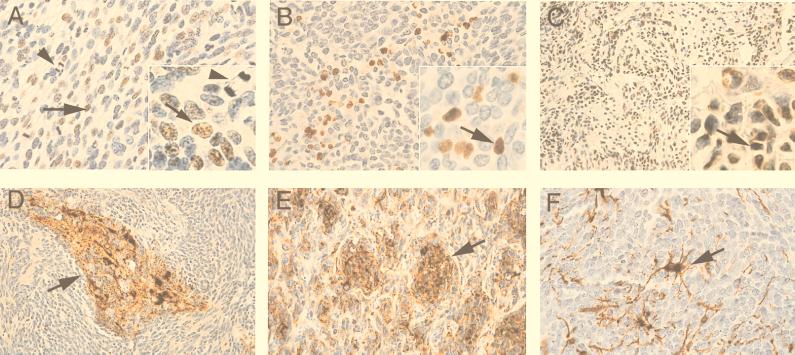
Immunohistochemistry then was performed on the samples to confirm their histological classification and to assess the samples for the expression of JCV T antigen. Because of the fairly stringent parameters required to perform immunohistochemistry for T antigen, only 16 tumors could be evaluated for production of the viral oncoprotein in tumor tissue (Table 1, 8–23). Conditions for T antigen immunostaining were optimized with sections of paraffin-embedded tumors derived from Syrian hamsters inoculated intracerebrally with JC virus. T antigen-positive nuclei were found by immunohistochemical analysis in 4 of the 16 specimens. Pathologically, this group consisted of one classical, one neuroblastic, and two desmoplastic medulloblastomas. Fig. 4 A–C illustrates results from immunostaining of classic (Fig. 4A), neuroblastic (Fig. 4B), and desmoplastic (Fig. 4C) medulloblastoma with antibody to T antigen. The number of nuclei that were immunopositive ranged from approximately 5% to 20% of the total tumor cells per high-power (×40) microscopic field. Notably, positive nuclei showed no predilection for cells with greater or lesser degrees of neuronal differentiation. This was demonstrated most clearly in the desmoplastic medulloblastoma, in which the less differentiated cells in compact, mantle-like areas and the more differentiated neuronal cells within the pale islands were both T antigen-positive. Synaptophysin and class III β-tubulin immunoreactivities provide evidence of the neuronal origin of these tumors (Fig. 4 D and E, respectively). Astrocytes, identified by the presence of glial fibrillary acidic protein, were morphologically reactive and likely not neoplastic (Fig. 4F). Of note, immunostaining for the JCV capsid protein, VP1, was negative in all four of these specimens. None of the control cerebellar specimens showed positive immunostaining for either T antigen or VP1 (data not shown).
Figure 4.
Immunohistological analysis of human of medulloblastomas. Immunostaining of tumor samples with anti-T antigen antibody (A, B, and C). Arrows indicate T antigen-positive cells. Arrowhead in A and A Inset show the presence of mitotic figures. Immunostaining of tumor samples with neuronal markers including synaptophysin (D), class III β-tubulin (E), and glial fibrillary acidic protein (GFAP, F) is shown [hematoxylin counterstain, original magnification: ×400 (A, B, E, and F); ×200 (C and D); ×1,000 (Insets)].
DISCUSSION
The use of highly sensitive molecular techniques has led to the detection of polyomavirus sequences, in particular, SV40, in a variety of human neoplastic tissues including choroid plexus tumors, ependymomas, medulloblastomas, osteosarcomas, and, more recently, in mesotheliomas (28). Although the presence of DNA sequences in tumor cells may not establish the etiological role of SV40 in the development of tumors in humans, results from animal studies have provided compelling evidence supporting the ability of this virus to cause cancer (29–31). Recently, the human polyomavirus, JCV, has received special attention because of the high incidence of PML, the fatal demyelinating disease induced by this virus, in AIDS patients (11). JCV also possesses oncogenic potential in experimental animals (18, 20, 21, 32–34), and its early protein, T antigen, has a transforming capability in cell culture (35, 36).
Because epidemiologic studies have revealed a surprisingly high percentage of the human population infected with JCV in early childhood, we sought to investigate the association of JCV with human pediatric neoplasia. We focused our attention on a malignancy of the CNS, more specifically, medulloblastoma, because results from earlier studies have demonstrated that cell type-specific enhancer/promoter activity of the JCV early gene restricts production of the viral early protein, T antigen, to CNS cells (for review, see ref. 37) and observations from animal studies have pointed to the ability of JCV to cause medulloblastomas (21). Moreover, results from transgenic animal studies further link the oncogenic potential of JCV to its early protein (18, 32, 34).
The results presented in this study provide evidence that the JCV genome can be detected in a significant fraction of human medulloblastomas and demonstrate further that JCV T antigen is produced in tumor cells. These observations raise several important questions including: (i) whether activation of the JCV early promoter, which results in the production of T antigen, is the first event in the development of human medulloblastomas; (ii) if so, how does the viral early promoter, which is silent in immunocompetent individuals, become activated in medulloblastoma patients who usually exhibit no sign of an impaired immune system; (iii) does the JCV T antigen produced in human tumors, similar to observations from tumors in experimental animals, form complexes with tumor suppressors including p53 and pRb and deregulate their function (17) and/or play a role in other events such as chromosomal damage (38) and activation of Pax (8) and, perhaps, other host regulatory genes. Finally, one also may question whether JCV functions as a cofactor or requires cofactors in inducing medulloblastomas.
As mentioned earlier, transcriptional activation of the JCV early promoter is mediated through the cooperative interaction of several ubiquitous and inducible regulatory proteins (37). Among these proteins, NFκB, c-jun/AP-1, and NF-1 are particularly interesting because their activity or expression is modified by the physiological condition of the cells. Through such modifications, the JCV early promoter can be activated and result in high levels of T antigen expression. In support of this notion, previous in vitro studies have demonstrated that activation of NFκB upon treatment of neural cells with PMA or cytokines causes enhancement of the JCV early promoter in CNS cells (39, 40). Thus, one may hypothesize that subtle and, perhaps, transient changes in the physiological condition of individuals harboring JCV sequences may cause alterations in the expression of cytokines and other cellular regulators, which, in turn, may trigger expression of the early promoter of the JCV genome in the CNS.
Interestingly, results from our immunohistochemical studies have shown that not all of the tumor cells produce T antigen, suggesting that expression of JCV T antigen may be extinguished in some, but not all, of the tumor cells. Consistent with this observation, our earlier studies in a transgenic animal model for medulloblastoma induced by expression of the JCV early genome have shown that not all tumor cells may produce T antigen (18). Moreover, our more recent in vitro studies demonstrate that expression of T antigen in cell lines derived from JCV-induced medulloblastomas in transgenic animals may be extinguished after several passages (B.K., unpublished observations). As such, one may speculate a hit-and-run role for T antigen in the development of medulloblastomas in which expression of T antigen may induce a cascade of events that leads to tumor formation. Once cells are programmed for uncontrolled proliferation, production of T antigen may no longer be essential and may be terminated. Currently, experiments are in progress to investigate the effect of transient expression of JCV T antigen in induction of CNS tumors in a whole animal system.
Sequencing of several clones derived from the PCR products amplified with primers corresponding to the N terminus of JCV T antigen revealed that five of the human medulloblastoma cases contained, in addition to the JCV DNA, sequences corresponding to another papovavirus, i.e., SV40. Of note, we found no DNA sequences corresponding to BK virus in the tested specimens. This is an interesting observation in light of seroepidemiological studies reporting that although 65% of children between the ages of 10 and 15 are seroprevalent to JCV, only 9% of them are positive for SV40 (28). Of note, the presence of the SV40 genome in human medulloblastomas has been reported previously (27, 41).
Interestingly, sequences for the N terminus of JCV T antigen were detected in 87% of the samples in this study, whereas the C-terminal region was detected in 56.5% of the samples. It is important to note that sequences within the N-terminal region of T antigen that were amplified correspond to the region of JCV T antigen that interacts with pRb. This region of T antigen, which is highly conserved among the primate polyomaviruses SV40, JCV, and BKV, has been shown to be tumorigenic in SV40 T antigen transgenic mice (42). As such, the significance of the detection of the N terminus of JCV T antigen in medulloblastomas should be studied further. Of note, sequences of the highly divergent C terminus of JCV were detected in fewer medulloblastoma samples, perhaps because of the high specificity and stringency of the experimental protocol.
The detection of JCV sequences in human brain tumors also renews an important question regarding the route of transmission and the site of latency for this virus. Although current opinion favors an upper respiratory route for JCV infection, more sensitive molecular techniques should be employed to investigate maternal and/or perinatal transmission of JCV. According to one model, after primary infection, JCV persists in renal tissue (43, 44), where it can replicate, although poorly, to generate progeny in urine (45). Under certain physiological conditions, the virus can be transferred to brain, presumably via B cells (for review, see ref. 46), and, after infection of CNS cells, actively express the viral genome. Recently, studies have provided evidence for frequent transmission of JCV from parent to child (47).
Thus, horizontal and perhaps vertical transmission of JCV in humans may result in expression of the viral early protein in the brains of children whose immune status permits transcription of the viral early promoter. Because infection with JCV occurs mainly in early childhood, the possibility that the detection of JCV within human medulloblastomas results from initial exposure to the virus also should be considered. Although the presence of JCV sequences in medulloblastomas may not establish a cause-and-effect relation to CNS malignancy, our observations, along with the established neurotropic nature of JCV and in vitro and in vivo animal studies demonstrating the transforming ability of JCV, invite future studies to reevaluate the role of this virus in the pathogenesis of pediatric medulloblastomas.
Acknowledgments
We thank past and present members of the Center for NeuroVirology and NeuroOncology for their support, insightful discussion, and sharing of ideas and reagents. We thank Cynthia Schriver for editorial assistance and preparation of this manuscript. This work was made possible by grants awarded by the National Institutes of Health to K.K.
ABBREVIATIONS
- PML
progressive multifocal leukoencephalopathy
- CNS
central nervous system
- JCV
human neurotropic JC virus
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Farwell J R, Dohrmann G J, Flannery J T. J Neurosurg. 1984;61:657–664. doi: 10.3171/jns.1984.61.4.0657. [DOI] [PubMed] [Google Scholar]
- 2.Roberts R O, Lynch C F, Jones M P, Hart M N. J Neuropathol Exp Neurol. 1991;50:134–144. doi: 10.1097/00005072-199103000-00005. [DOI] [PubMed] [Google Scholar]
- 3.Bigner S H, McLendon R E, Fuchs H E, McKeever P E, Friedman H S. Cancer Genet Cytogenet. 1997;97:125–134. doi: 10.1016/s0165-4608(96)00404-9. [DOI] [PubMed] [Google Scholar]
- 4.Griffin C A, Hawkins A L, Packer R J, Rorke L B, Emanuel B S. Cancer Res. 1988;48:175–180. [PubMed] [Google Scholar]
- 5.Cogen P H, McDonald J D. J Neurooncol. 1996;29:103–112. doi: 10.1007/BF00165523. [DOI] [PubMed] [Google Scholar]
- 6.Adesina A M, Nalbantoglu J, Cavenee W K. Cancer Res. 1994;54:5649–5651. [PubMed] [Google Scholar]
- 7.Ohgaki H, Eibl R H, Schwab M, Reichel M B, Mariani L, Gehring M, Petersen I, Holl T, Wiestler O D, Kleihues P. Mol Carcinog. 1993;8:74–80. doi: 10.1002/mc.2940080203. [DOI] [PubMed] [Google Scholar]
- 8.Kozmik Z, Sure U, Ruedi D, Busslinger M, Aguzzi A. Proc Natl Acad Sci USA. 1995;92:5709–5713. doi: 10.1073/pnas.92.12.5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Padgett B L, Walker D L. J Infect Dis. 1973;127:467–470. doi: 10.1093/infdis/127.4.467. [DOI] [PubMed] [Google Scholar]
- 10.Padgett B L, ZuRhein G, Walker D L, Echroade R, Dessel B. Lancet. 1971;i:1257–1260. doi: 10.1016/s0140-6736(71)91777-6. [DOI] [PubMed] [Google Scholar]
- 11.Berger J R, Concha M. J Neurovirol. 1995;1:5–18. doi: 10.3109/13550289509111006. [DOI] [PubMed] [Google Scholar]
- 12.Frisque R J, White F A. In: Molecular Neurovirology. Roos R P, editor. Towana, NJ: Humana; 1992. pp. 25–158. [Google Scholar]
- 13.Dyson N, Buchkovich K, Whyte P, Harlow E. Cell. 1989;58:249–255. doi: 10.1016/0092-8674(89)90839-8. [DOI] [PubMed] [Google Scholar]
- 14.Dyson N, Bernards R, Friend S H, Gooding L R, Hassell J A, Major E O, Pipas J M, VanDyke T, Harlow E. J Virol. 1990;64:1353–1356. doi: 10.1128/jvi.64.3.1353-1356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ludlow J W. FASEB J. 1993;7:866–871. doi: 10.1096/fasebj.7.10.8344486. [DOI] [PubMed] [Google Scholar]
- 16.Staib C, Pesh J, Gerwig R, Gerber J-K, Brehm U, Stangl A, Grummt F. Virology. 1996;219:237–246. doi: 10.1006/viro.1996.0241. [DOI] [PubMed] [Google Scholar]
- 17.Krynska B, Gordon J, Otte J, Franks R, Knobler R, Giordano A, DeLuca A, Khalili K. J Cell Biochem. 1997;67:223–230. doi: 10.1002/(sici)1097-4644(19971101)67:2<223::aid-jcb7>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 18.Krynska B, Otte J, Franks R, Khalili K, Croul S. Oncogene. 1999;18:39–46. doi: 10.1038/sj.onc.1202278. [DOI] [PubMed] [Google Scholar]
- 19.Small J A, Scangos G A, Cork L, Jay G, Khoury G. Cell. 1986;46:13–18. doi: 10.1016/0092-8674(86)90855-x. [DOI] [PubMed] [Google Scholar]
- 20.London W T, Houff S A, Madden D I, Fucillo D A, Gravell M, Wallen W C, Palmer A E, Sever J L, Padgett B L, Walker D L, et al. Science. 1978;201:1246–1248. doi: 10.1126/science.211583. [DOI] [PubMed] [Google Scholar]
- 21.ZuRhein G M. In: Polyomaviruses and Human Neurological Disease. Sever J L, Madden D M, editors. New York: Liss; 1983. pp. 205–221. [Google Scholar]
- 22.Gallia G L, Gordon J, Khalili K. J Neurovirol. 1998;4:175–181. doi: 10.3109/13550289809114517. [DOI] [PubMed] [Google Scholar]
- 23.Boldoroni R, Caldarelli-Stefano R, Monga G, Zocchi M, Mediati M, Tosoni A, Ferrante P. J Neurovirol. 1998;4:242–245. doi: 10.3109/13550289809114524. [DOI] [PubMed] [Google Scholar]
- 24.Rencic A, Gordon J, Otte J, Zoltick P, Khalili K, Andrews D. Proc Natl Acad Sci USA. 1996;93:7352–7357. doi: 10.1073/pnas.93.14.7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frisque RJ, Bream G L, Cannella M T. J Virol. 1984;51:458–469. doi: 10.1128/jvi.51.2.458-469.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Testa J R, Carbone M, Hirvonen A, Khalili K, Krynska B, Linnainmaa K, Pooley F D, Rizzo P, Rusch V, Xiao G H. Cancer Res. 1998;58:4505–4509. [PubMed] [Google Scholar]
- 27.Huang H, Rels R, Yonekawa Y, Lopes J M, Kleihues P, Ohgaki H. Brain Pathol. 1999;9:33–44. doi: 10.1111/j.1750-3639.1999.tb00207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Butel J S, Lednicky J A. J Natl Cancer Inst. 1999;91:119–134. doi: 10.1093/jnci/91.2.119. [DOI] [PubMed] [Google Scholar]
- 29.Brinster R, Chen H, Messing A, van Dyke T, Levine A, Palmiter R D. Cell. 1984;37:367–379. doi: 10.1016/0092-8674(84)90367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cicala C, Pompetti F, Carbone M. Am J Pathol. 1993;142:1524–1533. [PMC free article] [PubMed] [Google Scholar]
- 31.Diamondopoulos G T. J Natl Cancer Inst. 1973;50:1347–1365. doi: 10.1093/jnci/50.5.1347. [DOI] [PubMed] [Google Scholar]
- 32.Franks R, Rencic A, Gordon J, Zoltick P, Knobler R, Khalili K. Oncogene. 1996;12:2573–2578. [PubMed] [Google Scholar]
- 33.Ohsumi S, Ikehara I, Motoi M, Ogawa K, Nagashima K, Yasui K. Jpn J Cancer Res. 1985;76:429–431. [PubMed] [Google Scholar]
- 34.Small J, Khoury G, Jay G, Howley P, Scangos G. Proc Natl Acad Sci USA. 1986;83:8288–8292. doi: 10.1073/pnas.83.21.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Neill F J, Frisque R J, Xu X, Hu Y X, Carney H. Oncogene. 1995;10:1131–1139. [PubMed] [Google Scholar]
- 36.Tavis J E, Trowbridge P W, Frisque R J. Virology. 1994;199:384–392. doi: 10.1006/viro.1994.1136. [DOI] [PubMed] [Google Scholar]
- 37.Raj G, Khalili K. Virology. 1995;213:283–291. doi: 10.1006/viro.1995.0001. [DOI] [PubMed] [Google Scholar]
- 38.Neel J V, Major E O, Awa A A, Glover T, Burgess A, Traub R, Curfman B, Satoh C. Proc Natl Acad Sci USA. 1996;93:2690–2695. doi: 10.1073/pnas.93.7.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayreddy R P R, Safak M, Razmara M, Zoltick P, Khalili K. J Virol. 1996;70:2387–2393. doi: 10.1128/jvi.70.4.2387-2393.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ranganathan P N, Khalili K. Nucleic Acids Res. 1992;8:1959–1964. doi: 10.1093/nar/21.8.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreig P, Amtmann E, Jonas D, Fischer H, Zang K, Sauer G. Proc Natl Acad Sci USA. 1981;78:6446–6450. doi: 10.1073/pnas.78.10.6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Tobin G J, Pipas J M, Van Dyke T. Oncogene. 1992;7:1167–1175. [PubMed] [Google Scholar]
- 43.Chesters P M, Heritage J, McCance D J. J Infect Dis. 1983;147:676–684. doi: 10.1093/infdis/147.4.676. [DOI] [PubMed] [Google Scholar]
- 44.Tominaga T, Yogo Y, Kitamura T, Aso Y. Virology. 1992;186:736–791. doi: 10.1016/0042-6822(92)90040-v. [DOI] [PubMed] [Google Scholar]
- 45.Kitamura T, Aso Y, Kuniyoshi N, Hara K, Yogo Y. J Infect Dis. 1990;161:1128–1133. doi: 10.1093/infdis/161.6.1128. [DOI] [PubMed] [Google Scholar]
- 46.Gallia G L, Houff S A, Major E O, Khalili K. J Infect Dis. 1997;176:1603–1609. doi: 10.1086/514161. [DOI] [PubMed] [Google Scholar]
- 47.Kunitake T, Kitamura T, Guo J, Taguchi F, Kawabe K, Yogo Y. J Clin Microbiol. 1995;33:1448–1451. doi: 10.1128/jcm.33.6.1448-1451.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]