Abstract
mAb OV569 was made by immunizing mice with ovarian carcinoma cells. It binds to cells from ovarian carcinomas and, to a lesser extent, to cells from certain other carcinomas whereas the binding to normal tissues is low to nondetectable. It also binds to soluble molecule(s) in culture supernatants from antigen-positive carcinomas. OV569 recognizes a protein(s) of 42–45 kDa with the same N-terminal amino acid sequence as the membrane-bound portion of mesothelin and megakaryocyte potentiating factor (MPF). Binding assays with fusion proteins comprising either the N-terminal part of mesothelin/MPF (D1Ig), reported to be easily cleaved off, or a noncleavable, membrane-associated part (D2Ig) showed that OV569 only binds to D2hIg. A new member of the mesothelin/MPF family was discovered, which has an 82-bp insert in the membrane-associated part, leading to a frameshift of 212 bp, and whose predicted molecular structure indicates that it is soluble. To test patient sera for soluble tumor antigen, antigen was isolated from cell-free tumor culture supernatants via immunoadsorption with OV569 and used to generate murine mAbs to an epitope different from the one to which OV569 binds, after which mAbs to two different epitopes were used to develop a “sandwich ELISA.” Using this assay, the level of circulating antigen was elevated significantly in 23 of 30 sera from patients with ovarian carcinoma, as compared with 0 of 68 sera from healthy controls, 0 of 3 sera from patients with nonneoplastic diseases, and 25 of 75 sera from patients with other tumors. Soluble molecules of the mesothelin/MPF family may provide useful new marker(s) for diagnosis of ovarian carcinoma and/or monitoring its response to therapy.
Keywords: ovarian cancer, serum assays, tumor markers, immunotherapy
Detection of tumor markers that are released into the circulation can aid in the diagnosis and/or monitoring of therapeutic responses of patients with various tumors, including carcinomas of ovary (1–4), prostate (5), the gastrointestinal tract (6, 7), or breast (8). CA125 is the most commonly used serum marker for patients with ovarian carcinoma (1–3). Although it has proven clinically valuable in monitoring the response of patients to therapy, some ovarian carcinomas do not express CA125, and CA125 often is increased in patients with inflammatory disease. Thus, there is a need for improvement, either in the form of a more specific and/or sensitive assay or an assay that uses a different marker and can be used to complement CA125 toward the goal to improve patient survival by improving diagnosis.
We originally made a mouse mAb, OV569, which, according to immunohistology, can bind to >95% of sections of ovarian carcinoma and to a fraction of other human carcinomas, including non-small-cell lung carcinomas. Its reactivity with normal human tissues, except for mesothelium, is very low. OV569 identifies a 42- to 45-kDa protein(s) with an N-terminal amino acid sequence identical to that of the membrane-bound portion of mesothelin (9–11) and megakaryocyte potentiating factor, MPF (12). The frequent expression of such proteins in ovarian carcinoma suggests that they are present in the normal cells from which the tumors originate and/or that they contribute to the malignant phenotype. However, the function of these molecules in the cancer cells is yet unknown.
Antigen(s) to which OV569 binds was found to be released into the cell-free culture supernatant from antigen-positive carcinomas and into cell-free malignant effusions of patients with antigen-positive carcinomas. This differs from the published firm association between mesothelin and the cell surface (9, 11). Furthermore, as reported in this paper, OV 569 does not bind to a fusion protein, D1hIg, comprising the N-terminal end of mesothelin/MPF and reported to be easily cleaved off (12), whereas it binds to a different fusion protein, D2Ig, comprising the membrane-associated part of the molecule. We, therefore, searched for new member(s) of the mesothelin/MPF family by cloning cDNAs from a carcinoma line releasing soluble proteins binding to OV569 into cell culture supernatants. As reported in this paper, one such member so far has been identified. It has a frame shift involving 98 aa and has a molecular structure compatible with solubility. We show further that soluble mesothelin/MPF-related protein(s) binding to OV569 are detectable in the circulation of patients with ovarian carcinomas by using a double-determinant (13) immunoassay (“sandwich ELISA”). The expression of mesothelin/MPF-related proteins in the vast majority of ovarian carcinomas, as well as in certain other tumors, suggests that they have biological functions in the neoplastic cells and that at least some of them provide clinically useful therapeutic targets and diagnostic markers.
MATERIALS AND METHODS
Establishment of Hybridomas.
A hybridoma of mouse origin that makes a mAb, OV569, was made first by using standard techniques (14) after repeated immunization of BALB/c mice with cells from the malignant ascites of a patient with ovarian carcinoma and screening hybridomas by an ELISA (15). This mAb, which is an IgG1, was used for all studies described in this paper except for the double-determinant immunoassays, which also used other mAbs to the antigen recognized by OV569 (see below).
A second set of hybridomas was made to the antigen to which OV569 binds but that recognizes different epitopes. For use as immunogen, the OV569-binding antigen was purified from cell-free supernatants of cultured, antigen-positive ovarian and lung carcinomas by using affinity chromatography via a column to which mAb OV569 had been coupled. Purified antigen (30 μg protein in 0.1 ml) was mixed with 0.1 ml of Ribi adjuvant (Ribi Immunochem), and the mixture was injected into BALB/c mice at two s.c. sites, followed 14 days later by the first boost, which was given i.p. For boosting, the Ribi adjuvant was mixed with antigen purified by affinity chromatography from the supernatant of H4013 lung carcinoma cultures. After three boostings, the mice were given a final boost by injecting the antigen i.v.
Three days after the final boost, spleens were removed and hybridizations were carried out similar to the one described above to obtain OV569. The supernatants were tested for binding to the immunizing antigen, to a fusion protein, D2hIg, which had been prepared based on the sequence of the membrane-binding portion of mesothelin and which incorporated an Ig tail, and to antigen purified via immunoadsorption with an OV569 affinity column from tumor culture supernatant and ascites. The supernatants also were tested for binding to tumor cells expressing (or lacking) the OV569-defined antigen. Positive supernatants subsequently were screened in a competition assay in which plated antigen was combined with supernatant and biotinylated OV569 to select for mAbs that defined epitopes different from the one recognized by OV569. Three hybridomas, 4H3, 3G3, and 1A6, were identified that made antibodies binding to D2hIg and to antigen purified from supernatant of cultures of OV569-positive carcinomas and that did not compete with the OV569 mAb. Hybridoma 4H3, which makes an IgG1 mAb with the same name, was used, together with OV569 to establish a double-determinant (sandwich ELISA) assay (see below).
Immunohistology.
The Vectastain ABC system was used on frozen sections after the protocol provided by the manufacturer (Vector Laboratories). Slides were evaluated under code and checked regularly by an independent investigator. The tumor data are presented as “positive” when at least one-third of the cells stained, and the normal tissue data are scored as “positive” when more than 5% of cells stained at the same mAb dilutions. Both neoplastic cells and stroma cells were observed in tumor samples, and only the former were stained.
Fluorescence-Activated Cell Sorting (FACS) Analysis.
mAb binding to cell surface antigens was identified by FACS by using a Coulter Epics C FACS as described (16). The mean fluorescence intensity was determined, and the linear fluorescence equivalence (LFE) was calculated from the mean fluorescence intensity. The LFE of each test sample divided by the LFE of a negative control gave a ratio of the brightness of cells stained by specific vs. control antibody. An LFE >1.3 was rated as positive.
Antigen Purification.
An affinity chromatography column was prepared by coupling 1.5 g of cyanogen bromide-activated Sepharose 4B (Sigma) with 9.2 mg of OV569 according to Sigma’s protocol. Material from which antigen was purified (ascites, tumor culture supernatant) was passed through the column, after which the column was washed with 0.02% NaN3 in PBS, pH 7.2, until there was no longer any flow-through detected at OD280. Subsequently, it was eluted with a pH 2.6 elution buffer, and the eluate was collected in 2 ml, neutralized with 2.5 M Tris⋅HCl buffer, pH 8.8, and measured at OD280 and OD260.
Western Blotting.
The sample was diluted 1:1 with SDS sample buffer, and 20 μl (300 ng/lane) was loaded on a 14% SDS/PAGE gel and then run with SDS running buffer at 125 V for about 1.5 hr, followed by blotting to NOVEX poly(vinylidene difluoride) membrane (NOVEX, San Diego) with Tris-glycine SDS transfer buffer.
Blotting to the poly(vinylidene difluoride) membrane was blocked with 5% nonfat milk in washing buffer (0.2% Tween 20/PBS) at room temperature for 1 hr, followed by washing with washing buffer once for 10 min and twice for 5 min. OV569 (3 μg/ml) diluted in washing buffer with 1% nonfat milk was incubated at room temperature for 1 hr, after which there were three washes as above. A horseradish peroxidase-labeled goat anti-mouse IgG antibody, diluted 1:5,000, was added in washing buffer with 1% nonfat milk, followed by incubation at room temperature for 1 hr, washing with washing buffer once for 10 min and four times each for 5 min. The chemiluminescence substrate (ECL; Amersham Pharmacia) was added onto the membrane for 1 min in a dark room, followed by exposure on film.
Sandwich ELISA.
The wells of Maxisorp Immuno plates (Nalge Nunc) were coated at 4°C with 50 ng of purified 4H3 Ig in 50 μl/well carbonate-bicarbonate buffer (C-3041; Sigma). After removal of the supernatant, the wells were blocked for 2 hr at room temperature with 200 μl/well GSC blocking buffer (Genetic Systems, Seattle). This was followed by four washes, 200 μl/well, with PBS containing 0.1% Tween. Subsequently, 100 μl/well was added of sera diluted, at 2-fold steps, between 1:40 and 1:1,280, in blocking buffer (Genetic Systems), and the plates were incubated for 1 hr at room temperature. Subsequently, the plates were washed four times, 200 μl/well, with PBS containing 0.1% Tween, after which 50 μl/well biotinylated OV569 was added at 200 ng/ml in conjugate diluent (Genetic Systems), followed by 1-hr incubation at room temperature. The plates were washed four times, 200 μl/well, with PBS containing 0.1% Tween, after which 50 μl/well was added of horseradish peroxidase-streptavidin diluted 1:1,000 in conjugate diluent (Genetic Systems), followed by 45-min incubation at room temperature. After four washes, 200 μl/well, with PBS containing 0.1% Tween, buffered substrate TMB plus 1% conjugate (Genetic Systems) was added and the plates were incubated for 15 min. The reaction was stopped by addition of 2 M H2S04, and the plates were read in an ELISA reader (Spectracount microplate photometer; Packard) at 460 nm. The assay was carried out by including, in all experiments, the same two control samples: one from a healthy volunteer and one from a patient with ovarian carcinoma (C+), which had been shown in pilot tests not to give a detectable signal at a serum dilution of 1:40 (negative control) and a signal at a dilution of 1:1,280 (positive control), respectively.
Construction of D1hIg and D2hIg Fusion Proteins.
Two fusion proteins, D1hIg and D2hIg, were constructed, corresponding to the published, 33-kDa, soluble domain of MPF (10) and to the published, 44-kDa, membrane-bound domain of MPF and mesothelin (10), respectively. Vectors were constructed by PCR, with the sequences encoding the cleavage site and linker synthesized as part of the PCR primer. PCR was performed by using as a template cDNA of 4013, a lung tumor cell line developed in the principal investigator’s previous laboratory. The two PCR products, D1 and D2, were verified by sequencing and inserted into pCDM7B fused to the hinge and two constant domains of human IgG1 as described (17). Proteins were produced from COS cells by transfection of the mammalian expression plasmids as described (18).
3′ Rapid Amplification of cDNA Ends Amplification.
Total RNA was isolated from HE1P prostatic carcinoma cells (which were selected because they release soluble protein into cell-free culture supernatant that binds to OV569) by using RNAgents total RNA isolation system (Promega) according to the manufacturer’s protocol. Poly(A)+ RNA was purified from the total RNA by using mRNA Separator (CLONTECH) according to the procedure recommended by the manufacturer. mRNA from HE1P cells was reverse-transcribed to double-stranded cDNA by using a Marathon cDNA Amplification Kit (CLONTECH). After ligation to a Marathon cDNA adapter, cDNA was amplified by using Expand high-fidelity PCR system (Boehringer Mannheim) with adapter primer and a mesothelin gene-specific primer (GSP1: GGAAGTGGAGAAGACAGCCTGTCCTTC) corresponding to the known N-terminal amino acid sequence of the antigen to which OV569 binds. The PCR product was ligated into pGEM-T vector (Promega), and the ligation mixture was transformed into DH5α-competent cells (Life Technologies, Gaithersburg, MD). Plasmids were isolated from individual colonies by using the QIAprep spin miniprep kit (Qiagen, Chatsworth, CA) and sequenced by using the BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems).
Inverse PCR.
A published procedure (19) was used. Five milligrams of RNA from cultured carcinoma cells was extracted by Trizol (GIBCO/BRL), and mRNA was purified by magnetic beads [Poly(A) Ttract, mRNA Isolation System III; Promega]. Single-strand cDNA was generated by using a specific primer binding into the insert sequence (insert 56–80: GCG CTC TGA GTC ACC CCT CTC TCT G), and the second strand was generated according to the protocol of Marathon Kit (CLONTECH). The cDNA was religated on itself in 200 μl for 24 hr at 15°C. Five microliters of this ligation mixture was used for PCR amplification with 5′ primer MFP r290 (GGG ACG TCA CAT TCC ACT TG) and 3′ primer MPF f735 (AGA AAC TTC TGG GAC CCC AC) and nested with 5′ primer GSP-2 (GAA GGA CAG GCT GTC TTC TCC ACT TCC C) and 3′ primer insert r80–54 (CAG AGA GAG GGG TGA CTC AGA GC).
RESULTS
Immunohistology.
Table 1 summarizes data from immunohistological studies on frozen sections of various malignant neoplasms (most of which were from metastatic sites) and normal tissues. As shown in the table, OV569 bound to 20 of 21 (>95%) ovarian carcinomas, as well as to a smaller fraction of other carcinomas, particularly carcinomas of the uterus and stomach. The staining was seen at the cell surface as well as in the cytoplasm, and it was commonly uniform. Normal tissues did not stain except for mesothelium.
Table 1.
Immunohistological staining of normal human tissues and tumors with mAb OV569
| Normal tissues | Positive tested | Normal tissues | Positive tested | Tumors | Positive tested |
|---|---|---|---|---|---|
| Adrenal | 0/6 | Mesothelium | 1/1 | Ca. ovary | 20/21 |
| Brain | 0/7 | Nerve | 0/6 | Ca. endometrium | 3/7 |
| Breast | 0/7 | Ovary | 0/6 | Ca. cervix uteri | 5/8 |
| Cecum | 0/3 | Pancreas | 0/6 | Ca. breast | 4/18 |
| Colon | 0/6 | Placenta | 0/2 | Ca. stomach | 3/7 |
| Endometrium | 1/6 | Prostate | 0/7 | Ca. colon | 2/15 |
| Esophagus | 0/5 | Benign prostatic hypetrophia | 0/5 | Ca. testis | 0/2 |
| Heart | 0/8 | Skin | 0/6 | Ca. lung (non-small-cell) | 5/13 |
| Ileum | 5/5† | Stomach | 0/6 | Ca. lung (small-cell) | 0/3 |
| Jejunum | 0/4 | Spleen | 0/8 | Ca. bladder | 0/6 |
| Kidney | 0/7 | Thyroid | 0/4 | Ca. prostatic | 0/14 |
| Liver | 0/8 | Testis | 0/12 | Melanoma | 0/8 |
| Lung | 0/6 | Tonsil | 0/4 | ||
| Lymph node | 0/1 |
Ca., carcinoma of.
In positive samples, >33% of the tumor cells were stained. Normal stroma cells were not stained. A normal sample was noted as positive if it contained >5% of cells that stained with OV569.
Weak staining of <10% of cells.
Fluorescence-Activated Cell Sorting Analyses of Live Cells.
Fluorescein-conjugated mAb OV569 was incubated with live cells from cultures of a variety of carcinomas. Cells from 10 of 14 cultured lines of ovarian carcinomas and five of six lung carcinoma lines bound OV 569 (LFE > 1.3). Antibody binding also was detected to two of seven breast carcinoma lines and two of seven colon carcinoma lines. However, whereas 10 of the 14 ovarian carcinoma lines had a binding ratio of >2, only one of six lines from lung carcinoma, 1 of 6 lines from breast carcinoma, and 0 of six colon carcinoma lines had a binding ratio of >2.
Antigen Characterization.
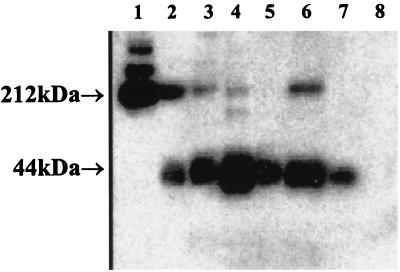
Either cell-free ascites from patients with ovarian carcinoma or cell-free supernatant from an antigen-positive ovarian carcinoma line was added on top of the affinity chromatography column, followed by washing and elution. The molecular mass of the OV569 antigen was determined by Western blotting, which demonstrated a consistent band at 42–45 kDa with ascites, supernatant, and tumor cell lysates, whereas additional, larger bands were detected in some samples. The Western blot shown in Fig. 1 illustrates the findings obtained.
Figure 1.
Western blot analysis of the antigen bound to OV569. Lanes: 1, D2hIg; 2, eluate of ovarian cancer ascites; 3, eluate of pleural effusion 3L; 4, eluate from supernatant of lung cancer line 4013; 5, lysate of 4013 cells; 6, eluate from supernatant of ovarian cancer line OVCAR-3; 7, lysate of OVCAR-3 cells; 8, lysate of kidney cancer 6K cells.
We determined the amino acid sequence of the N-terminal end of the molecule recognized by mAb OV569. OV569 was used to immunoprecipitate 35S-methionine-labeled glycoproteins of approximately 42–45 kDa from cells established in culture from lung carcinoma H4013. An initial antigen isolation was performed by passing the supernatant from cultures of H4013 cells over an affinity chromatography column that had been prepared by coupling OV569 to cyanogen bromide-activated Sepharose 4B according to the protocol of Sigma. The subsequently eluted material was alkylated to derivatize possible cysteine residues, purified on 7.5% SDS-polyacrylamide gel, and then electroblotted for sequence analysis. A diffuse band of approximately 42–45 kDa was detected.
N-terminal sequence analysis of this band yielded EVEKTACPSGKKAREIDES. An identical sequence has been identified (amino acids 294–312) within mesothelin and MPF (10, 11), two closely related differentiation antigens expressed on ovarian carcinomas. The same sequence subsequently was found when the 42- to 45-kDa molecule was isolated from the cell-free supernatant of cultured H4013 cells.
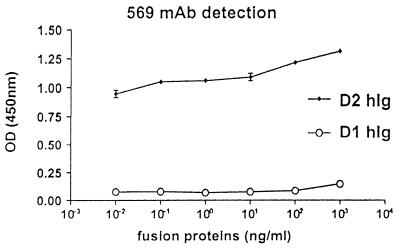
To confirm this finding, two fusion proteins were constructed, D1hIg and D2hIg, corresponding to the published, 33-kDa soluble domain of MPF (10, 12) and to the published, 44-kDa, membrane-bound domain of MPF (44 kDa) (10), respectively. D1hIg and D2hIg were used to test the specificity of OV569 by detection ELISA. Fig. 2 demonstrates that OV569 binds to an epitope expressed on the membrane-bound portion of MPF (D2hIg) and not on its published, soluble part (D1hIg). Because OV569 binds to a soluble protein, which is not D1hIg and which is expressed on MPF and mesothelin, we initiated experiments to search for new member(s) of the MPF/mesothelin family.
Figure 2.
mAb OV 569 specifically recognizes D2hIg, a fusion protein comprising the described membrane-bound portion of mesothelin/MPF (10), and does not bind to D1hIg, a fusion protein comprising the described soluble portion of MPF (10, 12).
Cloning of a cDNA Related to MPF and Mesothelin.
First, we amplified by 3′ rapid amplification of cDNA ends PCR the mRNA from a prostatic tumor cell line (He1P). Ten candidate clones were isolated and sequenced. Of these, eight exhibited a sequence identical to MPF, one exhibited a sequence identical to mesothelin, and one had a sequence related to MPF and mesothelin but also contained an 82-bp insert (GGT GGG CGG GGC GGC CAG GCC AGG GCT GGG GGC AGA GCT GGG GGC GTG GAG GTG GGC GCT CTG AGT CAC CCC TCT CTC TGT A) inserted in position 1874 of MPF. The molecule encoded by this gene is referred to as SMR (soluble, mesothelin/MPF-related).
As another approach to the same end, mesothelin/MPF-related cDNA from colon carcinoma 3719 was cloned by inverse PCR. Three PCR clones were sequenced and revealed an identical sequence to that described above plus an addition on the 5′ end that codes for 3 aa upstream of the cleavage site between D1 and D2. The sequence encoding those 3 aa is identical to the published sequences of MPF and mesothelin (as described in references cited above).
Characterization of SMR.
The 82-bp insert in position 1874 of MPF induces a frame shift of 212 bp. This frame shift codes for a new C terminus, which, in contrast to MPF and mesothelin, shows a hydrophilic tail, suggesting that it is soluble. According to the toppred2 transmembrane modeling program (Department of Biochemistry, Stockholm University), the structure indicates that it is soluble. The same conclusion is supported by other modeling programs, such as tmpred—Prediction of Transmembrane Regions and Orientation (20). The new clone is more closely related to MPF than to mesothelin in that it does not have a 24-bp insertion found in the central portion of the mesothelin protein but not in MPF, and it is identical to MPF in two places at which single base differences are found between MPF and mesothelin (the sequence is being deposited with GenBank). However, more work is needed (particularly expression of the gene) before we can conclude that this member of the MPF/mesothelin family, and not one or more relatives, is the one released into culture supernatants and patient serum (as described below). Importantly, the differences between mesothelin and MPF, both of which are expressed in normal (nonmalignant) tissue, do not lead to marked changes in downstream sequence, solubility, or subcellular location.
Assay for the OV569-Defined Antigen in Malignant Effusions and Sera.
A sandwich ELISA was constructed by using mAbs OV569 and 4H3, which bind to different epitopes on the mesothelin-related antigen. Antigen–antibody binding was measured as OD in an ELISA.
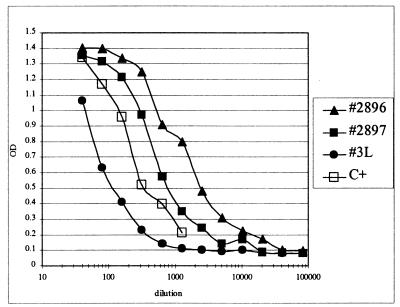
Fig. 3 demonstrates a signal (increased OD) at a dilution of 1:10,000 of ascites from one patient with ovarian carcinoma, at a dilution of 1:1,280 of ascites from another ovarian cancer patient, and at a dilution of 1:640 of a pleural effusion from a patient, 3L, with lung carcinoma. A positive control, C+ (serum from a patient with ovarian carcinoma), is included for comparison.
Figure 3.
Detection of the OV569-defined antigen in malignant effusions.
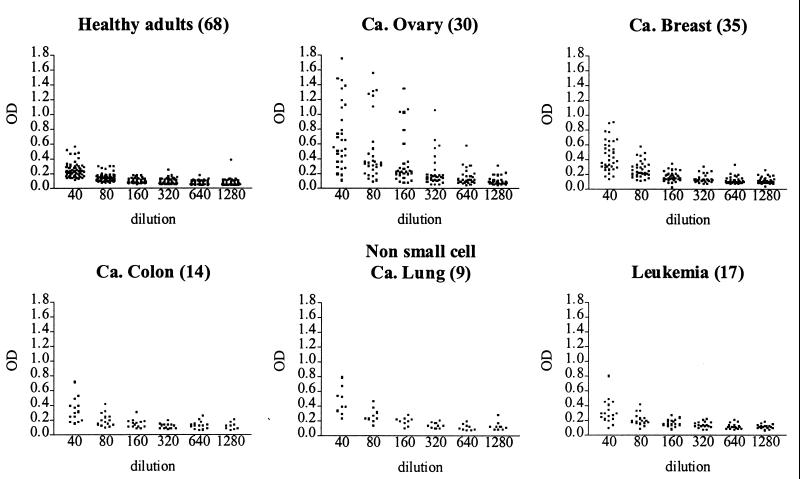
Subsequently, sera were assayed from patients with ovarian carcinoma, as well as from patients with various other tumors or nonneoplastic diseases and from healthy volunteers (Fig. 4). At a dilution of 1:160, 23 of 30 sera from patients who had ovarian carcinoma in stage 3 or 4 had circulating antigen levels that were significantly elevated (>3 SD in comparison with the negative control sample), as compared with 0 of 68 sera from healthy volunteers. Using the same criteria, 25 of 75 sera from patients with tumors other than ovarian carcinoma were positive, with the highest frequency of positive sera (66%) being seen among patients with lung carcinoma (Table 2). Sera from three patients with nonneoplastic diseases were negative, as was serum from one patient with a benign gynecological tumor.
Figure 4.
Detection of antigen (as increased OD) in sera of patients with ovarian cancer or other malignancies, as compared with sera from healthy adults. Each dot represents a patient. OD is plotted at various serum dilutions (1:40–80–160–320–640–1,280). Number of samples tested of each type of serum is given within parentheses.
Table 2.
Detection, by Sandwich ELISA, of the mesothelin-related antigen in sera from patients with various malignancies
| Diagnosis | Positive/tested |
|---|---|
| Healthy donors | 0/68 |
| Inflammatory disease | 0/3 |
| Benign tumor | 0/1 |
| Ca. ovary | 23/30 |
| Ca. breast | 11/35 |
| Ca. lung | 6/9 |
| Ca. colon | 2/14 |
| Leukemias | 6/17 |
Ca., carcinoma of.
DISCUSSION
We initially made a mAb, OV569, which binds to an antigen that is present in >95% of human ovarian carcinomas and in a smaller fraction of other carcinomas and is not detected in normal tissues, except for mesothelial cells and a small subpopulation of cells from intestines. According to N-terminal amino acid sequencing, the epitope to which OV569 binds is expressed on two closely related different antigens, mesothelin (10) and MPF (12). Mesothelin has been reported to be stably present at the cell surface (11), the published gene sequence indicates that the membrane-associated part of mesothelin/MPF is not soluble, and the epitope to which OV569 binds is located to that part (Fig. 2). Nevertheless, the molecule to which OV569 binds was found to be released to the cell-free culture supernatants from tumors expressing the antigen to which OV569 binds, suggesting that OV569 binds to an epitope that also is expressed on previously unknown, soluble member(s) of the MPF/mesothelin family. Experiments, therefore, were performed in which cDNAs were cloned from a tumor, He1P, which releases soluble OV569-binding protein into cell-free culture supernatants. One of 10 such cDNA clones was found to have an 82-bp insert in position 1874 of MPF, which induces a frame shift of 212 bp. This frame shift codes for a new C terminus, which, in contrast to MPF and mesothelin, shows a hydrophilic tail, suggesting that it is soluble.
To test whether soluble mesothelin/MPF-related protein(s) to which OV569 binds might provide marker(s) for serum assays for diagnosis and/or monitoring the response of patients to therapy, antigen was purified by immunoadsorption from cell-free medium of tumors binding mAb 569 and used to obtain murine mAbs that were directed to epitopes different from the one recognized by OV569. A combination of one of these mAbs, 4H3, with OV569 was used to develop a double-determinant (sandwich ELISA) assay.
Using this assay, we showed that there were high levels of free antigen in the ascites of two patients with ovarian carcinoma and the pleural effusion of one patient with lung carcinoma. Subsequently, we studied sera from 30 patients with advanced carcinoma of the ovary (stages III and IV), 75 patients with other tumors, three patients with nonneoplastic, inflammatory diseases, one patient with a benign gynecological tumor, and 68 healthy controls. The majority of sera from the ovarian carcinoma patients (23/30 = 77%) displayed ODs higher than any of 68 sera from healthy controls when compared at a dilution of 1:160 and counting as positive an OD >3 SD above the negative control included in all experiments. Because no sera were tested from patients with ovarian carcinoma stages I or II, no conclusions can be drawn as to the utility of the assay for early diagnosis.
Relatively few samples of sera were tested from groups of patients with tumors different from ovarian carcinoma. However, among nine lung cancer patients tested, six were positive. It will be important to evaluate more sera from additional patients with lung carcinoma, as well as patients with pancreatic carcinoma, which are known to express the related molecule, MPF (12).
Because most of the cancer patients whose sera were tested were receiving combination chemotherapy, we cannot exclude the possibility that such therapy influenced the results observed. That a lower reactivity was seen with sera from patients who had leukemia or carcinoma of the colon or breast, and also received chemotherapy, excludes the possibility that the findings obtained in the ovarian cancer group are caused by release of the mesothelin-related antigen from normal cells as a result of chemotherapy. It also makes it unlikely that the higher serum reactivity in the ovarian cancer group is a consequence of the chemotherapy the patients are receiving, a conclusion also supported by evidence that CA125 levels do not normally increase as patients are undergoing chemotherapy.
The data in Figs. 3 and 4 and Table 2 are presented as OD at various dilutions of ascites or serum. Because the relationship between OD and the amount of antigen is probably not stoichiometric, the data do not permit quantitation of the amount of antigen (as ng/ml) in the various samples until the completion of studies by using highly purified antigen for the construction of standard curves. Studies also have been initiated on serial samples from patients collected at diagnosis, after surgery, and during and after therapy, as well as on sera from patients with less advanced ovarian carcinoma (stages I and II). These studies will help define the clinical usefulness of MPF/mesothelin-related antigens as serum marker, either alone or in combination with CA125 (1–3) or other antigens (21), for monitoring ovarian carcinoma patients for responses to therapy and for establishing earlier diagnosis. Further work also should establish whether patients form antibodies to epitopes belonging to the MPF/mesothelin family by using, e.g., D2Ig as a probe, and whether the newly discovered molecule with an 82-bp insert provides a tumor-selective target for immunotherapy.
Acknowledgments
We acknowledge our former Bristol-Myers Squibb colleagues H. Marquardt, A. Wallace, and J. Sundstrom (amino acid sequencing), U. Garrigues and H. Wan (collaborators on the isolation of the OV569 hybridoma and immunohistological analysis), and A. Aruffo (preparation of a mesothelium fusion protein, D2, used in the initial screening of hybridomas), as well as Dr. C. Drescher (Swedish Hospital and Medical Center) and N. Urbain (Fred Hutchinson Cancer Research Center) for obtaining patient sera and Dr. W. Hagopian (Pacific Northwest Research Institute), who provided 52 of the control sera and also gave very helpful advice. We also want to acknowledge helpful discussions with, and support from, Dr. Y. Guo, Sidney Kimmel Cancer Center, La Jolla, CA, and the Second Military Medical University, Shanghai, People’s Republic of China. This work was initiated when three of the authors (N.S., K.E.H., and I.H.) worked at the Bristol-Myers Squibb Pharmaceutical Research Institute in Seattle. It was continued with support by a grant to I.H. from Bristol-Myers Squibb, as well as from the Pacific Northwest Research Institute in Seattle.
ABBREVIATIONS
- MPF
megakaryocyte potentiating factor
- LFE
linear fluorescence equivalence
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF180951).
References
- 1.Jacobs I, Bast R C. Hum Repro. 1989;4:1–12. doi: 10.1093/oxfordjournals.humrep.a136832. [DOI] [PubMed] [Google Scholar]
- 2.Mazurek A, Niklinski J, Laudanski T, Pluygers E. Eur J Cancer Prev. 1998;7:23–35. [PubMed] [Google Scholar]
- 3.Berek J S, Bast R C., Jr Cancer. 1995;76:2092–2096. doi: 10.1002/1097-0142(19951115)76:10+<2092::aid-cncr2820761331>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 4.Guadagni F, Roselli M, Cosimelli M, Ferroni P, Spila A, Cavaliere F, Casaldi V, Wappner G, Abbolito M R, Greiner J W, et al. Cancer Invest. 1995;13:227–238. doi: 10.3109/07357909509011692. [DOI] [PubMed] [Google Scholar]
- 5.Pannek J, Partin A W. Semin Urol Oncol. 1998;16:100–105. [PubMed] [Google Scholar]
- 6.Hunerbein M. Recent Results Cancer Res. 1998;146:48–55. doi: 10.1007/978-3-642-71967-7_4. [DOI] [PubMed] [Google Scholar]
- 7.Posner M R, Mayer R J. Hematol Oncol Clin North Am. 1994;8:533–553. [PubMed] [Google Scholar]
- 8.Rubach M, Szymendera J J, Kaminska J, Kowalska M. Int J Biol Markers. 1997;12:168–173. doi: 10.1177/172460089701200406. [DOI] [PubMed] [Google Scholar]
- 9.Chang K, Pai L H, Batra J K, Pastan I, Willingham M C. Cancer Res. 1992;52:181–186. [PubMed] [Google Scholar]
- 10.Chang K, Pastan I. Proc Natl Acad Sci USA. 1996;93:136–140. doi: 10.1073/pnas.93.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowdhury P S, Viner J L, Beers R, Pastan I. Proc Natl Acad Sci USA. 1998;95:669–674. doi: 10.1073/pnas.95.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kojima T, Oh-eda M, Hattori K, Taaniguchi Y, Tamura M, Ochi N, Yamaguchi N. J Biol Chem. 1995;270:21984–21990. doi: 10.1074/jbc.270.37.21984. [DOI] [PubMed] [Google Scholar]
- 13.Brown J P, Woodbury R G, Hart C E, Hellström I, Hellström K E. Proc Natl Acad Sci USA. 1981;78:539–543. doi: 10.1073/pnas.78.1.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yeh M Y, Hellström I, Brown J P, Warner G A, Hansen J A, Hellström K E. Proc Natl Acad Sci USA. 1979;76:2927–2931. doi: 10.1073/pnas.76.6.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douillard J Y, Hoffman T. Methods Enzymol. 1983;92:168–174. doi: 10.1016/0076-6879(83)92016-5. [DOI] [PubMed] [Google Scholar]
- 16.Hellström I, Horn D, Linsley P, Brown J P, Brankovan V, Hellström K E. Cancer Res. 1986;46:3917–3923. [PubMed] [Google Scholar]
- 17.Hollenbaugh D, Douthwright J, McDonald V, Aruffo A. J Immunol Methods. 1995;188:1–7. doi: 10.1016/0022-1759(95)00194-8. [DOI] [PubMed] [Google Scholar]
- 18.Hollenbaugh D, Aruffo A. In: Current Protocols in Immunology. Coligan J E, Kruisbeek A M, Margulies D H, Shevach E M, Strober W, editors. New York: Wiley; 1994. p. 10.19.1. [Google Scholar]
- 19.Zeiner M, Gehring U. BioTechniques. 1994;17:1051–1053. [PubMed] [Google Scholar]
- 20.Hofmann K, Stoffel W. Biol Chem Hoppe-Seyler. 1993;374:166. doi: 10.1515/bchm3.1993.374.7-12.507. [DOI] [PubMed] [Google Scholar]
- 21.Kudoh K, Kikuchi Y, Kita T, Tode T, Takano M, Hirata J, Mano Y, Yamamoto K, Nagata I. Gynecol Obstet Invest. 1999;47:52–57. doi: 10.1159/000010062. [DOI] [PubMed] [Google Scholar]