Abstract
Molluscum contagiosum virus (MCV) is a common, human poxvirus that causes small papular skin lesions that persist for long periods without signs of inflammation. Previous studies revealed that MCV encodes a family of proteins with homology to mammalian IL-18 binding proteins. IL-18 is a proinflammatory cytokine that induces synthesis of interferon γ, activates NK cells, and is required for a T-lymphocyte helper type 1 response. We expressed and purified the proteins encoded by the MC53L and MC54L genes of MCV, as well as their human and murine homologs. All four recombinant proteins were able to bind with high affinity to human and murine IL-18 molecules and inhibited IL-18 mediated interferon γ production in a dose-dependent manner. The pirating of IL-18 binding proteins by poxviruses and their use as decoy receptors is consistent with the critical role of IL-18 in defense against virus infections and provides a mechanism for evasion of the immune system by MCV.
Molluscum contagiosum virus (MCV), a human poxvirus with a worldwide distribution, commonly causes small papular skin lesions in children and young adults and a more extensive disease in patients with immunodeficiency states, including AIDS (1, 2). The virus-filled skin lesions frequently persist for many months without signs of inflammation, even in immunocompetent individuals (3). Research on MCV has been hindered by the inability to replicate the virus in tissue culture and the absence of useful animal models (4, 5). Analysis of the MCV genome, however, revealed >70 putative genes that are not present in other poxviruses and that might contribute to evasion of the human immune response, as well an additional 100 genes that are orthologs of genes in other poxviruses (6, 7). The former included a viral homolog of cellular chemokines with chemokine antagonist activity (8, 9), a major histocompatibility complex class I homolog that binds β2 microglobulin (10), a selenocysteine-containing glutathione peroxidase that inhibits peroxide and UV-mediated apoptosis (11), and death effector domain proteins that inhibit tumor necrosis factor (TNF) α and Fas ligand-mediated apoptosis (12–14). Another family of MCV genes—MC51L, MC53L, and MC54L—are homologs of recently discovered human and mouse IL-18 binding proteins (huIL-18BP and muIL-18BP, respectively) (15–17). IL-18BP homologs are also present in some other poxviruses, including the smallpox virus, but they have not been identified in the genomes of other virus families (7).
IL-18 is a pro-inflammatory cytokine that induces interferon (IFN) γ production in T cells and natural killer cells and plays a critical role in the T-lymphocyte helper type 1 response (18, 19). Thus, IL-18-deficient mice produce little IFN-γ and have low natural killer cell activity and impaired T lymphocyte helper type 1 responses despite the presence of IL-12 and other cytokines (20). IL-18 is related to IL-1β in structure (21–23) and is synthesized as a biologically inactive precursor that is cleaved by the intracellular cysteine protease IL-1β-converting enzyme, also known as caspase-1 (24). Secreted IL-18 binds to its specific membrane receptor, a member of the IL-1R family, and activates signal transduction pathways leading to the activation of NF-κB and synthesis of cytokines including IL-8, IL-1β, TNF-α, and Fas ligand (25, 26). Thus, the activity of IL-18 is distinct from that of IL-12, with which it is synergistic.
As its name implies, huIL-18BP binds IL-18 and consequently blocks induction of IFN-γ and other cytokines (15, 16). Similar binding, however, has not yet been reported for any of the viral IL-18BP homologs, which are ≈20–30% identical to huIL-18BP in amino acid sequence. Previously, we expressed MC51L, MC53L, and MC54L in mammalian cells and demonstrated that they were glycosylated and secreted (17). Here, we show that MC53L and MC54L bind IL-18 with high affinity and prevent IFN-γ production, suggesting that these viral proteins antagonize the development of an inflammatory response to MCV infection in humans.
MATERIALS AND METHODS
Viruses.
Recombinant vaccinia viruses expressing 6-histidine tagged MC53L, MC54L, muIL-18BP, and huIL-18BP were constructed essentially as described previously for the HA-tagged versions (17). Viral genes and huIL-18BP or muIL-18BP cDNAs were amplified by PCR with a primer that appended the coding sequence for a 6-histidine tag to the C terminus of each ORF. The reaction products were subcloned into the pMC02 transfer vector (27), and the resulting plasmids were transfected into BS-C-1 cells that were infected with vSC20, a highly attenuated growth factor-deficient vaccinia virus mutant (28). The recombinant viruses were selected for loss of thymidine kinase expression and were screened for the synthesis of β-glucuronidase (27). The correct DNA sequence of each insert was confirmed.
Protein Purification.
Roller bottles containing monolayers of BS-C-1 cells in serum free Optimem (GIBCO/BRL) medium were infected with 10 infectious units per cell of a recombinant virus. After 30 h, the medium was harvested, and suspended cells were removed by centrifugation at 1,000 × g for 10 min and 15,000 × g for 30 min. The supernatant was adjusted to contain 0.1% Triton X-100 and 10 mM imidazole and was incubated overnight at 4°C with Ni–nitrilotriacetic acid resin (Qiagen, Chatsworth, CA). The resin then was packed into a column and was washed consecutively with 15 and 30 mM imidazole in PBS containing 150 mM NaCl. The recombinant protein was eluted with 5 column volumes of 200 mM imidazole in PBS. Protein concentrations were determined by the Bradford assay.
IL-18 Binding to Membrane-Bound Proteins.
Approximately 80 ng of partially purified recombinant protein was denatured in SDS/PAGE loading buffer with or without 2-mercaptoethanol and was applied to a SDS 4–20% polyacrylamide gradient gel (Owl Separation Systems, Portsmouth, NH). After electrophoresis, the proteins were transferred to a nitrocellulose membrane and were incubated for 1 h with 200 nM recombinant human IL-18 (rhIL-18) (R & D Systems) in PBS with 5% powdered milk. The rhIL-18 that remained bound to the membrane after extensive washing was detected by chemiluminescence after incubation with anti-hIL-18 mAb (R & D Systems). The same membrane was stripped and reprobed with a mAb that recognizes five consecutive histidines (Qiagen).
IL-18 binding also was performed with unpurified recombinant proteins as follows. BS-C-1 cells in one well of a six-well plate were infected with a recombinant vaccinia virus at a multiplicity of 10. The medium was collected and centrifuged at 15,000 × g for 10 min to remove any suspended cells. Proteins in the supernatant were concentrated by ultrafiltration by using a Centricon 10 device (Millipore). The subsequent steps were identical to those used for purified protein.
Chemical Cross-Linking.
Approximately 100 ng of partially purified recombinant protein or 400 ng of ovalbumin or BSA were incubated with 200 ng of rhIL-18 in total volume of 15 μl at room temperature for 0.5 h. The chemical cross-linker bis(sulfosuccinimidyl) suberate (BS3; Pierce) was added to a final concentration of 1 mM. The reaction was terminated after 0.5 h by adding 1 μl of 1 M Tris⋅HCl (pH 8.0). The reaction mixtures were treated with SDS and 2-mercaptoethanol and were subjected to SDS/PAGE. IL-18 and proteins cross-linked to IL-18 were detected by Western blotting using polyclonal anti-rhIL-18 antibody (Ab) (R & D Systems).
Surface Plasmon Resonance.
A BIAcore 2000 biosensor, CM5 research grade chips, N-hydroxysuccinimide, N-ethyl-N′-(3-diethylaminopropyl)carbodiimide, and Tween 20 were obtained from BIACORE (Piscataway, N.J.). Partially purified recombinant huIL-18BP, MC54L, and MC53L were each immobilized onto a CM5 chip by using an amine-coupling procedure described by the manufacturer. Specifically, flow cells 1 and 2 on one CM5 chip were activated with N-hydroxysuccinimide/N-ethyl-N′-(3-diethylaminopropyl)carbodiimide for 7 min. The recombinant protein then was injected into flow cell 2. The remaining activated groups on both flow cells were blocked with a 7-min injection of 1 M ethanolamine (pH 8.0). Flow cell 1 served as a control surface for flow cell 2 during the kinetic experiments. To prevent mass transport effects, the level of immobilization was controlled so that Rmax was <120 resonance units (RU).
For kinetic experiments, various concentrations of rhIL-18 (R & D system) or recombinant murine IL-18 (rmIL-18) (PeptroTech, Rocky Hill, NJ) were injected in a buffer containing 0.01 M Hepes (pH 7.4), 0.15 M NaCl, 3 mM EDTA, and 0.1% Tween 20. A total of 250 μl of IL-18 proteins was injected at a flow rate of 20 μl/min. Dissociation was monitored for 20 min to 1 h, followed by a 10-μl injection of 10 mM glycine (pH 1.5) to regenerate the surface. To determine whether the binding was limited by mass transport, the flow rate at which IL-18 was delivered to the surface was altered from 5 μl/min to 80 μl/min.
Sensorgrams were analyzed with biaevaluation software (BIACORE). To correct for refractive index changes, the binding responses generated in the control surface were subtracted from the responses generated in the surface with immobilized ligands. The binding data from the injection of at least six different concentrations of analyte were globally fitted to a 1:1 binding model. Analyses with the same concentration series were repeated at least three times.
IL-18 Bioassay.
KG-1 cells (0.25 ml at 1.2 × 106 per ml) in Iscove’s modified Dulbecco’s medium containing 20% FBS (GIBCO/BRL) were seeded in wells of a 96-well plate and were stimulated with 20 ng/ml TNF-α and either 10 ng/ml rhIL-18 or rhIL-18 premixed with a purified recombinant protein. After stimulation at 37°C for 24 h, clarified supernatants of three-times frozen-thawed cells were assayed for human IFN-γ by ELISA using the Quantikine kit (R & D Systems).
RESULTS
Expression and Purification of Recombinant Proteins.
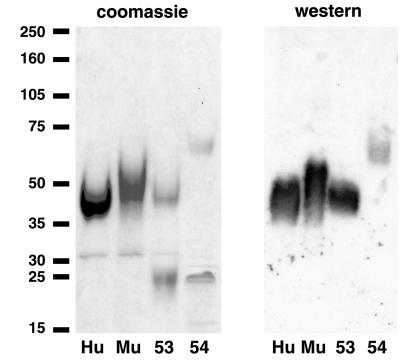
Of the three related MCV proteins, MC53L and MC54L most closely resembled huIL-18BP and muIL-18BP in sequence (17). Because the viral and eukaryotic proteins are highly glycosylated with conserved N-glycosylation sites, we used a mammalian cell system for expression. Recombinant vaccinia viruses that express MC53L, MC54L, huIL-18BP, or muIL-18BP with 6-histidine tails were constructed by homologous recombination. Advantage was taken of their 6-histidine tags to purify the secreted recombinant proteins by metal affinity chromatography. After the one-step procedure, the recombinant proteins were resolved by SDS/PAGE and were detected by staining with Coomassie blue and binding to an antipolyhistidine mAb (Fig. 1). In each case, the major band corresponded to the size expected for the glycosylated recombinant protein (17). The smaller proteins that did not react with the mAb could represent impurities or degradation products of the recombinant proteins.
Figure 1.
Purification of recombinant proteins. Recombinant MC53L, MC54L, huIL-18BP, and muIL-18BP were purified by metal affinity chromatography, were analyzed by SDS/PAGE, and were detected by Coomassie blue staining (Left) or by chemiluminescence after Western blotting with a mAb to the polyhistidine tag (Right). Hu, huIL-18BP; Mu, muIL-18BP; 53, MC53L; 54, MC54L. The positions and masses in kilodaltons of protein markers are shown on the extreme left.
Binding of IL-18 to SDS/PAGE-Purified Recombinant Proteins.
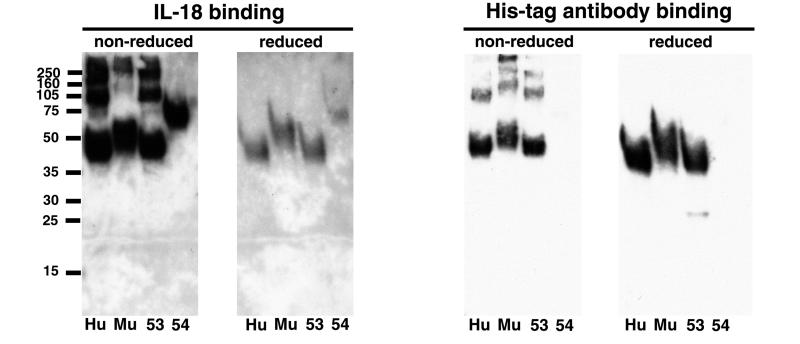
Binding of the recombinant proteins to IL-18 was demonstrated by several methods. Concentrated proteins from the total medium or after metal affinity chromatography were denatured with SDS in the presence or absence of reducing agent, were subjected to SDS/PAGE, and were transferred to a membrane. The membranes were incubated successively with human IL-18 and a mAb to IL-18. After washing, the bound mAb was detected by chemiluminescence. After this procedure, the membranes were stripped and reprobed with antipolyhistidine mAb to visualize the recombinant proteins. When the proteins were treated with a reducing agent before electrophoresis, IL-18 and antipolyhistidine mAb bound to single bands of the expected sizes but not to the smaller contaminating bands present in the metal affinity purified preparation (Fig. 2) nor to the larger number of contaminating proteins present in the concentrated total medium (not shown). In addition, neither ovalbumin nor BSA bound IL-18 when examined similarly (data not shown). For unknown reasons, the antipolyhistidine mAb reacted less strongly with MC54 than with the other recombinant proteins (Fig. 2). Greater IL-18 binding, but lower antipolyhistidine mAb binding, occurred when the recombinant proteins were unreduced (Fig. 2). Under the latter conditions, however, both monomeric and multimeric species were detected (Fig. 2). The greater IL-18 binding of the unreduced recombinant proteins may be attributable to more efficient renaturation when the disulfide bonds remained intact or because some disulfide bonds are near the binding site. With regard to the latter, there are conserved cysteines in the viral and cellular IL-18BPs (17). In contrast, the better binding of the antipolyhistidine mAb to the reduced proteins may result from better exposure of the tag in the unfolded species. By using similar methods, rmIL-18 also was shown to bind huIL-18BP, muIL-18BP, MC53L, and MC54L that was transferred to a membrane (data not shown).
Figure 2.
Binding of IL-18 to SDS/PAGE-purified recombinant proteins. Metal affinity resin-purified recombinant proteins were treated with SDS in the presence or absence of 2-mercaptoethanol and were analyzed by SDS/PAGE. The proteins were transferred to a membrane and were incubated successively with rhIL-18 and an hIL-18 mAb. Bound mAb was detected by chemiluminescence (Left). The membranes then were stripped and incubated with a mAb to the histidine tag (Right). His-tag, 6-histidine tag; other abbreviations are as in Fig. 1.
Cross-Linking of IL-18 to Native Recombinant Proteins.
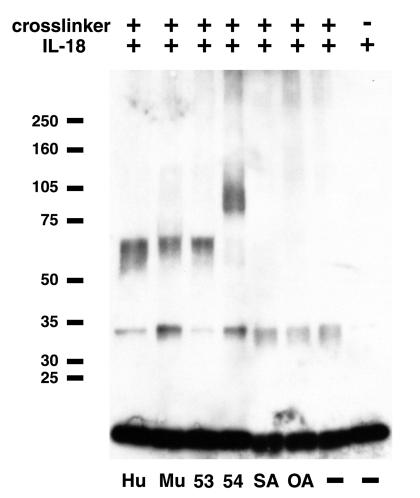
The binding of IL-18 to native viral and cellular IL-18BPs was demonstrated by chemical cross-linking followed by SDS/PAGE and immunoblotting with polyclonal Ab to IL-18. The Ab bound to free IL-18, which is 18 kDa, as well as to small amounts of a 34-kDa protein that presumably represents cross-linked IL-18 dimers (Fig. 3). Importantly, the Ab bound to protein species with electrophoretic mobilities that corresponded to the sizes of each recombinant protein cross-linked to IL-18 (Fig. 3). No cross-linking of IL-18 with ovalbumin or BSA was detected (Fig. 3). There was also no cross-linking between MC53L, MC54L, huIL-18BP, and muIL-18BP with human IL-1β, which is related in structure to IL-18 (data not shown).
Figure 3.
Cross-linking of IL-18 to native recombinant proteins. Metal affinity resin-purified recombinant proteins were incubated with rhIL-18 in the presence of bis(sulfosuccinimidyl) suberate and then were analyzed by SDS/PAGE. The proteins were transferred to a membrane and then were incubated with polyclonal Ab to hIL-18. The Ab was detected by chemiluminescence. SA, bovine serum albumin; OA, ovalbumin; other abbreviations are as in Fig. 1.
Kinetic Analysis of IL-18 Binding to Recombinant Proteins.
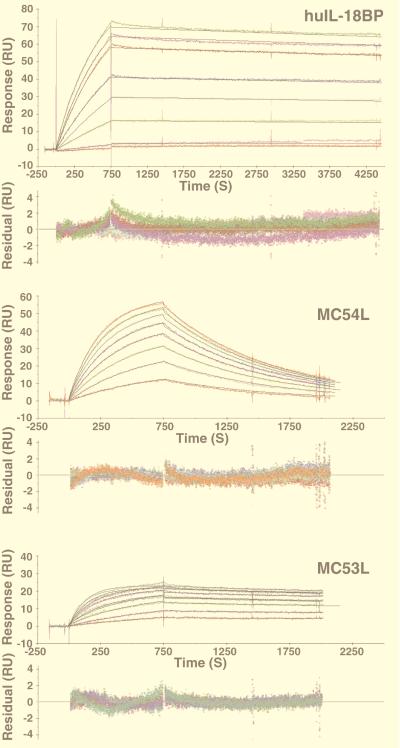
A surface plasmon resonance sensor was used to measure the binding of IL-18 to immobilized protein ligands. Recombinant huIL-18BP, MC53L, and MC54L were individually immobilized on CM5 chips by cross-linking primary amine residues to the dextran surface. Real-time association and dissociation of rhIL-18 and rmIL-18 with the immobilized proteins were monitored in a BIAcore 2000 biosensor. The binding of rhIL-18 to huIL-18BP was characterized by a sharp increase in response during the association phase of the sensorgram and an extremely slow decrease in response during the dissociation phase (Fig. 4). A sharp association phase also was noted for MC54L, but the dissociation phase was steeper than for huIL-18BP (Fig. 4). The sensorgram for MC53L was similar to that of huIL18BP except for a lower response maximum (Fig. 4), which may be attributable to coupling of some molecules through a lysine near the IL-18 binding site. The binding curves for rmIL-18 were similar in shape to those for rhIL-18 (data not shown).
Figure 4.
Detection of IL-18 binding to recombinant proteins by using surface plasmon resonance. Metal affinity resin-purified recombinant proteins were individually immobilized on a CM5 chip with the standard amine coupling procedure. Injection of rhIL-18 started at 0 seconds and stopped at 750 seconds. For huIL-18BP, rhIL-18 concentrations used were 0.35, 0.7, 3.5, 7, 10.5, 17.5, 21, and 24.5 nM. For MC54L, rhIL-18 concentrations used were 3.5, 7, 10.5, 14, 17.5, 21, 24.5, and 28 nM. For MC53L, rhIL-18 concentrations used were 7,14, 28, 42, 56, 70, 84, 112, and 140 nM. The colored and black lines are the actual responses in RU and globally fitted curves, respectively. The residual responses, below each set of curves, represent deviations of the actual responses from the fitted curves. The rms deviations were 0.749, 0.369, and 0.236 for huIL-18BP, MC54L, and MC54L, respectively.
The sensorgram data from at least three sets of experiments with rhIL-18 and rmIL-18 were globally fitted to a 1-to-1 binding model. The Kon rates of rhIL-18 were similar for all three ligands (Table 1). The Koff rates, however, varied and were highest for MC54L (Table 1), as expected from the relatively steep dissociation phase (Fig. 4). The Kd values were 0.4 nM for huIL18-BP, 2.1 nM for MC54L, and 10 nM for MC53L. The same order was obtained with rmIL-18, although the absolute Kd values were lower, ranging from 0.09 nM to 7.2 nM (Table 1).
Table 1.
Kinetic and affinity constants
| Ligand | rhIL-18
|
rmIL-18
|
||||
|---|---|---|---|---|---|---|
| KOn, 104/Ms | KOff, 10−4/s | Kd, nM | KOn, 105/Ms | KOff, 10−4/s | Kd, nM | |
| hIL-18BP | 7.23 ± 0.08 | 0.31 ± 0.09 | 0.4 ± 0.1 | 2.0 ± 0.3 | 0.19 ± 0.08 | 0.09 ± 0.03 |
| MC54L | 9.7 ± 0.2 | 10 ± 1 | 10 ± 1 | 2.9 ± 0.6 | 20 ± 2 | 7.2 ± 0.9 |
| MC53L | 7 ± 1 | 1.5 ± 0.4 | 2.1 ± 0.7 | 4.2 ± 0.2 | 0.4 ± 0.3 | 0.9 ± 0.7 |
Inhibition of IFN-γ Induction.
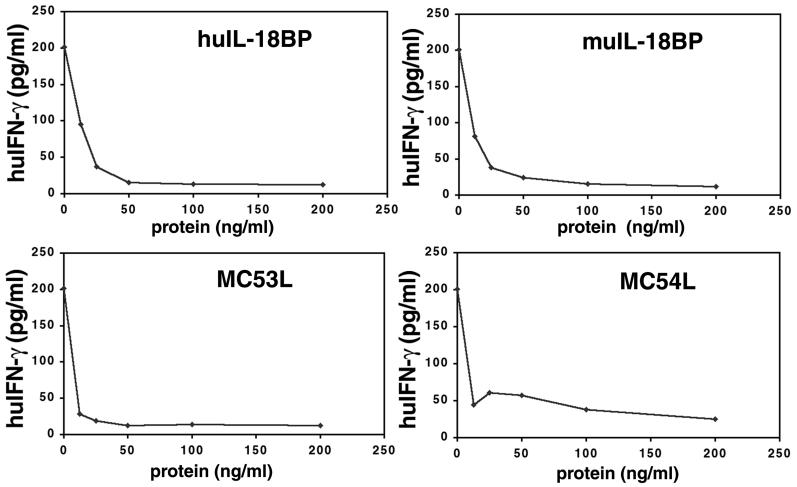
Numerous studies have indicated that the induction of IFN-γ is a biologically important property of IL-18. Recently, the ability of huIL-18BP to inhibit IFN-γ induction was shown by incubating human KG-1 monocytic cells with IL-18 or IL-18 plus TNF-α (15, 16). We confirmed the inhibitory effects of huIL-18BP and muIL-18BP (Fig. 5) and further demonstrated that MC53L and MC54L blocked IFN-γ production at nanogram levels in a dose-dependent manner (Fig. 5). MC53L was especially potent in this assay whereas MC54L did not reduce IFN-γ production to the base level, perhaps reflecting its lower affinity for IL-18.
Figure 5.
Inhibition of IFN-γ induction. KG-1 cells were incubated with 20 ng/ml TNF-α and either 10 ng/ml of rhIL-18 or rhIL-18 premixed with the indicated amounts of metal affinity resin-purified recombinant protein. After 24 h at 37°C, clarified supernatants were assayed for human IFN-γ.
DISCUSSION
The apparent homology of MC53L and MC54L with huIL-18BP led us to investigate the IL-18 binding properties of the viral proteins. The absence of any structure-function data for huIL-18BP and the relatively low, 23–34%, amino acid sequence identity between the viral and human proteins, however, made the outcome uncertain. Gratifying initial experiments confirmed that both MC53L and MC54L could bind and be cross-linked to IL-18. For further comparisons, we determined the affinities of MC53L, MC54L, and huIL-18BP for human and mouse IL-18 molecules. huIL-18BP bound to rhIL-18 and rmIL-18 with Kd values of 0.4 and 0.09 nM, respectively. Thus, the affinity between IL-18 and IL-18BP is much higher than that of IL-18 to its known receptor IL-18Rα (29) and equivalent to that of a high affinity receptor complex on T cells (30). The high affinity between huIL-18BP and rhIL-18 indicates that such interactions can occur at physiologically relevant concentrations. The association rates of MC54L and MC53L proteins for IL-18 were similar to that of huIL-18BP, but they had faster dissociation rates. The Kd values for rhIL-18 and rmIL-18 binding to MC53L were 2.1 and 0.9 nM, respectively. For MC54L, the values were 10 nM and 7.2 nM, respectively. It will be important to confirm these values by using other methods for immobilizing the ligands to the sensor chip. With MC53L in particular, the maximal response to IL-18 was only ≈20 RU, compared with 80 RU and 120 RU for MC54L and huIL18BP, respectively. This difference was not attributable to lower amounts of MC53L bound to the chip, but probably stems from the fact that MC53L has only two lysine residues available for cross-linking to the dextran surface, and both are in regions that are conserved among all IL-18 binding proteins. We suggest, therefore, that the cross-linking of at least one of the lysine residues inactivated MC53L. The latter idea is consistent with the much higher level of IL-18 binding achieved when the MC53L protein was indirectly captured via a mAb to its polyhistidine tag (Y.X., unpublished work). However, this indirect capture method did not allow an accurate determination of kinetic values because the dissociation of MC53L from the mAb was much faster than its dissociation from IL-18. We are currently exploring other indirect capturing methods to measure the affinities of the IL-18 binding proteins.
Because human and mouse IL-18 molecules are only 64% identical and the human IL-18 receptor binds preferentially to huIL-18 (31), we were surprised to find that huIL-18BP as well as MC54L and MC53L bound at least as well to rmIL-18 as to rhIL-18. Although the slightly higher affinities of the latter proteins for rmIL-18 over rhIL-18 may merely reflect the specific activities of the rhIL-18 and rmIL-18 proteins, which were obtained from different commercial sources, there would seem to be no doubt of the broader specificity of the binding proteins compared with that of the human IL-18 receptor. One could speculate that the broader specificity of the IL-18BPs compared with the IL-18 receptor reflects differences in the binding sites on the IL-18 molecules. Of more interest than species-specificity was the possibility that MC53L and MC54L could bind additional cytokines that are structurally related to IL-18, such as IL-1β. Precedence for this comes from the broad specificity that was found for an MCV-encoded chemokine homolog (9). However, neither the mammalian nor the MCV IL-18BPs bound to IL-1β.
Having found that MC54L and MC53L bound to IL-18, we needed to determine whether such binding interfered with IL-18 activity. Previous studies had shown that huIL-18BP blocked the binding of IL-18 to KG-1 cells and prevented IFN-γ induction (15, 16). We confirmed the latter results and demonstrated that both MC54L and MC53L potently inhibited IFN-γ induction.
Since the initial discovery of a vaccinia virus complement-binding protein (32, 33), a multitude of putative immune evasion molecules encoded by large DNA viruses have been described (34, 35). In many cases, the viruses seem to have acquired these genes by lateral transfer from their hosts and then modified and retained them by mutagenesis and natural selection, respectively. The MCV IL-18BP homologs are good examples of this phenomenon. MC53L and MC54L have structural features of huIL-18BP and muIL-18BP, including signal peptides, three conserved cysteines, and glycosylation sites. Yet, the viral genes have no introns, a high GC/AT ratio, and are only 34–23% identical to huIL-18BP. Some members of the Orthopoxvirus genus that have natural hosts (e.g., variola virus, cowpox virus, and ectromelia virus) also have a full-length IL-18BP homolog whereas the gene is shortened or deleted from vaccinia virus, an attenuated virus used as a vaccines (7). Similarly, swinepox virus, a member of the Suipoxvirus genus, has an IL-18BP homolog. Of these proteins, evidence for IL-18 binding has been demonstrated for the ectromelia virus homolog (S. Calderara, Y.X., and B.M., unpublished work).
IL-18 has been shown to protect mice against herpes simplex virus infection by IFN-γ-dependent and -independent pathways (36). The role of IL-18 on poxvirus pathogenesis has not been directly determined, although it should be possible to do so with ectromelia (mousepox) virus in a recently constructed IL-18 knock out mouse (20). Nevertheless, a critical role for IL-18 can be inferred from evidence that IFN-γ is important for host recovery from some poxviruses (37, 38). The finding that MCV encodes at least two IL-18 antagonists supports recent evidence indicating a key role for this cytokine in mediating innate and acquired immune responses to infectious agents (18). In the context of an MCV infection of the skin, it is relevant that IL-18 is expressed by stimulated epidermal keratinocytes as well as macrophages (39). Unfortunately, it will be difficult to test the roles of MC53L and MC54L in vivo because there is no animal model for MCV.
MCV lacks many of the immunomodulatory genes present in other poxviruses (6). One such protein is crmA, an intracellular inhibitor of caspase 1 that blocks the processing of IL1-β (40) and presumably IL-18, which is activated by the same caspase (24). However, in the context of an MCV infection, an intracellular caspase inhibitor would not prevent the activation of IL-18 by cells of the monocyte lineage because MCV is specific for keratinocytes. By contrast, MC53 and MC54L proteins are secreted and therefore could antagonize IL-18 produced by uninfected cells. Another poxvirus immunomodulatory protein absent from MCV is a soluble INF-γ receptor homolog (41–43). The reliance of MCV on IL-18 inhibitors supports other evidence that IL-18 is critical for IFN-γ induction by microbial organisms and viruses (19). Moreover, inhibition of IFN-γ alone would not block the induction of other cytokines by IL-18. Thus, by targeting IL-18, MCV can inhibit the cascade of downstream effects that follow activation of the IL-18 receptor.
Acknowledgments
We thank Tatiana Koonina for many helpful discussions and William Paul for a critical reading of the manuscript.
ABBREVIATIONS
- Ab
antibody
- BP
binding protein
- IFN
interferon
- MCV
molluscum contagiosum virus
- rhIL-18
recombinant human IL-18
- rmIL-18
recombinant mouse IL-18
- TNF
tumor necrosis factor
- RU
resonance unit
References
- 1.Gottlieb S L, Myskowski P L. Int J Dermatol. 1994;33:453–461. doi: 10.1111/j.1365-4362.1994.tb02853.x. [DOI] [PubMed] [Google Scholar]
- 2.Konya J, Thompson C H. J Infect Dis. 1999;179:701–704. doi: 10.1086/314620. [DOI] [PubMed] [Google Scholar]
- 3.Heng M C Y, Steuer M E, Levy A, McMahon S, Richman M, Allen S G, Blackhart B. Am J Dermatopathol. 1989;11:248–254. doi: 10.1097/00000372-198906000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Fife K H, Whitfeld M, Faust H, Goheen M P, Bryan J, Brown D. Virology. 1996;226:95–112. doi: 10.1006/viro.1996.0631. [DOI] [PubMed] [Google Scholar]
- 5.Buller R M L, Chen J B W, Kreider J. Virology. 1995;213:655–659. doi: 10.1006/viro.1995.0037. [DOI] [PubMed] [Google Scholar]
- 6.Senkevich T G, Bugert J J, Sisler J R, Koonin E V, Darai G, Moss B. Science. 1996;273:813–816. doi: 10.1126/science.273.5276.813. [DOI] [PubMed] [Google Scholar]
- 7.Senkevich T G, Koonin E V, Bugert J J, Darai G, Moss B. Virology. 1997;233:19–42. doi: 10.1006/viro.1997.8607. [DOI] [PubMed] [Google Scholar]
- 8.Krathwohl M D, Hromas R, Brown D R, Broxmeyer H E, Fife K H. Proc Natl Acad Sci USA. 1997;94:9875–9880. doi: 10.1073/pnas.94.18.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Damon I, Murphy P M, Moss B. Proc Natl Acad Sci USA. 1998;95:6403–6407. doi: 10.1073/pnas.95.11.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Senkevich T G, Moss B. Virology. 1998;250:397–407. doi: 10.1006/viro.1998.9390. [DOI] [PubMed] [Google Scholar]
- 11.Shisler J L, Senkevich T G, Berry M J, Moss B. Science. 1998;279:102–105. doi: 10.1126/science.279.5347.102. [DOI] [PubMed] [Google Scholar]
- 12.Bertin J, Armstrong R C, Ottilie S, Martin D A, Wang Y, Banks S, Wang G-H, Senkevich T G, Alnemri E S, Moss B, et al. Proc Natl Acad Sci USA. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Nelpel F, Mattman C, Burns K, Bodmer J-L, Schröter M, et al. Nature (London) 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 14.Hu S, Vincenz C, Buller M, Dixit V M. J Biol Chem. 1997;272:9621–9624. doi: 10.1074/jbc.272.15.9621. [DOI] [PubMed] [Google Scholar]
- 15.Novick D, Kim S-H, Fantuzzi G, Reznikov L L, Dinarello C A, Rubinstein M. Immunity. 1999;10:127–136. doi: 10.1016/s1074-7613(00)80013-8. [DOI] [PubMed] [Google Scholar]
- 16.Aizawa Y, Akita K, Taniai M, Torigoe K, Mori T, Nishida Y, Ushio S, Nukada Y, Tanimoto T, Ikegami H, et al. FEBS Lett. 1999;445:338–342. doi: 10.1016/s0014-5793(99)00148-9. [DOI] [PubMed] [Google Scholar]
- 17.Xiang Y, Moss B. Virology. 1999;257:297–302. doi: 10.1006/viro.1999.9676. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura K, Okamura H, Wada M, Nagata K, Tamura T. Infect Immun. 1989;57:590–595. doi: 10.1128/iai.57.2.590-595.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dinarello C A. J Allergy Clin Immunol. 1999;103:11–24. doi: 10.1016/s0091-6749(99)70518-x. [DOI] [PubMed] [Google Scholar]
- 20.Takeda K, Tsutsui H, Yoshimoto T, Adachi O, Yoshida N, Kishimoto T, Okamura H, Nakanishi K, Akira S. Immunity. 1998;8:383–390. doi: 10.1016/s1074-7613(00)80543-9. [DOI] [PubMed] [Google Scholar]
- 21.Ushio S, Namba M, Okura T, Hattori K, Nukada Y, Akita K, Tanabe F, Konishi K, Micallef M, Fujii M, et al. J Immunol. 1996;156:4274–4279. [PubMed] [Google Scholar]
- 22.Okamura H, Tsutsi H, Komatsu T, Yutsudo M, Hakura A, Tanimoto T, Torigoe K, Okura T, Nukada Y, Hattori K, et al. Nature (London) 1995;378:88–91. doi: 10.1038/378088a0. [DOI] [PubMed] [Google Scholar]
- 23.Bazan J F, Timans J C, Kastelein R A. Nature (London) 1996;379:591. doi: 10.1038/379591a0. [DOI] [PubMed] [Google Scholar]
- 24.Ghayur T, Banerjee S, Hugunin M, Butler D, Herzog L, Carter A, Quintal L, Sekut L, Talanian R, Paskind M, et al. Nature (London) 1997;386:619–623. doi: 10.1038/386619a0. [DOI] [PubMed] [Google Scholar]
- 25.Robinson D, Shibuya K, Mui A, Zonin F, Murphy E, Sana T, Hartley S B, Menon S, Kastelein R, Bazan F, O’Garra A. Immunity. 1997;7:571–581. doi: 10.1016/s1074-7613(00)80378-7. [DOI] [PubMed] [Google Scholar]
- 26.Puren A J, Fantuzzi G, Gu Y, Su M S, Dinarello C A. J Clin Invest. 1998;101:711–721. doi: 10.1172/JCI1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carroll M W, Moss B. BioTechniques. 1995;19:352–355. [PubMed] [Google Scholar]
- 28.Buller R M, Chakrabarti S, Cooper J A, Twardzik D R, Moss B. J Virol. 1988;62:866–877. doi: 10.1128/jvi.62.3.866-874.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torigoe K, Ushio S, Okura T, Kobayashi S, Taniai M, Kunikata T, Murakami T, Sanou O, Kojima H, Fujii M, et al. J Biol Chem. 1997;272:25737–25742. doi: 10.1074/jbc.272.41.25737. [DOI] [PubMed] [Google Scholar]
- 30.Yoshimoto T, Takeda K, Tanaka T, Ohkusu K, Kashiwamura S, Okamura H, Akira S, Nakanishi K. J Immunol. 1998;161:3400–3407. [PubMed] [Google Scholar]
- 31.Taniguchi M, Nagaoka K, Ushio S, Nukada Y, Okura T, Mori T, Yamauchi H, Ohta T, Ikegami H, Kurimoto M. J Immunol Methods. 1998;217:97–102. doi: 10.1016/s0022-1759(98)00098-2. [DOI] [PubMed] [Google Scholar]
- 32.Kotwal G J, Moss B. Nature (London) 1988;335:176–178. doi: 10.1038/335176a0. [DOI] [PubMed] [Google Scholar]
- 33.Kotwal G J, Isaacs S N, Mckenzie R, Frank M M, Moss B. Science. 1990;250:827–830. doi: 10.1126/science.2237434. [DOI] [PubMed] [Google Scholar]
- 34.McFadden G, Lalani A, Everett H, Nash P, Xu X. Semin Cell Dev Biol. 1998;9:359–368. doi: 10.1006/scdb.1998.0245. [DOI] [PubMed] [Google Scholar]
- 35.Ploegh H L. Science. 1998;280:248–253. doi: 10.1126/science.280.5361.248. [DOI] [PubMed] [Google Scholar]
- 36.Fujioka N, Akazawa R, Ohashi K, Fuji M, Ikeda M, Kurimoto M. J Virol. 1999;73:2401–2409. doi: 10.1128/jvi.73.3.2401-2409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, Vilcek J, Zinkernagel R M, Aguet M. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 38.Karupiah G, Frederickson T N, Holmes K L, Khairallah L H, Buller R M L. J Virol. 1993;67:4214–4226. doi: 10.1128/jvi.67.7.4214-4226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoll S, Muller G, Kurimoto M, Saloga J, Tanimoto T, Yamauchi H, Okamura H, Knop J, Enk A H. J Immunol. 1997;159:298–302. [PubMed] [Google Scholar]
- 40.Ray C A, Black R A, Kronheim S R, Greenstreet T A, Sleath P R, Salvesen G S, Pickup D J. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 41.Upton C, Mossman K, McFadden G. Science. 1992;258:1369–1373. doi: 10.1126/science.1455233. [DOI] [PubMed] [Google Scholar]
- 42.Alcami A, Smith G L. J Virol. 1995;69:4633–4639. doi: 10.1128/jvi.69.8.4633-4639.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mossman K, Upton C, Buller R M, McFadden G. Virology. 1995;208:762–769. doi: 10.1006/viro.1995.1208. [DOI] [PubMed] [Google Scholar]