Abstract
C3H/HeJ inbred mice are defective in that they are highly resistant to endotoxic shock as compared with normal responder mice. Their B cells and macrophages do not respond significantly when exposed to lipopolysaccharide (LPS), whereas cells from the responder mice do. Using a functional assay, we previously isolated a cDNA, which encodes for Ran/TC4 GTPase. We now show that this gene is mutated in C3H/HeJ mice, which accounts for their resistance to endotoxin stimulation. Sequence analysis of independent mutant Lpsd/Ran cDNAs isolated from splenic B cells of C3H/HeJ mice reveals a consistent single base substitution at position 870, where a thymidine is replaced with a cytidine. In situ hybridization maps the Lpsd/Ran cDNA to mouse chromosome 4. By retroviral gene transfer, the wild-type Lpsn/Ran cDNA but not the mutant Lpsd/Ran cDNA can restore LPS responsiveness of C3H/HeJ cells. Adenoviral gene transfer in vivo with the mutant Lpsd/Ran cDNA but not the wild-type Lpsn/Ran cDNA rescues endotoxin-sensitive mice from septic shock. Thus Lps/Ran is an important target for LPS-mediated signal transduction, and the Lpsd/Ran gene may be useful as a therapeutic sequence in gene therapy for endotoxemia and septic shock.
Keywords: gene therapy, septic shock, GTPase, retrovirus, adenovirus
The underlying genetic basis for the multiple responses of the host to lipopolysaccharide (LPS) endotoxins was initially defined with the discovery of the C3H/HeJ mutant mouse strain in the late 1960s (1). This strain is hyporesponsive to the immunostimulatory and pathophysiological effects of the lipid A component of LPS and, therefore, is considered to have a fundamental deficiency in its reactions to LPS as compared with closely related normal responder strains. This defect is expressed in a variety of ways. For example, C3H/HeJ B cells do not proliferate or differentiate with LPS, their T cells are not stimulated by LPS to enhance polyclonal B cell responses, and thymocyte proliferation in response to Con A is not enhanced by LPS (2, 3). Macrophage phagocytosis and cytotoxicity are not stimulated by LPS and the cytokines such as tumor necrosis factor (TNF), interleukin 1 (IL-1), colony-stimulating factor (CSF), interferon (IFN), and prostaglandins stimulated by LPS in responder cells are not induced in C3H/HeJ macrophages (4). Furthermore, the C3H/HeJ mouse is highly resistant to lethal endotoxic shock as compared with normal endotoxin-sensitive strains (1).
From the results of classical breeding experiments with the C3H/HeJ strain and various responder strains, it has been concluded that the mitogenic response to LPS is governed by a single gene locus composed of codominant alleles (4–8). Watson and Riblet determined that the locus (Lps) was on chromosome 4 and linked to the major urinary protein locus (Mup-1), but downstream from Mup-1 and the Lyb-2, -4, and -6 genes that control B cell activation (9). As a consequence of this appreciation of the genetics of endotoxin responsiveness, the C3H/HeJ strain has provided a powerful analytical tool to investigate the mechanism(s) of endotoxin-initiated events (9, 10). Indeed, if the product and function of the Lps gene can be found, the key to a clearer understanding of how LPS works will be at hand. Because LPS is the inducer of endotoxic shock, identification of the gene product should facilitate the development of clinical strategies to combat septic shock.
Recently, we have established a functional screening method in which cDNA from LPS-stimulated splenic B cells from C3H/HeOuJ responder mice was introduced into splenic B cells of C3H/HeJ LPS-nonresponder mice (11). From this screening effort, we were able to isolate one clone from the cDNA library, whose expression in C3H/HeJ splenic B cells could correct their hyporesponsiveness to LPS. Sequence analysis showed that this LPS-responsive gene encodes Ran/TC4, a GTPase important for nuclear transport (12–14). Our earlier report suggests that Ran/TC4 is important for LPS signal transduction. Whether this gene is defective in the C3H/HeJ genome has not been studied or documented. In this report, we now provide four pieces of direct evidence to suggest that the Ran (or Lps/Ran) of C3H/HeJ cells is defective and it accounts for their endotoxin resistance. First, there is a point mutation at the 3′ untranslated region (UTR) of Lpsd/Ran cDNA from C3H/HeJ mice. Second, the Lps/Ran gene maps to mouse chromosome 4. Third, expression of the Lpsn/Ran cDNA but not the Lpsd/Ran cDNA can restore LPS responsiveness of C3H/HeJ cells. Finally, adenoviral transfer of the Lpsd/Ran cDNA in vivo, but not the Lpsn/Ran cDNA, confers endotoxin resistance to sensitive mice, a phenotype similar to that of C3H/HeJ mice.
MATERIALS AND METHODS
Animals.
Inbred strains of mice BALB/c, C3H/OuJ, and C3H/HeOuJ were obtained from the Jackson Laboratory; CD1 outbred mice, from Charles River Breeding Laboratories. Mice used in the experiment were 6–8 weeks old.
Construction of N2-Lpsn and N2-Lpsd Retrovirus Vectors.
The Lpsn cDNA and Lpsd cDNA were isolated from the XhoI restriction fragment from pCD-LPSn and pCD-LPSd plasmids (11) and ligated into the XhoI restriction site of N2 retrovirus vector to produce pN2-Lpsn and pN2-Lpsd. Recombinant retroviruses were obtained after transfection into GP/E packaging cells (15–17).
To increase retroviral titer, we prepared concentrated virus stocks. Six hundred milliliters of viral supernatant was used for virus precipitation in 0.4 M NaCl and 8.5% PEG (Sigma) for 1–1.5 hr at 4°C with continuous stirring. After centrifugation at 7,500 rpm in a Jouan MR 1812 for 10 min, the pellet was dissolved in NTE buffer (100 mM NaCl/10 mM Tris⋅HCl, pH 7.4/1 mM EDTA) in 1% of the original volume. The sample was then passed through a 10-ml preswollen Sepharose CL-4B column at 4°C. Several fractions were collected and pooled. Each pooled fraction was filter-sterilized and tested for viral titer by virtue of G418 resistance in NIH/3T3 cells (15).
MTT Assay on Splenic B Cells Infected with N2-Lpsn or N2-Lpsd Retrovirus Vectors.
Ten million C3H/HeJ Splenic cells, depleted of red blood cells, were placed in a 100-mm cell culture plate containing 10 ml of S17CM (RPMI medium 1640 containing 10% fetal calf serum and 50 μM 2-mecaptoethanol incubated in S17 cells for 24 hr) for 6 hr. After incubation, nonadherent cells were collected and infected in a 100-mm plate with 10 ml of infection mixture. The infection mixture contained 3 ml of the concentrated viral supernatant with a titer of 2 × 106 G418-resistant colony-forming units/ml, 1 ml of S17CM, and 10 μg/ml Polybrene. Sixteen hours later, the cells were collected and reinfected with fresh infection mixture for another 16 hr. Finally, cells were harvested and stimulated with 10 μg/ml or 100 μg/ml hot-phenol-extracted LPS from Salmonella typhimurium in 2 ml of S17CM and incubated in a 24-well plate for 0, 12, 24, or 48 hr. Subsequently, 100 μl of the retrovirus-infected and LPS-stimulated cells were placed in a 96-well plate. Ten microliters of 5 mg/ml 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added and mixed with cells. After a 4-hr incubation, 100 μl of 0.04 M HCl in isopropyl alcohol was added to the wells and mixed thoroughly. OD at 570/690 nm in each well was read by a Bio-Tek microplate reader.
Reverse Transcriptase (RT)-PCR Amplification.
Total RNA was extracted from retrovirus-infected splenic B cells by using 4 M guanidine thiocyanate (15) and phenol/chloroform. Avian myeloblastosis virus RT was used to generate cDNA. Two primers were used for PCR, 5′ primer from the pCD portion of the XhoI fragment and 3′ primer from the Lps cDNA, to generate a 907-bp fragment (5′ primer: TCTAAAAGCTGCTGCAGG, 3′ primer: GTACACGATCTGCTTAGC).
Fluorescent in Situ Hybridization (FISH) Analysis.
Mouse chromosomes were prepared as described (18). Briefly, lymphocytes were isolated from mouse spleen and cultured at 37°C in RPMI 1640 supplemented with 15% fetal calf serum, 3 μg/ml Con A, 10 μg/ml LPS, and 50 μM 2-mercaptoethanol. After 44 hr, the cultured lymphocytes were treated with 0.18 mg/ml BrdUrd for an additional 14 hr. The synchronized cells were washed and recultured at 37°C for 4 hr in α-MEM with thymidine (2.5 μg/ml). The procedure for FISH detection was performed according to Heng et al. (18, 19). Briefly, the slides were baked at 55°C for 1 hr. After RNase A treatment, the slides were subjected to denaturation in 70% formamide in 2× SSC for 2 min at 70°C, followed by dehydration with ethanol. Lps/Ran-specific DNA probe was biotinylated with dATP by using the BRL BioNick labeling kit at 15°C for 1 hr (18). The probes were denatured at 75°C for 5 min in a hybridization mix consisting of 50% formamide and 10% dextran sulfate and mouse Cot I DNA and were prehybridized for 15 min at 37°C. This mixture was then loaded onto the “denatured” slides. After an overnight hybridization, the slides were subjected to washing, detection, and amplification using a published method (18). The FISH signals and the 4′,6-diamidino-2-phenylindole (DAPI) banding pattern were recorded separately by photographs. Chromosomal assignment was achieved by superimposing the FISH signals on a particular chromosome with the same chromosome banded with DAPI, with the same results obtained in photographs of ten different fields (19, 20).
Construction of Ad5-Lps Adenovirus Vectors and Virus Production.
Lpsn and Lpsd cDNAs were excised with BamHI at sites 3249 and 35 from pCD-Lpsn and pCD-Lpsd (the latter provided by StemCell Therapeutics). They were separately inserted in the E1 region of the adenovirus 5 (Ad5) genome and driven by cytomegalovirus (CMV) promoter. The green fluorescent protein (GFP) gene from pEGFP plasmid (CLONTECH) was excised with BamHI and NotI and inserted at the same site and driven by CMV promoter as well. The recombinant adenoviruses are replication defective because they are E1 deleted. However, they can replicate in 293 cells containing E1 gene. For the production of recombinant viruses, 293 cells were transfected with recombinant Ad5 DNA. Ten days after transfection, we collected the cells by scraping them off culture flasks. After three cycles of freezing in an ethanol/dry ice bath and rapid thawing at 37°C, lysate was collected and used to infect 293 cells in twenty 150-mm plates. Three days later, viruses were harvested as described above and purified by CsCl banding and dialyzed three times against PBS. To determine virus titer, 293 cells were seeded in six-well plates. Twenty-four hours after plating, they were infected with serial dilutions of the virus stocks. Two hours after infection, the medium was removed and cells were overlaid with medium containing 2% FBS and 1% agarose. Ten days later, the number of virus plaques was recorded. The Ad5 vectors have a titer of 1012 plaque-forming units (pfu)/ml.
In Vivo Adenoviral Lps/Ran Transfer.
The LD50 and LD100 of endotoxin were determined to be 0.6 mg and 1 mg, respectively, using outbred CD1 mice purchased from Charles River Breeding Laboratories. After determining that, we inoculated each mouse intraperitoneally with 1 mg of endotoxin, equivalent to LD100. One hour later, 0.5 ml of Ad5-Lpsd, Ad5-Lpsn, or Ad5-GFP virus containing 1010 pfu was administered into each mouse intravenously. Life and death of these experimental animals were then observed and recorded at 12-hr intervals.
RT-PCR Analysis for the Expression of Adenovirus in Peripheral Blood Cells.
CD1 mice were inoculated through the tail vein with approximately 1010 pfu of infectious recombinant Ad5 particles. After 2, 4, 6, 10, and 24 hr, whole blood samples for RT-PCR detection of Ad5 mRNA in transduced cells were collected into hematocrit tubes and centrifuged, and the buffy coat layer was lysed in GTC buffer for total RNA extraction (15). A 10-μl volume of each RNA sample was used for reverse transcription in a 20-μl mixture (15). Two of the 20 μl were used for nested PCR amplification (15). For each reaction, 1.5 unit of Taq DNA polymerase (Sigma) was used in a 50-μl reaction volume. After the samples had been denatured at 95°C for 5 min, each was subjected to 20 cycles of denaturation (94°C for 1 min), annealing (60°C for 1 min), and primer extension (72°C for 1 min). The last cycle included an additional step of primer extension at 72°C for 10 min. The primers for the first round of PCR amplification were 5′ primer sequence (T7 promoter, sense) = 5′-TAATACGACTCACTATAGGGAGA-3′, 3′ primer sequence (D2 antisense) = 5′-GAAATTCAGAAAGGAAACAACTCTGTTCCA-3′, which is an antisense sequence specific for Lps/Ran cDNA (starting at position 810). The expected size of the amplified DNA fragment was 1020 bp. For the second round of the nested PCR amplification, 2 μl of the PCR product from the first-round PCR amplification was used. The 5′ primer was pcDNA3 (Invitrogen) sense sequence 5′-ACTATAGGGAGACCCAAGCT-3′, which is specific for pcDNA3, the vector sequence attached to the Lps/Ran cDNA in the adenovirus vectors during construction. The 3′ primer (R1 antisense) sequence was 5′-AGCAGTCGTCTGAGCAACCT-3′, which is specific for Lps/Ran cDNA (starting at position 606). The expected size of the PCR-amplified product would be 793 bp. The samples were first denatured at 95°C for 5 min, then subjected to 30 cycles of 94°C for 1 min, 60°C for 1 min, and then 72°C for 1 min, followed by an extended primer-extension reaction of 72°C for 10 min.
RESULTS
Isolation of Mutant Lps/Ran cDNA from C3H/HeJ Mice.
To examine whether Lps/Ran DNA is defective in the genome of C3H/HeJ mice, which may explain their hyporesponsiveness to LPS stimulation, we constructed an Okayama–Berg cDNA expression library, pCD-HeJ, from mRNA of LPS-stimulated splenic B cells of C3H/HeJ mice (21). After screening 8 dishes of 5.8 × 104 bacterial colonies from the pCD-HeJ cDNA library per 150-mm dish, totaling about 4.5 × 105 colonies, we isolated 25 positive clones while using a 0.9-kb BamHI–PstI fragment of the wild-type Lpsn/Ran cDNA as a probe (11). Eight of the 25 clones contained the full-length cDNA based on the overall size of the wild-type Lps/Ran cDNA. DNA sequencing was then performed on plasmids of four different clones (Lpsd-4, -10, -15, and -17). The DNA sequence of each of these four clones was compared with that of the wild-type Lps/Ran cDNA (labeled as Lpsn in Fig. 1). Each clone showed a consistent point mutation at position 870 of the Lpsn cDNA (residue numbering begins at A of the ATG start site), where a thymidine (T) residue was replaced by a cytidine (C) residue (Fig. 1). Sequence analysis of the Ran cDNA also revealed that the plasmid DNAs must have derived from at least two independent clones of bacteria, as the 5′ Ran cDNA sequence started at different points among these four plasmids. These data strongly suggest that the point mutation present in the mutated Lpsd/Ran gene may account for the hyporesponsiveness of C3H/HeJ cells to LPS.
Figure 1.
DNA sequences of independent clones of Lpsd cDNA. The numbering starts at A (position 1) of the translation start site, ATG, indicated by boldface underlined type.
The four Lpsd clones analyzed all gave strongest positive hybridization signals with the Lpsn probe. This result probably explained why we obtained only one species of cDNA instead of other Ran isoform cDNAs, which were present when we sequenced multiple cDNA fragments obtained from various RT-PCR amplifications (A.K. and P.M.C.W., unpublished data).
Lps/Ran Gene Maps to Chromosome 4.
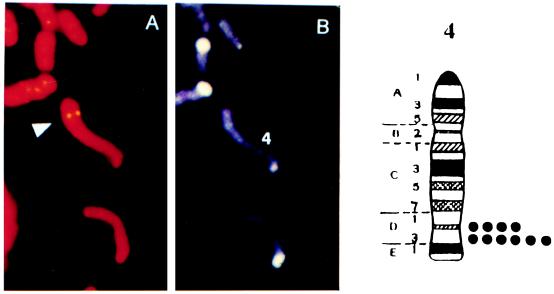
Classical breeding experiments with the C3H/HeJ strain and responder strains suggest that the mitogenic response to LPS is governed by a single genetic locus composed of codominant alleles (4), and the gene has been mapped to chromosome 4. To determine the location of the Lps/Ran gene, FISH analysis was carried out with the Lps/Ran cDNA sequence we isolated (11). Under the experimental condition detailed in Materials and Methods, there was a high background of nonspecific FISH signals, suggesting that the DNA probe may contain either highly repetitive sequences or homologous sequences. Accordingly, more stringent washing was applied so that the hybridization efficiency was estimated to be 42%, which was based on 42 positive signals on one pair of the chromosomes among 100 mitotic figures. DAPI banding was used for chromosome identity. Based on identical results on ten separate areas, the strongest signals were found to be on chromosome 4 (Fig. 2). Weaker but also strong signals were also found on chromosome 7. The high background signal is probably related to the fact that Lps/Ran is a member of the superfamily GTPase genes and that Ran GTPase has several isoforms (12, 22, 23). Drivas et al. (24) used DNA restriction fragment length variants associated with four ras-like cDNAs in recombinant inbred and backcross mice to define novel mouse ras-like genes and showed that there are 12 genetic loci, 9 of which were mapped to chromosomes 2, 4, 7, 8, 9, and 17.
Figure 2.
Chromosomal localization of Lps/Ran by in situ hybridization. (A) FISH signals. (B) Same mitotic figure stained with DAPI. The assignment of the FISH mapping data with chromosomal bands was achieved by superimposing FISH signals with DAPI-banded chromosomes. Each dot in the ideogram on the Right represents the double FISH signals detected on mouse chromosome 4.
Restoration of LPS Responsiveness After Retroviral Transfer of Lpsn but Not Lpsd Gene into C3H/HeJ Cells.
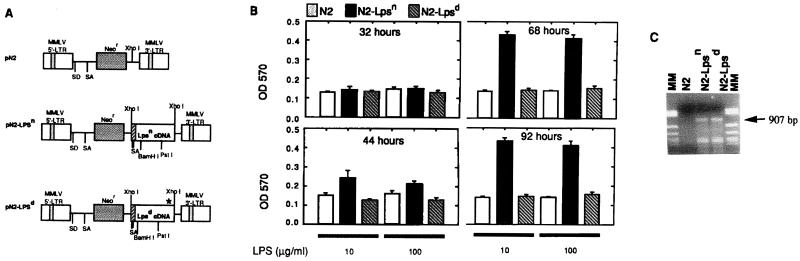
To compare the biological functions of the two cDNAs, we inserted the Lpsn and the Lpsd cDNAs into retroviral vectors (15, 18, 19). Each vector DNA was separately introduced into GP/E virus packaging cells (25), and recombinant retroviruses were produced as described previously (15, 18). Splenic B cells from C3H/HeJ mice were depleted of red blood cells and subjected to retroviral infection and LPS stimulation. Viral titers from concentrated stocks of the N2 parental vector, N2-Lpsn, or N2-Lpsd were adjusted to 2 × 106 G418-resistant colony-forming units/ml (Fig. 3A). At various times after retroviral infection and LPS stimulation, cells were harvested and subjected to MTT proliferation assay (26). At 32 hr, no significant difference in cell proliferation was found among all cultures (Fig. 3B). At 44 hr, N2-Lpsn-infected cells, but not N2-Lpsd- or N2-infected cells, showed a clear increase in cell proliferation (Fig. 3B). At 68 hr, there was a maximal 4-fold OD difference in the LPS response between the N2-Lpsn-infected culture and the N2- or N2-Lpsd-infected culture. This response was maintained at 92 hr.
Figure 3.
(A) N2, N2-Lpsn, and N2-Lpsd retrovirus vectors. (B) MTT assay on splenic B cells cultured with the above recombinant retroviruses. (C) RT-PCR to show expression of Lps/Ran cDNAs (907 bp) from retrovirus-infected splenic B cells. MM, molecular weight markers.
To further show that the response, or the lack of response, to LPS stimulation is caused by the successful retroviral transfer of the corresponding wild-type or mutant Ran cDNA, we performed RT-PCR on cells from all cultures. We used a 5′ oligoprimer specific for a sequence of simian virus 40 promoter that is present within the retroviral vector, and a 3′ primer specific for the Lpsn or Lpsd cDNA. Fig. 3C shows the presence of a predicted 907-bp band in cultures transduced with N2-Lpsn and with N2-Lpsd, but not with N2 virus, confirming the successful transfer and expression of the transduced genes. This primer is specific for Lps RNA because when the genomic DNA was used for PCR, the 907-bp band shown in Fig. 3C could not be observed. This was because the 5′ primer was specific for the vector sequence upstream of the Lps cDNA construct. Thus we showed that introduction and expression of Lpsn cDNA, but not Lpsd cDNA, in splenic B cells from C3H/HeJ mice resulted in restoration of a significant response to LPS stimulation.
Development of Endotoxin Resistance After Adenoviral Transfer of the Lpsd Gene but Not the Lpsn Gene into Endotoxin-Sensitive Mice.
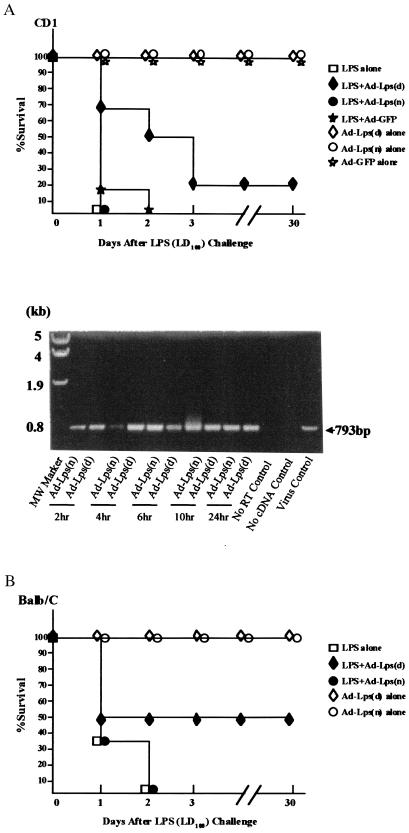
As an extension of our comparative studies between Lpsn and Lpsd cDNA, we performed in vivo gene transfer studies. Mice were induced to undergo septic shock and were treated with vectors carrying the Lpsd, Lpsn, or GFP cDNA. As shock occurs within 3 days, we chose to construct recombinant adenoviruses which could produce a titer of 1012 infectious virus particles per ml. Lpsd/Ran, Lpsn/Ran, and GFP genes were cloned into and replaced the E1 region of the Ad5 vector (Fig. 4). The titer of each recombinant adenovirus stock was determined and appropriately adjusted, and 1010 pfu of infectious virus particles were injected intravenously into each mouse 1 hr after endotoxin treatment. In this first series of experiments, we used outbred CD1 mice because we have gene therapy application in mind and wanted to be able to observe the degree of heterogeneity in the experimental outcome. The mice were monitored over time. All mice treated with the LD100 dose of 1 mg of LPS died within 36 hr, as did mice treated with Ad5-Lpsn or with Ad5-GFP (Fig. 5A). Half of the mice treated with Lpsd died within the same period of time, but the other half had extended life or survived (Fig. 5). Heterogeneity of survival after gene transfer treatment correlated with our use of outbred CD1 mice. One mouse treated with Ad5-Lpsd and surviving the endotoxin challenge manifested cachexia initially but became healthy after recovery. None of the mice treated with Ad5 alone died, indicating absence of toxicity. When inbred BALB/c mice were used for the same experimental design, the percentage of mice treated with Ad5-Lpsd that survived increased to 50%, whereas none of the mice treated with Ad5-Lpsn or Ad5-GFP survived more than 2 days (Fig. 5B).
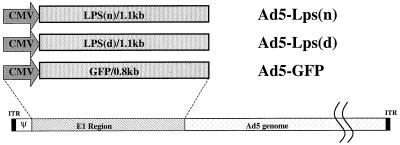
Figure 4.
Construction of Ad5-Lps adenovirus vectors. Black boxes represent viral invert terminal repeat (ITR); Ψ, the packaging sequence; CMV, cytomegalovirus promoter sequence. In the constructs, the E1 region of Ad5 was replaced with Lpsn/Ran cDNA (1.1 kb), Lpsd/Ran cDNA (1.1 kb), or the GFP gene (0.8 kb).
Figure 5.
Endotoxin resistance after adenoviral transfer of Lpsd/Ran cDNA into LPS-sensitive mice. (A) (Upper) Survival curves. Six CD1 mice were used in each group treated with endotoxin plus Ad5-GFP, Ad5-Lps(n), or Ad5-Lps(d); three mice were used in each group with virus alone or LPS alone. Infectious virus particles inoculated per mouse are 1 × 1010. (Lower) Expression of adenoviral genes in peripheral blood mononuclear cells by nested RT-PCR on total RNA from mice infected with Ad5-Lps(n) or Ad5-Lps(n) at various times. The expected size of the amplified fragment is 793 bp. (B) Survival curves. For BALB/c mice, 5 × 108 to 5 × 109 pfu of adenovirus was injected into each mouse 1 hr before injection of 1 mg of LPS per mouse. Eight, 6, 4, and 6 mice were used in the groups treated with LPS plus Ad5-Lps(d), with LPS plus Ad5-Lps(n), with virus alone, and with LPS alone, respectively. □, Mice treated with LPS alone; ⋄, Ad5-Lps(d)/Ran alone; ♦, LPS with Ad5-Lps(d)/Ran; ○, Ad5-Lps(n)/Ran alone; ●, LPS with Ad5-Lps(n)/Ran; ⋆, Ad5-GFP alone; and ★, LPS with Ad5-GFP.
To verify the presence of adenoviral gene transfer, peripheral blood mononuclear cells of mice transduced with Ad5-Lpsd or Ad5-Lpsn at different times after in vivo gene transfer were collected for RNA extraction. After nested RT-PCR amplification using specific primers, RNA of cells as early as 2 hr after in vivo gene transfer contained a 793-bp band, suggesting the presence of adenoviral mRNA. No such band was observed in the absence of reverse transcription or cDNA template (Fig. 5A Lower). These data suggest that as early as 2 hr after intravenous inoculation of the vector, adenoviral gene expression could occur, and vectors carrying the Lpsd gene prolonged the survival of mice receiving LD100 doses of LPS.
DISCUSSION
In this study, we have shown that Lpsd/Ran of C3H/HeJ mice is different from Lpsn/Ran of C3H/HeOuJ mice. The genetic and biological differences between the two molecules correlate with the differences in the responsiveness of the cells from these mice to endotoxin. In addition to these findings, we also found that there are major RNA structural differences and biochemical differences between Lpsn/Ran and Lpsd/Ran (P.M.C.W. and H.C., unpublished results). These data suggest that the Lpsn/Ran gene may be an Lpsn gene and the Lpsd/Ran, the defective Lpsd counterpart deduced from various studies using endotoxin-resistant C3H/HeJ inbred mice.
The result of our FISH analysis shows that the Lps/Ran gene is on chromosome 4 in the mouse, a finding consistent with that of Coutavas et al. (22). A recent genetic and physical mapping report identifies the Toll-like receptor (Tlr-4) as a candidate gene in the critical region of the Lps locus on chromosome 4 in the mouse (27). Poltorak et al. have further identified a missense mutation in the third exon of the Tlr-4 gene of C3H/HeJ mice, although no functional comparative studies were carried out in that study (28). Another gene, Toll-like receptor-2 (Tlr-2), encodes a signaling receptor that is activated by LPS but a response depends on the LPS-binding protein (LBP) and is enhanced by CD14, which is not involved in LPS-mediated response in B lymphocytes (29, 30). Toll-like receptor genes (Tlr) belong to a family of IL-1 receptor-like genes that are related but distinct genes found to be located at different chromosomes in humans (31).
Genetic experiments from a number of laboratories, which involved the breeding of the C3H/HeJ mouse to various inbred strains, have led to a consensus that the polyclonal activation of B cells by LPS is governed by a single locus (Lps) composed of codominant alleles located on chromosome 4 and linked to the major urinary protein locus (Mup-1) (5–8). In another study of recombinant strains of mice, additional endotoxin responses, including hypothermia and the elevation of a serum colony-stimulating factor and a mouse amyloid A protein, were claimed to be regulated by the Lps locus. However, there were some exceptions in that the response in some responder strains were quantitatively different than other strains in the amyloid A protein, suggesting to the authors another locus that is distinct from Lps (32). Indeed, even with murine B cells, Glode and Rosenstreich (33) presented data from 11 closely related C3H mouse strains supporting their conclusions that three additional genes or three possible alleles at one or more autosomal loci determine the response of B cells from C3H mice to LPS. Furthermore, of even greater interest is the early report that resistance to endotoxemia, which in reality is the hallmark of the host response to LPS, is governed by multiple genes as determined by the measuring of LD50 doses of LPS in parental strains and F1 and backcross hybrids (34). Consequently, these results are consistent with the concept that many reactions to LPS are complex phenomena undoubtedly involving different cell types and cell products. Therefore, LPS responses may be subject to gene control independent of the Lps locus as presently defined, so multiple genes may be involved in the overall host response to endotoxin.
The single base substitution in Lpsd/Ran mRNA at the 3′ UTR suggests the presence of a unique mRNA domain to which regulatory protein(s) may bind and impose a posttranscriptional regulation, affecting the translational efficiency or mRNA stability, transport, or intracellular localization (35–40). During Drosophila development 3′ UTRs are important in the spatial and temporal regulation of nanos, bicoid, and hunchback mRNA (41–43). They also define localization sites of tra-2, fem-3, and glp-1 mRNAs of Caenorhabditis elegans for translational regulation (44). In macrophages, LPS induces the production of tumor necrosis factor α (TNFα); it also induces the production of tristetratprolin (TTP), a CCCH (Cys-Cys-Cys-His) class of zinc finger protein, which negatively regulates TNFα (45, 46). This LPS-inducible negative-feedback regulation is achieved by TTP binding to the AU-rich region of the TNFα mRNA and destabilizing it (46). Other RNA-binding proteins have been reported to regulate TNFα production by binding to its 3′ UTR as well (47, 48). Interestingly, activation of TTP by LPS also appears to be mediated through the 3′ UTR of TTP mRNA, again by an AU-rich instability sequence (49). The fact that Lps/Ran mRNA also contains two AUUUA motifs, and that Lps/Ran is an LPS response gene, suggests that Lps/Ran, like TNFα and TTP genes, is regulated by means of its mRNA AU-rich motif located at the 3′ UTR. Regulation of Lps/Ran mRNA by its 3′ UTR is consistent with our finding that a point mutation at position 870 affected cellular response to LPS stimulation (Fig. 1). This single base change is only 24 bases downstream of the second AUUUA motif of Lps/Ran mRNA (845–849). The single base substitution from a T to a C at position 870 could result in a drastic change in secondary structure, which could lead to a significant biological change such as a reduced LPS responsiveness.
The magnitude of Lpsn/Ran cDNA in restoring the endotoxin response of C3H/HeJ splenic B cells was about one-third that of splenic B cells from responder mice (11). This moderate response would be consistent with the codominant characteristics of the Lps alleles, as was suggested by analysis of F1 hybrid from crosses between the LPS-responder inbred mice and the C3H/HeJ hyporesponder mice (5, 9, 32). Therefore, introduction of a single copy of Lpsn/Ran cDNA into C3H/HeJ cells, which contain 2 copies of Lpsd/Ran gene, would result in restoration of an LPS response significantly less than that of LPS-responder cells.
Our results for adenoviral transfer of the Lpsd/Ran gene and the Lpsn/Ran gene into LPS-sensitive mice are also consistent with the codominant phenotype of the two alleles. Mice that received Ad5-Lpsd/Ran, like C3H/HeJ, developed some resistance to endotoxin challenge, whereas mice that received Ad5-Lpsn/Ran remained sensitive to endotoxin (Fig. 5A Upper). Transfer and expression of the Lps/Ran gene in both liver and peripheral blood mononuclear cells were confirmed in recipient mice (Fig. 5B and data not shown). The mouse that was treated with a lethal endotoxin dose and Ad5-Lpsd/Ran actually exhibited cachexia but after recovery became as active as normal mice. The nature of this resistance may be related to reduced production of endotoxin-induced cytokines in LPS-responder cells expressing the Lpsd/Ran cDNA, because our preliminary data showed that ANA1 cells, a macrophage cell line established from LPS-responder mice, when transduced with Lpsd/Ran produced lower amount of LPS-induced TNFα compared with control cells (P.M.C.W., unpublished work). When inbred BALB/c mice were used, four of eight mice inoculated with Ad5-Lpsd/Ran survived LPS challenge but others did not. This first series of gene therapy experiments provides a good base upon which to perform more experiments to define the optimal gene therapy protocol for endotoxemia and septic shock.
Acknowledgments
We thank Dr. Rachael Sheppard and Dr. Stephen Pielder for their helpful discussions, and Dr. Henry Heng at SeeDNA Biotech (York University, Ontario), who performed the FISH analysis in this study. P.M.C.W. especially thanks Dr. Daniel Rotrosen (National Institute of Arthritis and Infectious Diseases) for his appreciation and support. This work was supported in part by grants from the National Institutes of Health (RO1AI39159 and RO1CA70854 to P.M.C.W.) and in part by grants from StemCell Therapeutics. Y.W.K. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- LPS
lipopolysaccharide
- UTR
untranslated region
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- RT
reverse transcriptase
- FISH
fluorescent in situ hybridization
- DAPI
4′,6-diamidino-2-phenylindole
- Ad5
adenovirus 5
- GFP
green fluorescent protein
- pfu
plaque-forming units
- TNFα
tumor necrosis factor α
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF159256).
References
- 1.Sultzer B M. Nature (London) 1968;219:1253–1254. doi: 10.1038/2191253a0. [DOI] [PubMed] [Google Scholar]
- 2.Sultzer B M. Abstr. Am. Soc. Microbiol. Abstr. M69; 1973. p. 85. [Google Scholar]
- 3.Tanabe M J, Nakano M. Microbiol Immunol. 1979;23:1097–1107. doi: 10.1111/j.1348-0421.1979.tb00541.x. [DOI] [PubMed] [Google Scholar]
- 4.Morrison D C, Ryan J L. Adv Immunol. 1979;28:293–450. doi: 10.1016/s0065-2776(08)60802-0. [DOI] [PubMed] [Google Scholar]
- 5.Sultzer B M. Infect Immun. 1976;13:1579–1584. doi: 10.1128/iai.13.6.1579-1584.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coutinho A. Scand J Immunol. 1976;5:129–135. doi: 10.1111/j.1365-3083.1976.tb02999.x. [DOI] [PubMed] [Google Scholar]
- 7.Watson J, Riblet R, Taylor B A. J Immunol. 1977;118:2088–2093. [PubMed] [Google Scholar]
- 8.Watson J, Kelly K, Largen M, Taylor B A. J Immunol. 1978;120:422–424. [PubMed] [Google Scholar]
- 9.Watson J, Riblet R. J Immunol. 1975;114:1462–1468. [PubMed] [Google Scholar]
- 10.Raetz C R H. Annu Rev Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 11.Kang A D, Wong P M C, Chen H, Castagna R, Chung S W, Sultzer B M. Infect Immun. 1996;64:4612–4617. doi: 10.1128/iai.64.11.4612-4617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bischoff F R, Krebber H, Kempf T, Hermes I, Ponstingl H. Proc Natl Acad Sci USA. 1995;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore M S. Science. 1996;272:47. doi: 10.1126/science.272.5258.47. [DOI] [PubMed] [Google Scholar]
- 14.Koepp D M, Silver P A. Cell. 1996;87:1–4. doi: 10.1016/s0092-8674(00)81315-x. [DOI] [PubMed] [Google Scholar]
- 15.Wong, B. Y., Chen, H., Chung, S. W. & Wong, P. M. C. (1994) 68, 5523–5531. [DOI] [PMC free article] [PubMed]
- 16.Wong P M C, Chung S W, Nienhuis A W. Genes Dev. 1987;1:358–365. doi: 10.1101/gad.1.4.358. [DOI] [PubMed] [Google Scholar]
- 17.Wong P M C, Chung S W, Chung S W, Dunbar C E, Bodine D M, Ruscetti S, Nienhuis A W. Mol Cell Biol. 1989;9:798–808. doi: 10.1128/mcb.9.2.798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heng H H Q, Squire J, Tsui L C. Proc Natl Acad Sci USA. 1992;89:9509–9513. doi: 10.1073/pnas.89.20.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heng H H Q, Tsui L C. Chromosoma. 1993;102:325–332. doi: 10.1007/BF00661275. [DOI] [PubMed] [Google Scholar]
- 20.Heng H H Q, Tsui L C. In: Methods in Molecular Biology. Choo K H A, editor. Clifton, NJ: Humana; 1994. pp. 35–49. [DOI] [PubMed] [Google Scholar]
- 21.Okayama H, Berg P. Mol Cell Biol. 1982;2:161–170. doi: 10.1128/mcb.2.2.161. . F. R.F. R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coutavas E E, Hsieh C M, Ren M, Drivas G T, Rush M G, D’Eustachio P D. Mamm Genome. 1994;5:623–628. doi: 10.1007/BF00411457. [DOI] [PubMed] [Google Scholar]
- 23.Drivas G T, Shih A, Coutavas E, Rush M G, D’Eustachio P. Mol Cell Biol. 1990;10:1793–1798. doi: 10.1128/mcb.10.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drivas G, Massey R, Chang H Y, Rush M G, D’Eustachio P. Mamm Genome. 1991;1:112–117. doi: 10.1007/BF02443787. [DOI] [PubMed] [Google Scholar]
- 25.Markowitz D, Goff S, Bank A. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han X D, Chung S W, Wong P M C. Proc Natl Acad Sci USA. 1995;92:11014–11018. doi: 10.1073/pnas.92.24.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poltorak A, Smirnova I, He X, Liu M Y, Van Huffel C, Birdwell D, Alejos E, Silva M, Du X, Thompson P, et al. Blood Cells Mol Dis. 1998;15:340–355. doi: 10.1006/bcmd.1998.0201. [DOI] [PubMed] [Google Scholar]
- 28.Poltorak A, He X, Smirnova I, Liu M Y, Huffel C V, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 29.Yang R, Mark M R, Gray A, Huang A, Xie M H, Zhang M, Goddard A, Wood W I, Gurney A L, Godowski P J, et al. Nature (London) 1998;395:284–288. doi: 10.1038/26239. [DOI] [PubMed] [Google Scholar]
- 30.Gerard C. Nature (London) 1998;395:217–219. doi: 10.1038/26104. [DOI] [PubMed] [Google Scholar]
- 31.Rock F L, Hardiman G, Timans J C, Kastelein R A, Bazan J F. Proc Natl Acad Sci USA. 1998;95:588–593. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson J, Largen M, McAdam K. J Exp Med. 1978;147:39–47. doi: 10.1084/jem.147.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glode L M, Rosenstreich D L. J Immunol. 1976;117:2061–2066. [PubMed] [Google Scholar]
- 34.Sultzer B M. Infect Immun. 1972;5:107–113. doi: 10.1128/iai.5.1.107-113.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klausner R D, Rouault T A, Harford J B. Cell. 1993;72:19–28. doi: 10.1016/0092-8674(93)90046-s. [DOI] [PubMed] [Google Scholar]
- 36.Belsaco J G, Brawerman G, editors. Control of Messenger RNA stability. San Diego: Academic; 1993. [Google Scholar]
- 37.Wilson T, Treisman R. Nature (London) 1996;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- 38.Dubnau J, Struhl G. Nature (London) 1996;379:694–699. doi: 10.1038/379694a0. [DOI] [PubMed] [Google Scholar]
- 39.Ostareck-Lederer A, Ostrareck D H, Standart N, Thiele B J. EMBO J. 1994;13:1476–1481. doi: 10.1002/j.1460-2075.1994.tb06402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaw G, Kamen R. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 41.Singer R H. Curr Biol. 1993;3:719–721. doi: 10.1016/0960-9822(93)90079-4. [DOI] [PubMed] [Google Scholar]
- 42.Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring W J, Jäckle H. Nature (London) 1996;379:746–749. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 43.Chan S K, Struhl G. Nature (London) 1997;388:634. doi: 10.1038/41692. [DOI] [PubMed] [Google Scholar]
- 44.Evans T C, Crittenden S L, Kodoyianni V, Kimble J. Cell. 1994;77:183–194. doi: 10.1016/0092-8674(94)90311-5. [DOI] [PubMed] [Google Scholar]
- 45.Taylor G A, Carballo E, Lee D M, Lai WS, Thompson M J, Patel D D, Schenkman D I, Gilkeson G S, Broxmeyer H E, Haynes B F, Blackshear P J. Immunity. 1996;4:445–454. doi: 10.1016/s1074-7613(00)80411-2. [DOI] [PubMed] [Google Scholar]
- 46.Carballo E, Lai W S, Blackshear P J. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 47.Hel Z, Shamene E, Radzioch D. Mol Cell Biol. 1996;16:5579–5590. doi: 10.1128/mcb.16.10.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hel Z, Marco S D, Radzioch D. Nucleic Acids Res. 1998;26:2803–2812. doi: 10.1093/nar/26.11.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai W S, Thompson J, Taylor G A, Liu Y, Blackshear P R. J Biol Chem. 1995;270:25266–25272. doi: 10.1074/jbc.270.42.25266. [DOI] [PubMed] [Google Scholar]