Abstract
Theiler’s virus and Mengo virus are representatives of the Cardiovirus genus within the picornavirus family. Their genome is an 8-kilobase long positive strand RNA molecule. This RNA molecule plays three roles in infected cells: It serves as a messenger RNA, acts as a template for genome replication, and is encapsidated to form progeny virions. We observed that a cis-acting signal required for replication of Theiler’s virus was contained within a 130-nt stretch of the region encoding the capsid protein VP2. This RNA sequence does not influence internal ribosome entry site-mediated translation initiation and thus likely acts directly as a signal for the replication complex. We found a similar signal in the VP2-coding sequence of Mengo virus, and both signals could be functionally exchanged. Within the replication element, a 9-nt sequence that is highly conserved among cardioviruses was shown to be essential for replication. This conserved sequence was contained in mostly unpaired regions of the RNA secondary structure predicted for the replication elements of the various cardioviruses. Interestingly, a similar replication element has been reported to occur in the distantly related human rhinovirus type 14, suggesting that such elements could be conserved throughout the picornavirus family. However, the different location of the replication elements in rhinovirus and cardioviruses, and the fact that they were not functionally exchangeable, is raising intriguing questions about the evolution of such signals in picornaviruses.
The genome of picornaviruses is an 8-kilobase long RNA molecule of positive polarity. This genomic RNA plays three distinct roles in the virus life cycle: (i) It serves as a messenger RNA molecule encoding a polyprotein that is processed to yield all of the viral proteins, (ii) it is used as a template for the replication process, and (iii) it is encapsidated to form mature virions (1). Discrete regulatory elements involved in these functions have been mapped on picornaviral genomes (2). An internal ribosome entry site accounts for the initiation of translation of the polyprotein (3, 4). This leaves the 5′ and 3′ ends of the genome free to function as replication signals. Nevertheless, some overlap exists between 5′ replication signals and the internal ribosome entry site (5). Such overlapping signals could ensure a time-controlled balance between the production of viral proteins and that of viral genomes. The genome of picornaviruses also contains a specific packaging signal, so far unidentified, that enables the capsids to discriminate between virus-specific and cellular RNA (6, 7).
Theiler’s murine encephalomyelitis virus (TMEV or Theiler’s virus) is a naturally occurring pathogen of the mouse belonging to the Picornaviridae family (8). The various TMEV isolates are generally classified into two subgroups on the basis of the pathology they induce in susceptible mice. The first subgroup contains two highly neurovirulent strains, GDVII and FA, that cause an acute lethal polioencephalomyelitis. The second subgroup contains persistent strains, including strain DA, that provoke a chronic demyelinating disease of the central nervous system considered as a model of multiple sclerosis (9–11).
Although they cause markedly different pathologies, neurovirulent and persistent strains of TMEV are closely related and share ≈95% identity at the amino acid sequence level (12–14). Theiler’s viruses are classified among cardioviruses, together with encephalomyocarditis virus (EMCV) and Mengo virus (an EMCV isolate), with which they share ≈60% identity.
In this work, we observed that the sequence coding for the VP2 protein of Theiler’s virus contains a cis-acting signal required for replication of the genome. We mapped and characterized this cis-acting replication element (CRE). It did not account for the difference of replication level observed between persistent and neurovirulent strains of Theiler’s virus. We identified a similar CRE in the genome of Mengo virus and showed that the CRE could be functionally exchanged between Mengo virus and Theiler’s virus replicons. Interestingly, a similar CRE was detected in the VP1-coding region of human rhinovirus type 14 (HRV-14) and was recently characterized (15, 16). The occurrence of replication elements in HRV-14 and in the distantly related cardioviruses suggests the presence of similar elements in other picornaviruses. However, although it probably plays the same role, the CRE of HRV-14 could not functionally replace that of TMEV.
MATERIALS AND METHODS
Construction of Recombinant Plasmids.
The replicons of viruses DA, GDVII, Mengo virus, and HRV-14 were derived from the plasmids containing the corresponding full length cDNA clones: pTMDA1 (17, 18), pTMGDVII (19), pMC24 (also named pM16.1; kindly provided by Ann Palmenberg, University of Wisconsin, Madison, WI) (20), and pHRV14-as3f (kindly provided by Tim Skern, University of Vienna) (21). Site-directed mutagenesis was performed as described by Kunkel (22). Pfu polymerase (Stratagene) was used for amplification of fragments to be cloned. Fragments subjected to site-directed mutagenesis or amplified by PCR and used in subsequent constructs were entirely sequenced on one strand to check the absence of undesired mutations. For pTM612 and pTM614, the PCR fragments were not sequenced, but two clones from independent PCR amplification were tested.
Oligonucleotides.
The sequence of the primers used for PCR amplification, for site-directed mutagenesis, or as linkers is published as supplemental data on the PNAS web site, www.pnas.org.
Cell Culture.
BHK-21 cells were cultured in Glasgow’s modified Eagle medium (GIBCO/BRL) supplemented with 10% heat-inactivated newborn calf serum (GIBCO/BRL), 100 units/ml penicillin, 100 μg/ml streptomycin, and 130 g/liter tryptose phosphate broth (GIBCO/BRL). HeLa cells were cultured in DMEM (GIBCO/BRL) supplemented with 5% heat-inactivated fetal bovine serum (GIBCO/BRL), 100 units/ml penicillin, and 100 μg/ml streptomycin (GIBCO/BRL).
Transfection of Cells, Dot Blot Hybridization, and Measure of Luciferase Activity.
Replication of viral RNA was measured by dot blot hybridization, after transfection of the viral genome in BHK-21 cells, for TMEV as described (18) or after transfection in HeLa cells, for Mengo virus, by using a similar method. Luciferase activity from BHK-21 transfected cells was measured by using the luciferase reporter gene assay (Boehringer Mannheim).
In Vitro Translation Experiments.
RNAs were transcribed in vitro from the various cDNA clones with T7 RNA polymerase (Boehringer Mannheim) and were used to program rabbit reticulocyte lysates (Promega) as recommended by the manufacturer. Coupled transcription translation reactions (TNT, Promega) were used to check the absence of frame shifts in constructs subjected to site-directed mutagenesis.
Measure of Replication by Incorporation of 3H-Uridine.
Cells cultured in 6-well plates were infected at a multiplicity of infection of 1 plaque-forming unit per cell. Forty-five min after infection, cells were washed and treated with 5 μg/ml of actinomycin D. One hour later, 3H-uridine was added to the culture medium at a final concentration of 20 μCi/ml (6 μCi per well). At various times postinfection, cells were washed three times and were lysed in a buffer containing 100 mM sodium chloride, 1 mM EDTA, 1% SDS, and 10 mM Tris hydrochloride (pH7.5). Nucleic acids were precipitated with 10% of trichloroacetic acid, and the radioactivity was counted. The replication rate was calculated by the ratio between 3H-uridine incorporation at 4 and 8 hours postinfection.
RESULTS
The Capsid-Encoding Region of Theiler’s Virus Is Required for Replication of the Genome.
Previous work from our laboratory showed that in-frame deletions could be introduced in the 5′ half of Theiler’s virus genome, in regions encoding the L, VP4, VP2, VP3, VP1, and 2A proteins without affecting replication of viral RNA (18).
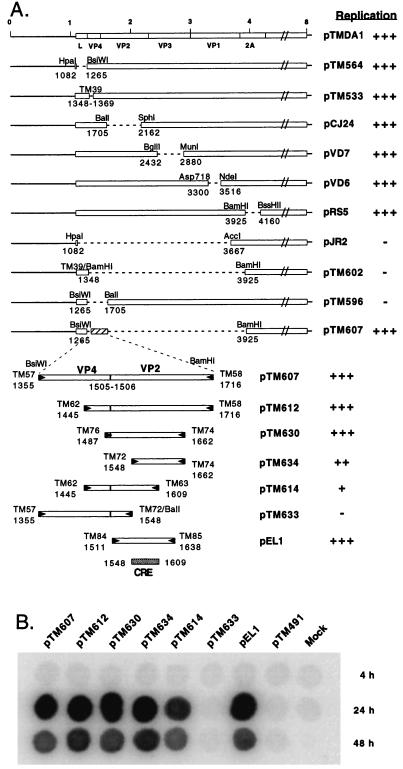
However, replicons pJR2 and pTM602, which contain large in-frame deletions extending from L to VP1 and from VP4 to 2A respectively, failed to replicate at a detectable level in transfected BHK-21 cells (Fig. 1). We analyzed a set of additional in-frame deletion mutants to map the region required for replication. Data presented in Fig. 1 show that the presence of a 362-nt sequence of the capsid-encoding region (coordinates 1,355–1,716 of the DA1 genome) was sufficient to restore the replication ability of a replicon containing an L to 2A deletion. To further map the minimal sequence required for replication, various subfragments of the 362-bp replication region were amplified by PCR and were cloned to replace, in pTMDA1, a large BsiWI-BamHI restriction fragment extending from the L to the 2A region. Such a minireplicon (pEL1) containing a 128-nt fragment of the VP2-coding region (nucleotides 1,511–1,638) replicated to almost wild-type levels. The essential replication element probably lies in a 62-nt sequence (nucleotides 1,548–1,609) because deletions of directly adjacent sequences (pTM614 and pTM634) decreased but did not abolish replication (Fig. 1).
Figure 1.
Localization of the replication element. (A) The various replicons derived from pTMDA1 are represented. Plasmids pTM564, pTM533, pCJ24, pVD7, pVD6, and pRS5 were described earlier (18). pJR2 was constructed by deleting a restriction fragment comprised between a HpaI site introduced by site-directed mutagenesis at nucleotide 1,082 and an AccI site at nucleotide 3,667. pTM602 was derived from pTM533 by the deletion of a BamHI restriction fragment and filling-in the restriction sites to restore the reading frame. pTM596 results from a BsiWI-BalI deletion of pTMDA1. pTM607 was obtained by replacing a BsiWI-BamHI restriction fragment of pTMDA1, extending from region L to region 2A, by a 362-bp PCR fragment (enlarged) containing the VP4-VP2 boundary. BamHI or BsiWI restriction sites were incorporated into the primers to facilitate the cloning of the PCR fragment between the corresponding sites of pTMDA1. The other constructs were obtained by cloning various PCR-amplified subfragments of this region between the same sites. pTM633 was derived from pTM607 by the deletion of fragment starting at a BalI site introduced at nucleotide 1,548 by site-directed mutagenesis with oligonucleotide TM72, and the BalI site contained in the genome at nucleotide 1,708. pTM634 has a deletion from the artificial BalI site introduced at nucleotide 1,548 and the BsiWI site filled-in with Klenow enzyme. Dotted lines indicate the deletions. RNAs transcribed in vitro from the various constructs were transfected into BHK-21 cells. Replication of the genomes was followed by dot blot hybridization. (+++, ++, +, −) indicate the replication levels from wild type (+++) to absence of detection (−). The dotted box identifies the 62-nt region deduced to contain the minimal replication element (CRE). (B) Dot blot hybridization of RNA extracted 4 hours, 24 hours, and 48 hours after transfection. RNA replication levels peaked ≈24 hours after transfection. The subsequent decrease in RNA levels probably reflects cell death caused by the accumulation of viral products. pTM491 was used as a negative control. It is a replication-deficient construct carrying an out-of-frame deletion extending from the 2C to the 3C region.
The Replication Element Is cis-Acting and Unidirectional.
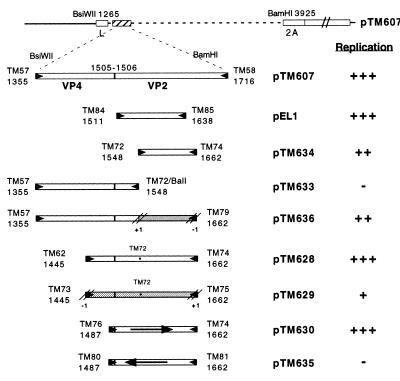
To discriminate between an effect caused by the RNA sequence itself and an activity of the VP2 peptide encoded by the corresponding region, we analyzed the replication abilities of two constructs (pTM629 and pTM636) in which the minimal CRE was inserted in an alternative ORF so that the peptide encoded by the region would have no homology to the VP2 peptide. The frame was restored at the end of the CRE region to ensure proper translation of the downstream P2 and P3 sequences, which are necessary for replication (Fig. 2).
Figure 2.
The CRE is a strand-specific RNA signal. The various plasmids were constructed by PCR as described in Fig. 1. The CRE sequence was cloned in two alternative reading frames (pTM629 and pTM636) (shaded boxes) to differentiate the role of the RNA sequence from that of the corresponding VP2 peptide. In pTM635, the CRE was inverted (reverse arrow). An asterisk indicates a point mutation introduced in some clones, by site-directed mutagenesis with oligonucleotide TM72, to eliminate a stop codon in an alternative reading frame and to introduce a BalI site. The replication ability of the different replicons in BHK-21 cells is indicated as in Fig. 1.
The RNA derived from pTM636, in which the minimal CRE is out of frame, replicated to nearly wild-type levels, suggesting that the RNA sequence rather than the peptide encoded by the CRE region was the element involved in replication. The other replicon (pTM629), in which the CRE was cloned out of frame, reproducibly replicated more efficiently than the replication negative controls, albeit at a very low level as compared with the positive control, pTM628. The fact that replication was more efficient for pTM629 than for the negative control confirms the role played by the RNA sequence. The reason for decreased replication of pTM629 as compared with that of pTM628 is not known. It could be attributable to the presence of low usage codons within the alternative frame or to an unpredicted alteration of the RNA secondary structure. In plasmid pTM635, the CRE corresponding to that of pTM630 was inverted without introducing any stop codon. Viral RNA transcribed from this construct failed to replicate, suggesting that the CRE is strand-specific.
The Replication Element Is not Involved in Translation Initiation.
A likely explanation of our results is that the RNA sequence in VP2 enhances either plus or minus strand synthesis during replication of the genome. On the other hand, it is also possible that this element is a downstream extension of the internal ribosome entry site that directs translation initiation of the viral polyprotein. To discriminate between these possibilities, we compared, in reticulocyte lysates, the efficiencies of internal ribosome entry site-driven translation of the genomes of replication-proficient and of replication-deficient minireplicons. pTM633, a replication-deficient construct, was translated with the same efficiency as the replication proficient constructs, such as pEL1 or pTM630, suggesting that the CRE does not influence translation efficiency but, rather, participates directly in the replication process (data not shown). None of the deletions in the L-P1–2A precursor affected polyprotein processing at the 2A-2B boundary, ruling out an indirect effect of polyprotein processing on replication.
Influence of the CRE on the Replication of Theiler’s Virus Isolates.
Theiler’s virus strains are divided into two subgroups according to the disease they produce in the mouse. Neurovirulent strains, such as strain GDVII, cause an acute lethal encephalitis. On the other hand, persistent strains like DA or BeAn induce a mild encephalitis followed by a chronic demyelinating disease. In vitro, in BHK-21 cells, virus GDVII was found to replicate to higher titers and to produce larger plaques than virus DA. Interestingly, replacing the capsid-encoding region of the neurovirulent strain GDVII by that of virus DA yielded a nonlethal virus with a persistent phenotype like virus DA. Conversely, grafting the capsid of a neurovirulent strain on a persistent virus greatly increased the neurovirulence of the virus (23–25). The plaque size of the viruses also was exchanged by capsid swapping (data not shown), confirming the major role played by the capsid in the virus phenotype. This has been interpreted as a possible difference in receptor recognition by capsids of neurovirulent and persistent strains. In the light of our data, we hypothesized that the CRE found in the VP2-coding region also could account for a difference in replication level, which could influence neurovirulence or persistence as well as plaque size. Thus, we compared the replication abilities of the previously constructed R2 and R3 recombinant viruses resulting from capsid swapping (25) (Table 1). First, we checked whether a CRE similar to that found in the DA strain also existed in virus GDVII, as was suggested by extensive nucleotide sequence conservation (≈90% identity). Therefore, we examined the replication capabilities of GDVII mutants in which either the 128- or 362-bp fragments encompassing the CRE, or a neighboring fragment, replaced a large L to 2A in-frame deletion. As expected, only the constructs containing the putative CRE replicated in transfected BHK-21 cells, showing that the function of the CRE is conserved among Theiler’s virus isolates (data not shown). In addition, the CRE of GDVII and DA replicons were found to be functionally exchangeable (data not shown).
Table 1.
Phenotype of chimeric viruses constructed between strains DA and GDVII
| Virus | Plaque size | Mortality | Persistence | Replic* |
|---|---|---|---|---|
| GDVII | Large | 100% | − | 274% |
| DA | Small | 0% | + | 100% |
| R2 (DA capsid) | Small | 0% | + | 186% |
| R3 (GDVII capsid) | Large | 90% | − | 126% |
Relative rates of replication: Data from a representative experiment are shown. The replication rate of virus DA was taken as 100%.
We then examined the replication efficiencies of viruses DA, GDVII, R2, and R3 in BHK-21 cells by following 3H-uridine incorporation in the viral genomes. Virus GDVII reproducibly replicated more efficiently than virus DA. To our surprise, but in agreement with data reported by Adami et al. (26), we found that replication of virus R3 (capsid of GDVII in a DA background) was closer to that of virus DA than to that of virus GDVII, although virus R3 was neurovirulent and produced large plaques in BHK-21 cells (Table 1). On the other hand, replication of virus R2 was consistently higher than that of virus R3 (albeit lower than that of virus GDVII) in spite of the fact that virus R2 is persistent and produces small plaques. Although they do not rule out an influence of the CRE, these data suggest a stronger impact of the capsid-receptor interaction on the phenotype of the viruses.
The CRE Is Conserved Among Cardioviruses.
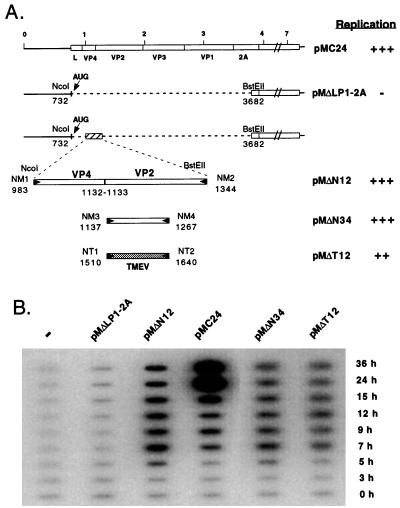
Sequence alignments reveal a high degree of nucleotide sequence identity in the VP2-coding region of the different cardioviruses. To check whether the CRE was functionally conserved among cardioviruses, a deletion was introduced in-frame between regions L and 2A in the genome of Mengo virus contained in plasmid pMC24. The construct was named pMΔLP1–2A. As expected, the subgenomic RNA transcribed in vitro from this clone failed to replicate in HeLa cells (Fig. 3). Again, cloning a 362-bp (pMΔN12) or a 131-bp (pMΔN34) sequence of Mengo virus corresponding to the CRE of TMEV restored replication of the Mengo virus replicon. As was observed for TMEV, replication generally appeared to be slightly decreased in some experiments involving a 131-bp CRE fragment as compared with the larger fragment. Interestingly, cloning the 131-bp CRE of TMEV DA strain in pMΔLP1–2A also restored replication, albeit at a slightly lower level than the corresponding Mengo virus sequence, showing that the CRE of Mengo and Theiler’s virus are functionally exchangeable.
Figure 3.
The CRE is functionally conserved in Mengo virus. (A) The various replicons derived from pMC24 are represented. A restriction fragment of pMC24 extending from a NcoI site (nucleotide 732) present 10 nucleotides downstream of the initiation codon of the polyprotein to a BstEII site present in the 2A region (nucleotide 3,682) was replaced by different PCR-amplified fragments containing the putative CRE of Mengo virus. NcoI or BstEII restriction sites were incorporated into the primers to facilitate cloning. The constructs were first obtained in plasmid pM5A1, which contains a ApaI-PstI (nucleotides 266–5,032) subgenomic fragment of the Mengo virus cDNA and were subsequently transferred into the pMC24 full length cDNA clone by cloning back a PflmI-BglII fragment (nucleotides 601–3,817 in the Mengo virus cDNA). Plasmid pMΔN12 contains a 362-bp PCR fragment spanning nucleotides 983–1,344 of the VP4/VP2 region of Mengo virus. pMΔN34 was obtained by cloning a PCR fragment containing nucleotides 1,137–1,267 of Mengo virus whereas pMΔT12 contains the homologous region (nucleotides 1,511–1,641) amplified from strain DA of TMEV. pMΔLP12A was obtained from pMΔN12 by deleting a XhoI-BstEII restriction fragment by using an XhoI-BstEII linker made with primers XBE1 and XBE2. Replication levels are indicated as in Fig. 1. (B) RNA was transcribed in vitro from the various Mengo virus derived replicons and was transfected into HeLa cells. Replication of the genome was followed by slot blot hybridization of total RNA extracted 0, 3, 5, 7, 9, 12, 15, 24, and 36 hours after transfection.
RNA Secondary Structure.
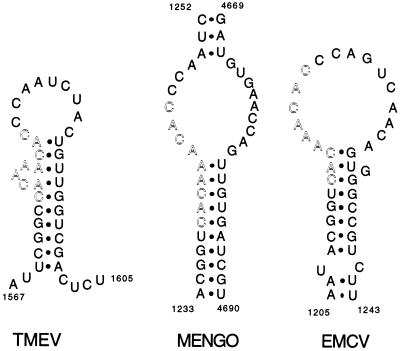
Predictions of the RNA secondary structures made for TMEV, EMCV, and Mengo virus failed to reveal any clearly conserved structure for the CRE. However, in the predictions made on the positive strand, either locally or on the entire genome (27), a 9-nt sequence, perfectly conserved in all three viruses, was detected in a consistently unpaired structure (a loop or a bulge) (Fig. 4). For all three viruses, the structure around the 9-nt conserved sequence was characterized by a low P-num value (27), which reflects a low propensity, for a nucleotide, to associate with alternative partners under suboptimal energy conditions (29).
Figure 4.
Predicted secondary structures around the 9-bp conserved sequence of the CRE region of Theiler’s virus, Mengo virus, and EMCV. The secondary structures shown are the structures predicted by Palmenberg and Sgro (27), by folding the full length viral RNA molecule. For Theiler’s virus (TMEV), the sequence of strain BeAn was replaced by the sequence of strain DA1, used in this study. Local structure predictions done with the Fold algorithm (28) predict an identical stem and loop structure for TMEV strains DA1, BeAn, and GDVII. Outlined residues are the nine nucleotides that are highly conserved among cardioviruses.
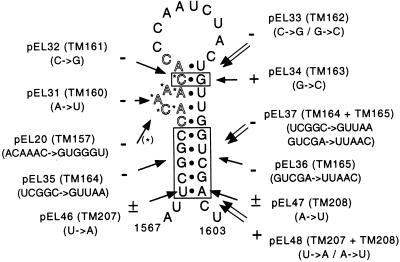
We used site-directed mutagenesis to assess the importance of the 9-nt conserved sequence and of the secondary structure of the RNA in that region. As can be seen in Fig. 5, all of the mutations affecting the 9-nt conserved sequence (pEL20, pEL31, pEL32) totally abrogated replication. In particular, the single A to U replacement at nucleotide 1,577 in pEL31 completely blocked replication, although it did not modify the amino acid sequence or the predicted secondary structure of the RNA. This clearly outlines the importance of the nucleotide sequence of this conserved region. On the other hand, point mutations destabilizing the 5′ (pEL46) or the 3′ (pEL47) part of the stem decreased replication. The combined mutations (pEL48) predicted to restore the secondary structure restored wild-type replication, indicating that the secondary structure of the CRE RNA is important for replication. When five nucleotides were mutated in the 5′ or 3′ part of the stem (pEL35, pEL36), replication was completely blocked. In this case, the combined mutation (pEL37) predicted to restore the secondary structure failed to restore replication, probably because the mutations in the 5′ part of the stem are directly adjacent to the 9-nt conserved sequence. The C to G point mutation of nucleotide 1,580 (pEL32) also inhibited replication. In contrast, a G to C mutation in the complementary sequence had only a weak influence on replication whereas the combined mutation (pEL33) failed to restore replication. This suggests that nucleotide 1,580, which lies in the 9-nt conserved sequence, must be conserved. This also might indicate that nucleotides 1,580 and 1,593, as well as the bulge containing the conserved 9-nt sequence, are part of a larger loop resembling that predicted for EMCV.
Figure 5.
Mutations introduced in the CRE of Theiler’s virus. The stem and loop structure of virus DA1 is presented as predicted by Palmenberg and Sgro (27). The conserved 9-nt sequence is outlined. Mutations were introduced by site-directed mutagenesis with the indicated oligonucleotides in a cloned subfragment containing the CRE. The mutated fragment was subsequently reintroduced into pEL8, a replicon containing a 374-nt fragment (amplified with primers TM57-TM86) encompassing the CRE. pEL8 is identical to pTM607 (See Fig. 1) except that it contains 12 additional viral nucleotides at the 3′ end of the CRE fragment. + and − indicate the replication abilities measured by dot blot after transfection of the replicon RNA in BHK-21 cells.
In conclusion, the mutations affecting either the RNA sequence of the 9-nt conserved motive or the secondary structure of the region inhibited the replication of viral RNA. The fact that the CRE of Mengo virus and TMEV are readily exchangeable indicates that the actual secondary structure of the Mengo virus CRE probably resembles that of TMEV or EMCV rather than involving long distance interactions (see Fig. 4).
CRE Exchange Between Theiler’s Virus and HRV-14.
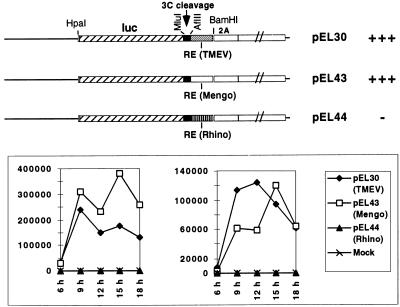
The CRE of cardioviruses likely fulfills the same role as the recently characterized cis-acting replication element of HRV-14 (15, 16), although these elements share no sequence homology and map at different places of the coding region. To test whether the CRE of HRV-14 could functionally replace that of Theiler’s virus, we constructed a TMEV replicon (pEL30) expressing the firefly luciferase as well as identical replicons in which the CRE of TMEV was replaced by that of Mengo virus (pEL43) or by that of HRV-14 (pEL44) (Fig. 6). On transfection of BHK-21 cells, luciferase activity produced by pEL30 and pEL43 was comparable, confirming that the CRE are exchangeable among cardioviruses. In contrast, the replicon containing the replication element of HRV-14 only produced background levels (1,000-fold less) of luciferase, indicative of an extremely low replication level, if any. Thus, in spite of the apparent conservation of their functions, the CRE of HRV-14 and cardioviruses were not exchangeable.
Figure 6.
CRE exchange between Theiler’s virus and HRV-14. pEL30 is a replicon derived from pTMDA1. The luciferase gene was introduced, in frame, 6 codons downstream of the beginning of the viral polyprotein, at the level of a HpaI site created at nucleotide 1,082. At its 3′ end, the stop codon of the luc gene was eliminated, and the sequence was fused to a 60-bp sequence containing a cleavage site for the viral 3C protease (from the L-VP4 boundary). The latter sequence was amplified by PCR with primers TM197 and TM198 and was cloned as a MluI-SnaBI fragment. In this construct, the CRE of TMEV is a 374-bp fragment corresponding to that of pEL8. It is linked, at its 5′ end, to an AflII site bording the 3C cleavage site and, at its 3′ end, to the BamHI site located in the 2A region. pEL43 was obtained by replacing the AflII-BamHI fragment containing the CRE of TMEV by a 368-bp PCR fragment containing the Mengo virus CRE (nucleotides 988–1,355 of pMC24, amplified with primers TM199-TM200). Similarly, the CRE of HRV-14 was amplified from plasmid pHRV14-as3f as a 369-bp fragment (nucleotides 2,175–2,543 of HRV-14) with primers TM201 and TM202 and was cloned between the AflII and BamHI sites to yield pEL44. RNA transcribed in vitro from these replicons was transfected in BHK-21 cells, and the luciferase activity (arbitrary units) was followed as a measure of replication. The data from two representative experiments are shown.
DISCUSSION
We detected, in the VP2-encoding region of cardioviruses (two Theiler’s virus isolates and Mengo virus), a 130-nt sequence involved in genome replication. The sequence did not influence translation efficiency and thus likely acts as a replication signal. This region was sufficient to impart almost wild-type replication levels to replicons carrying a large deletion extending from the leader (L) region to region 2A. The actual replication signal is probably contained in a smaller, 62-nt fragment. In this region, we identified a 9-nt conserved sequence that is part of a putative loop (EMCV) or bulge (Mengo virus or TMEV) and that is characterized by a low P-num value, a property proposed to be characteristic of biologically active sequences (27). Point mutations affecting the sequence of the 9-nt motive or the secondary structure of the region inhibited replication.
How this sequence acts is still unknown. It might interact with some other parts of the RNA molecule, to form tertiary structures that guide the replication complex, as was proposed for an internal replication signal found in the genome of bacteriophage Qβ (30). Alternatively, it could act as the binding site for a viral or a cellular factor involved in the initiation or elongation steps of replication.
In the CRE of HRV-14, a stem and loop structure present on the positive strand was proposed to act as a signal for the initiation of negative strand synthesis. No obvious sequence or structure similarity could be found between the replication element of HRV-14 and that of cardioviruses, nor could the replication element of HRV-14 functionally replace that of Theiler’s virus. Nevertheless, the replication elements of cardioviruses and rhinovirus likely play the same role. The fact that they are found in distantly related picornaviruses suggests that similar elements might occur in all of the picornaviruses. It is surprising, then, to find that a conserved function might have evolved from different coding regions of the picornavirus genome. In poliovirus and coxsackievirus, artificially created minireplicons lacking the entire capsid-encoding region were found to replicate efficiently, ruling out the involvement of the capsid region in the replication process (31, 32). If these viruses contain equivalent replication signals, they should be located elsewhere in the P2 or P3 coding regions.
The genome of picornaviruses is a trifunctional RNA molecule: On entry into the target cell cytoplasm, it must be translated to yield the viral proteins; subsequently, it serves as a template for negative strand RNA synthesis; finally, it interacts with capsid components to form progeny viral particles. The CRE of picornaviruses has at least a dual role: It acts as an RNA signal for replication and as a coding sequence. It is tempting to speculate that the CRE might be a component ensuring the balance between translation and replication of the genomic RNA, although other mechanisms could account for this regulation (33).
An internal replication signal also was reported to occur in the 1a coding region of the mouse hepatitis virus, a coronavirus (34, 35). Interestingly, as in the case of picornaviruses, the genomic RNA molecule of coronaviruses also serves as a messenger RNA (encoding region 1 proteins). This suggests that the presence of cis-acting replication signals in coding sequences may be a common feature of positive strand RNA viruses and supports the idea that such signals could participate in the balance between translation and replication of the genomic RNA.
Supplementary Material
Acknowledgments
We thank Sylvie van der Werf for helpful discussions and Pierre Rensonnet for technical assistance. We are grateful to Ann C. Palmenberg and Tim Skern, who kindly provided plasmids containing the cDNA of Mengo virus and rhinovirus respectively. T.M. is Senior Research Associate with the Belgian National Fund for Scientific Research (FNRS). We are indebted to family de Spoelberg, who supported this work through a “Haas-Teichen” postdoctoral fellowship to P.-E.L. This work was supported by convention 3.4573.94F of the Fonds de la Recherche Scientifique Médicale, crédit aux chercheurs FNRS 1.5.185.96F, and Fonds de Développement Scientifique (FDS/FSR) of the University of Louvain.
ABBREVIATIONS
- CRE
cis-acting replication element
- TMEV
Theiler’s murine encephalomyelitis virus
- EMCV
Encephalomyocarditis virus
- HRV-14
human rhinovirus type 14
References
- 1.Rueckert R R. In: Virology. Fields B N, Knipe D M, editors. New York: Raven; 1996. pp. 609–654. [Google Scholar]
- 2.Xiang W, Paul A V, Wimmer E. Semin Virol. 1997;8:256–273. [Google Scholar]
- 3.Jang S K, Kräusslich H-G, Nicklin M J H, Duke G M, Palmenberg A C, Wimmer E. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelletier J, Sonenberg N. Nature (London) 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 5.Borman A M, Deliat F G, Kean K M. EMBO J. 1994;13:3149–3157. doi: 10.1002/j.1460-2075.1994.tb06613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nomoto A, Kitamura N, Golini F, Wimmer E. Proc Natl Acad Sci USA. 1977;74:5345–5349. doi: 10.1073/pnas.74.12.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Novak J E, Kirkegaard K. J Virol. 1991;65:3384–3387. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theiler M, Gard S. J Exp Med. 1940;72:49–67. doi: 10.1084/jem.72.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lipton H L. Infect Immun. 1975;11:1147–1155. doi: 10.1128/iai.11.5.1147-1155.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez M, Oleszak E, Leibowitz J. CRC Crit Rev Immunol. 1987;7:325–365. [PubMed] [Google Scholar]
- 11.Monteyne P, Bureau J-F, Brahic M. Immunol Rev. 1997;159:163–176. doi: 10.1111/j.1600-065x.1997.tb01014.x. [DOI] [PubMed] [Google Scholar]
- 12.Ohara Y, Stein S, Fu J, Stillman L, Klaman L, Roos R P. Virology. 1988;164:245–255. doi: 10.1016/0042-6822(88)90642-3. [DOI] [PubMed] [Google Scholar]
- 13.Pevear D C, Borkowski J, Calenoff M, Oh C K, Ostrowski B, Lipton H L. Virology. 1988;165:1–12. doi: 10.1016/0042-6822(88)90652-6. [DOI] [PubMed] [Google Scholar]
- 14.Michiels T, Jarousse N, Brahic M. Virology. 1995;214:550–558. doi: 10.1006/viro.1995.0066. [DOI] [PubMed] [Google Scholar]
- 15.McKnight K L, Lemon S M. J Virol. 1996;70:1941–1952. doi: 10.1128/jvi.70.3.1941-1952.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKnight K L, Lemon S M. RNA. 1998;4:1569–1584. doi: 10.1017/s1355838298981006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McAllister A, Tangy F, Aubert C, Brahic M. Microb Pathog. 1989;7:381–388. doi: 10.1016/0882-4010(89)90041-7. [DOI] [PubMed] [Google Scholar]
- 18.Michiels T, Dejong V, Rodrigus R, Shaw-Jackson C. J Virol. 1997;71:9549–9556. doi: 10.1128/jvi.71.12.9549-9556.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tangy F, McAllister A, Brahic M. J Virol. 1989;63:1101–1106. doi: 10.1128/jvi.63.3.1101-1106.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duke G M, Palmenberg A C. J Virol. 1989;63:1822–1826. doi: 10.1128/jvi.63.4.1822-1826.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skern T, Torgersen H, Auer H, Kuechler E, Blaas D. Virology. 1991;183:757–763. doi: 10.1016/0042-6822(91)91006-3. [DOI] [PubMed] [Google Scholar]
- 22.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calenoff M A, Faaberg K S, Lipton H L. Proc Natl Acad Sci USA. 1990;87:978–982. doi: 10.1073/pnas.87.3.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fu J, Stein S, Rosenstein L, Bodwell T, Routbort M, Semler B L, Roos R P. Proc Natl Acad Sci USA. 1990;87:4125–4129. doi: 10.1073/pnas.87.11.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McAllister A, Tangy F, Aubert C, Brahic M. J Virol. 1990;64:4252–4257. doi: 10.1128/jvi.64.9.4252-4257.1990. , and correction (1993) 67, 2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adami C, Pritchard A E, Knauf T, Luo M, Lipton H L. J Virol. 1998;72:1662–1665. doi: 10.1128/jvi.72.2.1662-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Palmenberg A C, Sgro J-Y. Semin Virol. 1997;8:231–241. [Google Scholar]
- 28.Zuker M. Methods Mol Biol. 1994;25:267–294. doi: 10.1385/0-89603-276-0:267. [DOI] [PubMed] [Google Scholar]
- 29.Jaeger J A, Turner D H, Zuker M. Proc Natl Acad Sci USA. 1989;86:7706–7710. doi: 10.1073/pnas.86.20.7706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuppli D, Barrera I, Weber H. J Mol Biol. 1994;243:811–815. doi: 10.1006/jmbi.1994.1682. [DOI] [PubMed] [Google Scholar]
- 31.Percy N, Barclay W S, Sullivan M, Almond J W. J Virol. 1992;66:5040–5046. doi: 10.1128/jvi.66.8.5040-5046.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barclay W, Li Q, Hutchinson G, Moon D, Richardson A, Percy N, Almond J W, Evans D J. J Gen Virol. 1998;79:1725–1734. doi: 10.1099/0022-1317-79-7-1725. [DOI] [PubMed] [Google Scholar]
- 33.Gamarnik A V, Andino R. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y-N, Makino S. J Virol. 1995;69:4963–4971. doi: 10.1128/jvi.69.8.4963-4971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Repass J F, Makino S. J Virol. 1998;72:7926–7933. doi: 10.1128/jvi.72.10.7926-7933.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.