Abstract
The interaction between two regulatory proteins plays a crucial role in the control of several biological events, including gene transcription. In this report, we demonstrate that the interaction between the cellular sequence-specific single-stranded DNA binding protein Purα and the HIV type 1 (HIV-1) Tat protein is mediated by specific ribonucleic acids. The region of Tat that is important for its interaction with Purα includes the region demonstrated to bind Tat’s viral RNA target, TAR. A 10-nucleotide GC-rich consensus sequence identified in RNAs associated with Purα derived from human U-87MG cells plays an important role in the Purα:Tat interaction as examined by an in vitro reconstitution assay. Furthermore, expression of the Purα-associated RNA in these cells enhances transcriptional activation of the HIV-1 promoter by Tat and Purα.
Purα is a single-stranded DNA binding protein that has specific affinity for the purine-rich sequence (GGN)n (1, 2). The Purα binding sites are present in viral and eukaryotic origins of DNA replication as well as in many promoter sequences. As such, this protein has been implicated in control of both viral (3–5) and cellular gene transcription (6–9) as well as viral DNA replication (10, 11). In addition, Purα has been reported to interact with several cellular and viral regulatory proteins (4, 10, 12–14, 30). In earlier studies, it was shown that Purα interacts with a purine-rich sequence, named upTAR, located in the regulatory region of the human neurotropic JC virus (JCV) (10, 15). JCV is the etiologic agent of progressive multifocal leukoencephalopathy, a fatal demyelinating disease of the central nervous system that usually affects immunosuppressed patients (for review, see ref. 16). The high incidence of progressive multifocal leukoencephalopathy among individuals with AIDS in comparison to other immunocompromised patients implies that the presence of HIV type 1 (HIV-1) in the brains of infected individuals may directly contribute to the pathogenesis of this disease. In support of this model, earlier in vitro studies have indicated direct intercommunication between HIV-1 and JCV through the HIV-1-encoded regulatory protein Tat (17, 19). Evidently, the upTAR region of JCV is important for HIV-1 Tat to stimulate transcription of the JCVL promoter (19, 20). Tat is a transcription transactivator that, on interaction with a cis-acting RNA sequence, termed TAR, located in the leader of HIV-1 RNA, stimulates transcription of the viral genome (for review, see ref. 21). To exert its activity, Tat also interacts with several cellular-specific proteins, some with the ability to interact with nucleic acids and/or modify other transcription regulators (22). The capacity of Tat to stimulate the JCVL promoter without being directly associated with upTAR led to the assumption that the association of Tat and Purα is important for activating the JCVL promoter. In support of this notion, results from protein–protein interaction studies revealed that a Purα and Tat complex can be detected in lysates of glial cell expressing the Tat gene and that purified Tat and Purα associate specifically and with high affinity (4). In subsequent studies, it was shown that overexpression of Purα results in a modest activation of HIV-1 long terminal repeat (LTR) transcription and that the loop structure within the TAR element is important for this event (5). All of these observations suggest that Purα may function as a cellular partner for Tat and, by associating with Tat, facilitates its transcriptional activity.
In this report, we provide evidence that the molecular interaction between Purα and Tat is RNA-dependent. The participant RNA that selectively associates with Purα is capable of reconstituting this Purα:Tat interaction in vitro, and its overexpression enhances the transcriptional activation of the HIV-1 promoter by Tat and Purα.
MATERIALS AND METHODS
Plasmids and Cell Culture.
The plasmid pEBV-His B-Purα contains the coding region of the Purα gene cloned downstream of a histidine epitope tag. pEBV-His B-Purα was constructed by first subcloning the Purα EcoRI fragment from pGEX1λT-Purα (13) into EcoRI-digested pCDNA3 (Invitrogen), generating pCDNA3-Purα. Subsequently, the BamHI/XhoI fragment from pCDNA3-Purα was cloned into BamHI/XhoI cleaved pEBV-His B (Invitrogen), generating pEBV-His B-Purα. HIV-1 glutathione S-transferase (GST)–Tat expression vectors were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program (Bethesda, MD). pCMV-Tat and pCMV-14.4 were created by placing the Tat cDNA and the 338-base cDNA representing the Purα-associated RNA, respectively, at the 3′ end of the CMV promoter in the pCDNA3 vector. The 338-base cDNA was also cloned in the antisense orientation in pCONA3. All plasmids were verified by terminal dideoxy DNA sequencing with Sequenase (United States Biochemical).
U-87MG cells were obtained from the American Type Culture Collection (no. HTB-14) and were maintained at 37°C in DMEM supplemented with 10% fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified incubator containing 5% CO2.
DNA Transfections and Preparation of Cell Extracts.
Transient transfections were carried out by the calcium phosphate technique as described (23). In brief, 1.5 × 106 cells were plated on 10-cm dishes and were grown overnight. Three hours before transfection, cells were fed with fresh growth media, and transfections were carried out with 30 μg of DNA. The precipitate was removed from the cells 3 h later, and cells were subsequently subjected to a glycerol shock. Forty-eight hours after transfection, cells were lysed for 20 min on ice in LB 150 (50 mM Tris, pH 7.4/150 mM NaCl/5 mM EDTA/0.1% Nonidet P-40) buffer containing 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 mM PMSF, and 50 mM NaF. Cell debris was pelleted by centrifugation at 14,000 rpm for 15 min at 4°C. The supernatant was assayed for protein content by Bradford analysis (Bio-Rad) and either was used immediately or was stored at −80°C. For luciferase assay, experiments were performed with 5 × 106 cells in 60-mm dishes. Cells were harvested 36 h posttransfection, and protein extracts were used to examine the level of luciferase activity (24).
Overexpression and Purification of Recombinant Proteins.
HIV-1 GST-Tat and GST-Purα fusion proteins were expressed and purified as described (25). In brief, bacteria harboring the plasmids pGST, pGST-Tat, pGST-Tat 72, pGST-Tat 48, pGST-Tat Δ2/36, or pGST-Purα were grown overnight at 37°C in LB supplemented with 100 mg/liter ampicillin. The following morning, cells were diluted 1:10 with fresh medium, were grown to an optical density at 600 nm of 0.6–0.8, and were induced for 90 min at 37°C with 0.35 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were pelleted at 6,500 × g at 4°C, were resuspended in NETN buffer (100 mM NaCl/1 mM EDTA/20 mM Tris, pH 8.0/0.5% Nonidet P-40) containing 1 μg/ml leupeptin, 1 μg/ml aprotinin, and 1 mM PMSF, and were sonicated on ice. The bacterial lysate was centrifuged at 30,000 × g at 4°C to remove insoluble material. Glutathione-Sepharose beads (Amersham Pharmacia) were added to the lysate, and binding of GST fusion proteins was allowed to occur at 4°C for 3 h. Beads were pelleted and washed three times with 100 volumes of NETN buffer, and the integrity and purity of the GST fusion proteins were analyzed by SDS/PAGE followed by Coomassie blue staining. Known amounts of BSA were included on the same gel for determination of the yield of the full length proteins. Radiolabeled Purα and Tat proteins were synthesized with the TNT-coupled wheat germ extract system according to the manufacturer’s recommendations (Promega).
In Vitro Protein–Protein Interactions (GST Pull-Down Assay).
For in vitro binding assays, either 3 μl of [35S]labeled in vitro translated Purα or Tat or 200 μg of cellular extract containing a histidine-tagged Purα was incubated with 5 μg of GST or fusion proteins GST-Tat 86, GST-Tat 72, GST-Tat 48, GST-Tat Δ2/36, or GST-Purα coupled to glutathione Sepharose beads in 300 μl of LB 150 for 1 h at 4°C with continuous rocking. After the incubation, the beads were pelleted and washed five times with LB 150 buffer. The bound proteins were eluted with Laemmli sample buffer, were heated to 95°C for 10 min, and were separated by SDS/PAGE. Purα and Tat were detected by either fluorography or immunoblot analysis with a T7 monoclonal antibody recognizing the histidine tag (Novagen). For experiments shown in Fig. 1 D–F, extracts containing histidine tagged Purα, [35S]labeled Purα, or [35S]labeled Tat were pretreated with 10 μg/ml of soluble DNase-free RNase (Boehringer Mannheim) or 50 units of RNase-free DNase (Boehringer Mannheim) for 30 min at room temperature before the pull-down assay as described above. In experiments in which it was necessary to selectively remove RNase from the binding reaction (Fig. 3), in vitro translated [35S]labeled Purα was treated with 10 units of insoluble RNase (RNase conjugated to agarose beads) (Sigma) per microliter of protein for 2 h at room temperature. After treatment with insoluble RNase, the RNA-free [35S]labeled Purα containing extract was separated from the RNase agarose bead conjugate first by microcentrifugation, followed by filtration through a 0.2-μm cellulose acetate filter Spin-X column (CoStar). Conversely, GST-Tat was treated with soluble RNase for 2 h at room temperature followed by extensive washing with LB 150 to remove any residual RNase. In experiments that used RNase-treated samples, all reactions were supplemented with 40 units of RNase inhibitor (Boehringer Mannheim) and 0.5 mM DTT. For reconstitution experiments, 9 μg of total mammalian RNA or tRNA (Fig. 3B), 5 μg of either a synthetic RNA oligonucleotide derived from the GC consensus sequence or a mutant variant with the putative Pur elements mutated (Fig. 3C), or 9 μg of in vitro transcribed RNA in the sense or antisense orientation (Fig. 3D) were included in the binding reactions.
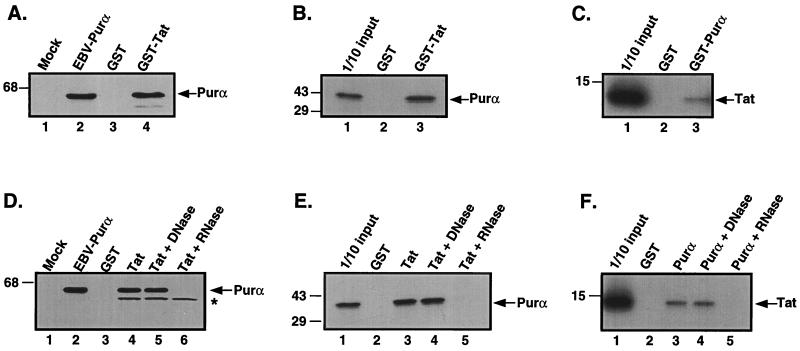
Figure 1.
HIV Tat and Purα associate with each other via RNA. (A) Extract containing a histidine-tagged Purα was incubated with either GST (lane 3) or GST-Tat fusion proteins (lane 4) attached to glutathione Sepharose resin. (B) In vitro translated [35S]labeled Purα was incubated with either GST (lane 2) or GST-Tat fusion proteins (lane 3). (C) In vitro translated [35S]labeled Tat protein was incubated with either GST (lane 2) or GST-Purα fusion proteins (lane 3). For A, B, and C, incubation was performed for 1 h at 4°C, and the resin was subsequently pelleted and washed with binding buffer. Bound proteins were eluted by boiling in Laemmli sample buffer and, after separation by SDS/PAGE, were detected by either immunoblot analysis with T7 antibody (A) or autoradiography (B and C). An amount equivalent to 10% of the material used for the assays was applied to the input lane in B and C. The position of the molecular mass markers are indicated on the left of the gels. The arrow indicates the position of histidine tagged Purα (A), [35S]labeled in vitro translated Purα (B), and [35S]labeled in vitro translated Tat (C). (D) Extract containing a histidine-tagged Purα was incubated with GST (lane 3), GST-Tat (lane 4), or GST-Tat in the presence of DNase (lane 5) and RNase (lane 6). (E) In vitro translated [35S]labeled Purα was incubated with GST (lane 2), GST-Tat (lane 3), or GST-Tat in the presence of DNase (lane 4) and RNase (lane 5). (F) In vitro translated [35S]labeled Tat was incubated with GST (lane 2), GST-Purα (lane 3), or GST-Purα in the presence of DNase (lane 4) and RNase (lane 5). The position of molecular mass markers are indicated on the left of each gel. Arrows indicate the position of histidine-tagged Purα (D), in vitro translated Purα (E), and in vitro translated Tat (F). The asterisk indicates a nonspecific cross reacting band.
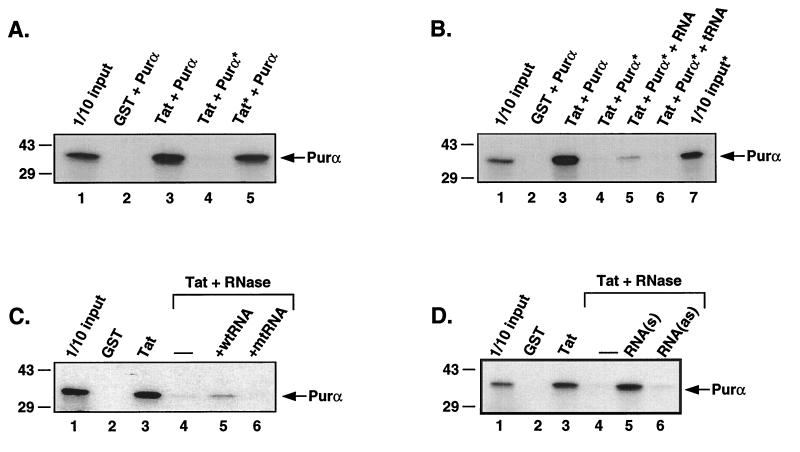
Figure 3.
Reconstitution of the interaction between Purα and Tat protein by the addition of RNA. (A) In vitro translated Purα (lane 4) or GST-Tat (lane 5) were selectively pretreated with RNase before incubation with each other. The asterisks indicate treatment with RNase. (B) In vitro translated Purα pretreated with agarose-RNase beads was incubated with GST-Tat in the presence of total cellular RNA (lane 5) or tRNA (lane 6). Lane 7 contains 1/10 of the amount of [35S]labeled Purα used in the binding reactions shown in lanes 4–6 (pretreated with RNase). The asterisks indicate treatment with RNase. The arrows indicate the position of the in vitro translated Purα protein. (C) In vitro translated Purα after RNase treatment (as indicated) was mixed with GST-Tat in the presence of the synthetic RNA representing the consensus GC-rich sequence (CCCGGCCGGU) (lane 5) or its mutant variant (AUGACUUGUC) (lane 6). (D) Binding reactions were carried out with in vitro translated Purα after RNase treatment (as indicated) and GST-Tat, in the presence of the in vitro transcribed 338-base fragment in the sense (lane 5) and antisense (lane 6) orientation.
Immunoblot Analysis.
For immunoblot analysis, protein samples were resolved by SDS/PAGE and were transferred to supported nitrocellulose membranes (Bio-Rad) in transfer buffer containing 25 mM Tris (pH 7.4), 193 mM glycine, and 20% methanol. Membranes were blocked for 1 h at room temperature with 10% nonfat dry milk in PBS-T (1× PBS/0.1% Tween-20). After three brief washes in PBS-T, membranes were incubated with a 1:3,000 dilution of T7 monoclonal antibody (Novagen) for 1 h at room temperature in 0.5% nonfat dry milk in PBS-T. The blots were subsequently washed three times for 15 min in PBS-T. Membranes were incubated with a 1:10,000 dilution of a goat-anti-mouse secondary antibody conjugated to horseradish peroxidase for 1 h at room temperature in PBS-T. After five washes, proteins were visualized with the enhanced chemiluminescence detection system according to the manufacturer’s instructions (Amersham Pharmacia).
RNA Isolation.
Total RNA was isolated from U-87MG cells by using an acid phenol extraction (26). Cells were washed twice with 1× PBS and were lysed with a solution containing 50 mM sodium acetate (pH 5.1) and 1% SDS. Cell lysates were scraped into tubes containing an equal volume of prewarmed acid phenol [equilibrated with 50 mM sodium acetate (pH 5.1)] and were incubated at 60°C for 15 min. The aqueous phase was extracted with neutral phenol/chloroform [equilibrated with 50 mM Tris (pH 7.4)], followed by chloroform extraction and ethanol precipitation. Integrity of the RNA was verified by electrophoresis in a 1.2%, 0.4% formaldehyde, 1× morpholinepropanesulfonic acid (Mops) gel. For reverse transcription–PCR, the RNAs from immunoprecipitated Purα were prepared and used in reverse transcriptase assays by using random hexamers according to the procedure detailed previously (27). The amplified cDNAs were placed in the TA cloning vector, and their nucleotide compositions were determined by direct DNA sequencing.
In Vitro Transcription and RNA Binding Assays.
TAR RNA was transcribed in vitro by using T7 RNA polymerase. Transcription reactions (20 μl) contained 500 ng of linearized DNA templates, 40 mM Tris⋅HCl (pH 7.5), 6 mM MgCl2, 10 mM DTT, 4 mM spermidine, 20 units of RNase inhibitor, 0.5 mM of ATP, GTP, and CTP, 50 μCi of [α32P]UTP (400 Ci/mmol), and 20 units of T7 RNA polymerase (Boehringer Mannheim). Reactions were incubated for 1 h at 37°C followed by the addition of 20 units of RNase-free DNase I and subsequently were incubated for 15 min at 37°C. Radioactively labeled TAR RNA was further gel purified on a 9% denaturing polyacrylamide gel. The 338-base RNA was transcribed in the sense and antisense orientations from Xba1 linearize pCDNA vectors containing the cDNA of the 338-base RNA in the sense and antisense orientations, respectively.
For RNA binding assays, 100,000 cpm of TAR RNA was incubated with GST or GST-Tat, GST-Tat 72, GST-Tat 48, or GST-Tat Δ2/36 in 300 μl of LB 150 for 1 h at 4°C with continuous rocking. After incubation, beads were pelleted, were washed five times with LB 150 buffer, and were phenol/chloroform-extracted. Bound RNA was precipitated and separated on a 12% denaturing polyacrylamide gel.
RESULTS AND DISCUSSION
To examine the association of Tat and Purα, human astrocytic U-87MG cells were transiently transfected with a control plasmid (pEBV-B) or a histidine-tagged Purα expression plasmid (EBV-HisB-Purα). Extract derived from cells transfected with the histidine-tagged Purα plasmid was incubated with GST or GST-Tat fusion protein coupled to glutathione Sepharose beads. As shown in Fig. 1A, the tagged Purα was bound by the GST Tat fusion protein (lane 4) but not by the GST protein alone (lane 3). In another series of experiments, in vitro translated [35S]labeled Purα was incubated with GST or GST-Tat coupled to glutathione Sepharose beads. As shown in Fig. 1B, [35S]labeled Purα was retained by the GST-Tat fusion protein (lane 3) but not by GST alone (lane 2). Reciprocally, in vitro translated [35S]labeled Tat protein was incubated with GST or GST-Purα coupled to glutathione Sepharose beads. As shown in Fig. 1C, [35S]labeled Tat was retained by the GST-Purα fusion protein (lane 3) but not by the GST protein alone (lane 2). These observations are consistent with previous studies (4) and demonstrate an association between the HIV-1 Tat protein and the cellular protein Purα.
Because Purα has been demonstrated to bind both DNA (1, 2, 4, 10, 13, 15) and RNA (5, 27, 28), we next examined the dependence of the interaction between Purα and Tat on nucleic acids. To address this question, we performed our binding assays in the presence of RNase and DNase. As shown in Fig. 1D, DNase had no effect on the interaction between GST-Tat with either transiently transfected histidine-tagged Purα (Fig. 1D, compare lanes 4 and 5) or in vitro translated Purα (Fig. 1E, compare lanes 3 and 4). Likewise, DNase had no effect on the interaction between GST-Purα and in vitro translated Tat protein (Fig. 1F, lanes 3 and 4). Incubation with RNase, however, abrogated the interaction between Tat and Purα (Fig. 1D, compare lanes 4 and 6; Fig. 1 E and F, compare lanes 3 and 5). This effect may not be attributed to a degradation of Purα and Tat during the incubation because RNase treatment had no effect on the stability of either protein (data not shown). Taken together, these results demonstrate that the interaction between the HIV-1 Tat protein and Purα is mediated by nucleic acids, more specifically RNA.
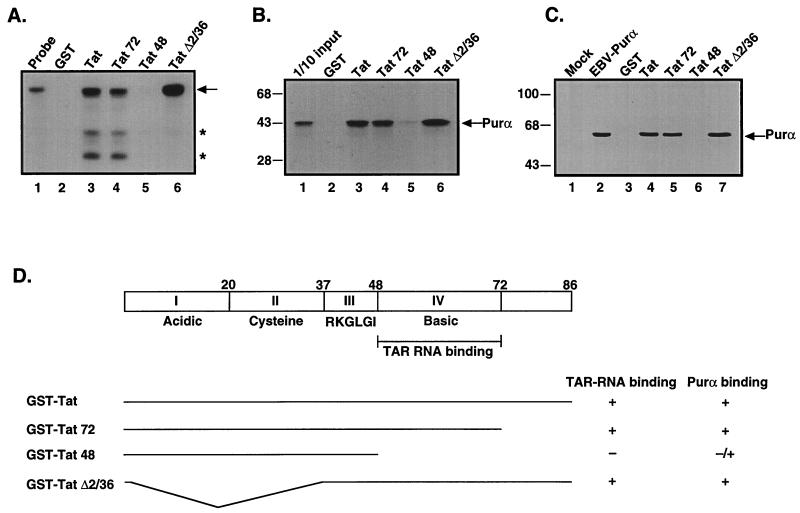
To further investigate the interaction between these two proteins, the region of Tat that is important for its interaction with Purα was determined by using a series of Tat mutants (Fig. 2D). Of note, one of these mutants, Tat 48, does not contain the TAR RNA binding region. To confirm that our recombinant GST-Tat deletion mutants retained or lacked TAR RNA binding, we incubated these Tat mutants with radiolabeled TAR RNA. As shown in Fig. 2A, full-length GST Tat was able to bind the TAR RNA (lane 3) whereas GST did not (lane 2). Mutants Tat 72 and Tat Δ2/36 also retained the ability to bind TAR RNA whereas Tat 48 was unable to bind to TAR RNA. We next examined the ability of these Tat mutants to bind to Purα. As shown in Fig. 2B, in vitro translated [35S]labeled Purα binds to full-length Tat, Tat 72, and Tat Δ2/36 (lanes 3, 4, and 6, respectively) to a much greater extent than to Tat 48. It is noteworthy that Purα weakly binds to Tat 48 (Fig. 2B, lane 5). Similar results were obtained by using extract containing transfected histidine tagged Purα (Fig. 2C). These results, summarized in Fig. 2D, demonstrate that the RNA binding region of Tat, specifically amino acids 48 to 72, is also the region of Tat involved in the interaction with Purα.
Figure 2.
The Purα binding domain localizes to the HIV-1 Tat RNA binding domain. (A) Radiolabeled TAR RNA probe was incubated with GST, GST-Tat, or various GST-Tat mutants, (GST-Tat 72, GST-Tat 48, and GST-Tat Δ2/36) for 1 h at 4°C. Beads were washed extensively, and bound RNA was separated by denaturing gel electrophoresis. The arrow indicates full-length TAR RNA probe; the asterisks indicate degraded RNA species observed in GST-Tat and GST-Tat 72. (B and C) In vitro translated [35S]labeled Purα protein (B) or cellular extract containing a histidine tagged Purα from U-87MG cells transfected with pEBV-His B-Purα (C) was incubated with GST (B, lane 2; C, lane 3), GST-Tat (B, lane 3; C, lane 4), or GST-Tat mutants (B, lanes 4–6; C, lanes 5–7). After incubation for 1 h at 4°C, the resins were pelleted and washed with binding buffer. Bound proteins were eluted by boiling in Laemmli sample buffer and, after separation by SDS/PAGE, were detected by either autoradiography (B) or immunoblot analysis with T7 antibody (C). (D) Schematic of HIV-1 Tat and the various GST-Tat deletion mutants. The binding of TAR RNA and Purα to these mutants is summarized on the right.
To determine which of the two proteins contained RNA species that were important for the interaction, each protein was selectively pre-treated with RNase. To this end, in vitro translated Purα was treated with RNase conjugated to agarose beads. After centrifugation to remove the agarose-RNase conjugate, the RNase-treated, in vitro translated Purα was filtered to eliminate any potential carry-over. Conversely, GST and GST-Tat Sepharose beads were treated with soluble RNase. After treatment, the Sepharose beads were washed extensively to remove the soluble RNase. As shown in Fig. 3A, pretreatment of the in vitro translated Purα protein abrogated the interaction between these two proteins (compare lanes 3 and 4) whereas pretreatment of the GST-Tat protein did not affect the interaction (compare lanes 3 and 5). These results suggest that the RNA important in the interaction between Purα and Tat is a Purα bound RNA molecule(s).
If the interaction between Purα and Tat is RNA-dependent, RNA should be able to reconstitute the interaction between these two proteins. In an attempt to reconstitute the interaction between these two proteins, RNase-treated, in vitro translated Purα was incubated with GST-Tat in the absence and presence of RNA. As shown in Fig. 3B, the addition of total cellular mammalian RNA reconstitutes the interaction between these two proteins (lane 5) whereas the addition of tRNA did not influence the interaction at all (lane 6). These results further demonstrate that the interaction between Purα and Tat proteins is mediated by RNA.
To identify Purα-associated RNA(s), immunopurified Purα from U-87MG cell nuclear extract was used to isolate RNA molecules and synthesize complementary DNAs by the reverse transcription–PCR technique. Sequence analysis of four randomly selected complementary DNAs revealed the presence of 250–500 nucleotide fragments, which all share a common decanucleotide motif (CCCGGCc/gc/gGG) and several GC/GT dinucleotides. To examine the importance of the decanucleotide GC-rich consensus in the Purα:Tat interaction, a synthetic 10-base ribonucleotide RNA fragment and its mutant variants with no Purα binding site were added to the binding reaction containing RNase-treated [35S]labeled Purα and GST-Tat. As shown in Fig. 3C, the addition of wild-type, but not mutant, RNA partially restored the Purα:Tat association (compare lane 3 with lanes 5 and 6). A similar experiment was carried out with a 338-base RNA that was associated with Purα in U-87MG cells. In addition to the consensus decanucleotide, the 338-base RNA encompasses multiple GGT/GGG trinucleotides. Results illustrated in Fig. 3D indicate that, in the sense orientation, the in vitro synthesized 338-base RNA effectively restored the Tat:Purα association in the binding reaction containing RNase-treated [35S]labeled Purα and GST-Tat (compare lane 3 with lane 5). Under similar conditions, the 338-base RNA in the antisense orientation showed no significant effect on the assembly of Purα:Tat (Fig. 3D, lane 6). Perhaps it should be noted that our recent Northern blot studies using the 338-base pair DNA probe have revealed detection of a highly abundant 2.0-kilobase RNA species in the human astrocytic cell line, U-87MG, suggesting that the 338-base RNA is indeed expressed in eukaryotic cells (data not shown). These results indicate that cellular RNA associated with Purα is able to reconstitute the interaction between Purα and Tat.
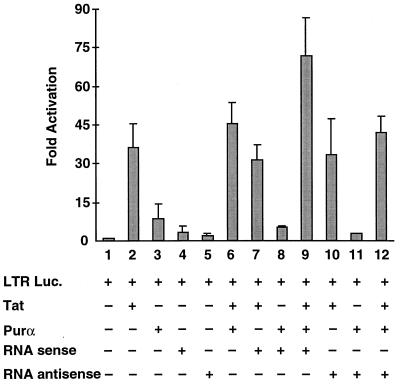
To determine the functional significance of the Purα-associated cellular RNA with respect to the function of Purα and Tat, U-87MG cells were transfected with an HIV-1 LTR-reporter plasmid in the absence and presence of expression plasmids for Tat, Purα, and the 338-base Purα-associated RNA. As shown in Fig. 4, coexpression of Purα and Tat in these astrocytic cells increased the transcriptional activation of the LTR to a level that was slightly higher than that seen with either Tat and Purα alone (compare lane 6 with lanes 2 and 3). Of interest, the 338-base RNA alone in either the sense or antisense orientation showed little effect on the LTR (Fig. 4, compare lane 1 with lanes 4 and 5, respectively) and did not increase the level of Tat-mediated transcriptional activity (compare lane 2 with lanes 7 and 10) or Purα-induced transcriptional activity (compare lane 3 with lanes 8 and 11). Expression of the 338-base RNA noticeably elevated the level of transactivation of the HIV-1 promoter by Tat plus Purα (Fig. 4, compare lane 6 with lane 9). Of interest, this enhancement was not observed when the 338-base RNA was produced in the antisense orientation (Fig. 4, compare lane 6 with lane 12). These results suggest that an RNA molecule that is capable of reconstituting the interaction between Purα and Tat increases the activity of these two proteins when coexpressed in cells. This is specific because overexpression of the RNA in the antisense orientation did not result in an increase in transcriptional activity.
Figure 4.
Functional importance of Purα-associated RNA in Tat and Purα effects on LTR activity. U-87MG cells were transfected with 0.5 μg of LTR-luciferase reporter construct in the absence or presence of 10 μg of expression plasmids for Tat, Purα, and the 338-base RNA in the sense or antisense orientation, as indicated at the bottom of the graph. Luciferase activity was determined 36 h after transfection, and the levels of promoter activation by Tat, Purα, and the 338-base RNA, alone or in combination, were determined and graphed as fold activation. In all cases, the final amount of DNA in the transfection mixture was brought to 35 μg with pCMV DNA. The results shown represent the mean of three independent experiments. Bars indicate standard deviation.
In this report, we further dissect the molecular interaction between Purα and Tat and demonstrate that this interaction depends on nucleic acids: more specifically, RNA (Fig. 1). Our results also suggest that the RNA that is important for this interaction is selectively bound to Purα. Moreover, total cellular RNA is able to reconstitute the association between these two proteins after pretreatment of Purα with RNase (Fig. 3). Analysis of RNA species isolated by virtue of their ability to be immunoprecipitated with Purα from mammalian cellular extract reveals molecules with GC-rich regions. These regions that contain putative Purα binding sites are capable of restoring the interaction between these two proteins. This RNA-facilitated reconstitution is specific because inclusion of a mutant oligonucleotide cannot restore the interaction. Moreover, the association between these two proteins also can be restored with a full-length 338-base molecule. This is also specific because inclusion of an antisense RNA cannot restore the interaction. This RNA molecule also has consequences with respect to the function of these two proteins. Coexpression of the RNA in the sense orientation results in increased transcriptional activity of Purα and Tat on the HIV-1 LTR. Thus, a cellular RNA molecule(s) capable of reconstituting the association of Purα and Tat enhances the transcriptional activation of the HIV-1 promoter by Tat and Purα.
Our results suggest that the RNA component important for the interaction between Purα and Tat associates directly with Purα. This is noteworthy in light of a previous study demonstrating that Purα associates with a cellular RNA with significant homology to 7SL (27). Results from those studies demonstrated that PU-RNA inhibits the interaction between Purα and a region of the myelin basic protein gene promoter, designated MB1. Although expressed during various stages of mouse brain development, PU-RNA is associated with Purα only at the early stages of brain development, before the time of maximum myelination. This is also the time when the association of Purα with MB1 is also low. Taken together, these results suggest that PU-RNA may function as a cofactor negatively regulating the interaction between Purα and the MBP promoter sequence. These results combined with the current study suggest that RNA molecules may not only affect the ability of Purα to form a complex with other proteins but also may influence the ability of Purα to interact with its target DNA.
Other studies have reported that RNA molecules can stimulate the interaction between a single-stranded DNA binding Pur factor with a GC-rich motif (28). Although both proteins target GC-rich sequences, the molecular mass of the protein described in that study (27 kDa) is less than that of Purα.
Our results suggest that RNA that is important for the Purα:Tat interaction is selectively bound to Purα. Recent studies (29) indicate that RNA can induce structure in a disordered or partially disordered protein. Perhaps RNA associates with Purα stabilizing its structure, allowing it to interact with its protein partners. Such models suggest that RNA can act as a regulatory molecule in the control of gene expression by affecting protein structure. For example, two proteins that interact at a very precise time in development or at various stages of the cell cycle could associate with one another because of an RNA-facilitated interaction. Thus, the physiologically regulated RNA species, by facilitating communication of the regulatory proteins, may have a major impact on a variety of biological events. The identification and functional characterization of the RNA species associated with Purα in cells at various stages may establish the role of RNA protein interactions in controlling cellular gene expression and differentiation.
Acknowledgments
We thank past and present members of the Center for NeuroVirology and NeuroOncology for their support and sharing of reagents. G.L.G. and S.A.A. are M.D./Ph.D. students in the Department of Biochemistry and Molecular Biology at Thomas Jefferson University. We also thank Cynthia Schriver for editorial assistance and preparation of this manuscript. This work was made possible by grants awarded by the National Institutes of Health to K.K.
ABBREVIATIONS
- JCV
JC virus
- GST
glutathione S-transferase
- LTR
long terminal repeat
References
- 1.Bergemann A D, Johnson E M. Mol Cell Biol. 1992;12:1257–1265. doi: 10.1128/mcb.12.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergemann A D, Ma Z W, Johnson E M. Mol Cell Biol. 1992;12:5673–5682. doi: 10.1128/mcb.12.12.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen N N, Khalili K. J Virol. 1995;69:5843–5848. doi: 10.1128/jvi.69.9.5843-5848.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Krachmarov C, Chepenik L G, Barr-Vagell S, Khalili K, Johnson E M. Proc Natl Acad Sci USA. 1996;93:14112–14117. doi: 10.1073/pnas.93.24.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chepenik L G, Tretiakova A P, Krachmarov C P, Johnson E M, Khalili K. Gene. 1997;210:37–44. doi: 10.1016/s0378-1119(98)00033-x. [DOI] [PubMed] [Google Scholar]
- 6.Du Q, Tomkinson A E, Gardner P D. J Biol Chem. 1997;272:14990–14995. doi: 10.1074/jbc.272.23.14990. [DOI] [PubMed] [Google Scholar]
- 7.Haas S, Thatikunta P, Steplewski A, Johnson E M, Khalili K, Amini S. J Cell Biol. 1995;130:1171–1179. doi: 10.1083/jcb.130.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelm R J, Jr, Elder P K, Strauch A R, Getz M J. J Biol Chem. 1997;272:26727–26733. doi: 10.1074/jbc.272.42.26727. [DOI] [PubMed] [Google Scholar]
- 9.Zambrano N, De Renzis S, Minopoli G, Faraonio R, Donini V, Scaloni A, Cimino F, Russo T. Biochem J. 1997;328:293–300. doi: 10.1042/bj3280293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang C-F, Gallia G L, Muralidharan V, Chen N N, Zoltick P, Johnson E M, Khalili K. J Virol. 1996;70:4150–4156. doi: 10.1128/jvi.70.6.4150-4156.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurk M, Weissinger F, Lottspeich F, Schwarz U, Winnacker E L. Nucleic Acids Res. 1996;24:2799–2806. doi: 10.1093/nar/24.14.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallia G L, Safak M, Khalili K. J Biol Chem. 1998;273:32662–32669. doi: 10.1074/jbc.273.49.32662. [DOI] [PubMed] [Google Scholar]
- 13.Johnson E M, Chen P L, Krachmarov C P, Barr S M, Kanovsky M, Ma Z W, Lee W H. J Biol Chem. 1995;270:24352–24360. doi: 10.1074/jbc.270.41.24352. [DOI] [PubMed] [Google Scholar]
- 14.Darbinian, N., Gallia, G. L., Kundu, M., Shcherbik, N., Tretiakova, A. & Khalili, K. (1999) Oncogene, in press. [DOI] [PubMed]
- 15.Chen N N, Chang C-F, Gallia G L, Kerr D A, Johnson E, Krachmarov C, Barr S M, Frisque R J, Bollag B, Khalili K. Proc Natl Acad Sci USA. 1995;92:1087–1091. doi: 10.1073/pnas.92.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger J R, Concha M. J Neurovirol. 1995;1:5–18. doi: 10.3109/13550289509111006. [DOI] [PubMed] [Google Scholar]
- 17.Chowdhury M, Taylor J P, Tada H, Rappaport J, Amini S, Wong-Staal F, Khalili K. Oncogene. 1990;5:1737–1742. [PubMed] [Google Scholar]
- 18.Tada H, Rappaport J, Amini S, Lashgari M, Wong-Stahl F, Khalili K. Proc Natl Acad Sci USA. 1990;87:3479–3483. doi: 10.1073/pnas.87.9.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chowdhury M, Kundu M, Khalili K. Oncogene. 1993;8:887–892. [PubMed] [Google Scholar]
- 20.Chowdhury M, Taylor J P, Chang C-F, Rappaport J, Khalili K. J Virol. 1992;66:7355–7361. doi: 10.1128/jvi.66.12.7355-7361.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones K A. Genes Dev. 1997;11:2593–2599. doi: 10.1101/gad.11.20.2593. [DOI] [PubMed] [Google Scholar]
- 22.Cullen B R. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 23.Graham F L, van der Eb A. Virology. 1973;52:456–457. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 24.Ausubel F, Brent R, Kingston R E, Moar D D, Siedman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1989. [Google Scholar]
- 25.Smith D B, Johnson K S. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 26.Queen C, Baltimore D. Cell. 1983;33:741–748. doi: 10.1016/0092-8674(83)90016-8. [DOI] [PubMed] [Google Scholar]
- 27.Tretiakova A, Gallia G L, Shcherbik N, Jameson B A, Johnson E M, Khalili K. J Biol Chem. 1998;273:22241–22247. doi: 10.1074/jbc.273.35.22241. [DOI] [PubMed] [Google Scholar]
- 28.Herault Y, Chatelain G, Brun G, Michel D. Gene Exp. 1995;4:85–93. [PMC free article] [PubMed] [Google Scholar]
- 29.Frankel A D, Smith C A. Cell. 1998;92:149–151. doi: 10.1016/s0092-8674(00)80908-3. [DOI] [PubMed] [Google Scholar]
- 30.Tretiakova A, Steplewski A, Johnson E M, Khalili K, Amini S. J Cell Physiol. 1999;181:160–168. doi: 10.1002/(SICI)1097-4652(199910)181:1<160::AID-JCP17>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]