Abstract
Mutants of Salmonella typhimurium lacking DNA adenine methylase are attenuated for virulence in BALB/c mice. LD50 values of a DNA adenine methylation (Dam)− mutant are at least 103- to 104-fold higher than those of the parental strain when administrated by oral or intraperitoneal routes. Dam− mutants are unable to proliferate in target organs but persist in low numbers in these locations. Efficient protection to challenge with the virulent parental strain is observed in mice infected with a Dam− mutant. Use of the ileal loop assay shows that Dam− mutants are less cytotoxic to M cells and fail to invade enterocytes. In the tissue culture model, lack of DNA adenine methylation causes reduced ability to invade nonphagocytic cells. In contrast, no effect is observed either in intracellular proliferation within nonphagocytic cells or in survival within macrophages. The invasion defect of Dam− mutants is correlated with a distinct pattern of secreted proteins, which is observed in both PhoP+ and PhoP− backgrounds. Altogether, our observations suggest a multifactorial role of Dam methylation in Salmonella virulence.
In the bacterial cell, DNA adenine methylation (Dam) modulates a variety of processes such as DNA replication, chromosome segregation, mismatch repair, and transcription of certain genes (1–3). Given these multiple roles, it is not surprising that dam mutations are highly pleiotropic; however, lack of Dam methylation does not impair viability (1, 3, 4). The existence of genes whose transcription is regulated by Dam methylation has been known for two decades in Escherichia coli and its phages (1–3), and novel cases have recently been described in Salmonella typhimurium (4–6). Genes regulated by Dam methylation usually contain GATC sites either in their promoter or in upstream regulatory sequences. The methylation state of these GATC sites controls the interaction between RNA polymerase or regulatory proteins and their cognate DNA-binding sites (1, 7). A paradigmatic case of transcriptional regulation by Dam methylation is the E. coli pap operon, which directs the synthesis of pili required for adhesion to uroepithelial cells. In uropathogenic E. coli, expression of pap is subjected to phase variation (8, 9). The ON and OFF stages are dictated by binding of the regulatory protein Lrp and accessory proteins to two GATC sites upstream of the transcription initiation site (8, 10). Recent work has shown that expression of other E. coli operons encoding virulence-related fimbriae, such as sfa, daa, and fae, are also regulated by Lrp binding (11, 12). As in the pap operon, Lrp mediates transcriptional control over these fimbrial operons depending on the methylation status of critical GATC sites (11, 12).
Dam− mutants of Salmonella typhimurium were recently described (4). The same study showed that Dam methylation regulates the expression of a locus in the S. typhimurium virulence plasmid (4). This finding and the fact that Dam methylation modulates the expression of virulence-related fimbriae in E. coli (10) led us to examine whether Dam methylation could play a role in S. typhimurium virulence. Tests in the BALB/c murine model were unambiguous: absence of DNA adenine methylation produces severe attenuation of S. typhimurium virulence. A partial defect in the capacity of S. typhimurium to invade nonphagocytic cells is also observed. Furthermore, Dam− mutants are unable to invade enterocytes and to cause cytotoxicity on M cells of ileal Peyer’s patches. Our conclusion that DNA adenine methylation plays a major role in S. typhimurium virulence is supported by the recent finding that Dam− mutants of Salmonella show altered expression of a number of in vivo-induced genes (6).
MATERIALS AND METHODS
Bacterial Strains, Plasmids, Growth Media, and Growth Conditions.
S. typhimurium SL1344 was used as the standard virulent strain (13). Strains SV1610 (SL1344 dam-228∷MudJ), SV4036 (mutL111∷Tn10), SV4056 (phoP7953∷Tn10) and SV4089 (dam-228∷MudJ phoP7953∷Tn10), all derived from SL1344, are described in this study. The allele mutL111∷Tn10 was obtained from G. C. Walker, Massachusetts Institute of Technology, Cambridge, MA. The allele phoP7953∷Tn10 was provided by E. A. Groisman, Washington University, St. Louis, MO. Plasmid pTP166, obtained from M. G. Marinus, University of Massachusetts, Worcester, MA, is a ColE1 derivative harboring the wild-type dam gene of E. coli under the control of a tac promoter (14). Noninvasive S. typhimurium strains SB220 (SL1344 sipC∷aph) and SB302 (SL1344 invJ∷aph) were provided by J. E. Galán, Yale University, New Haven, CT (15, 16). Strains were grown overnight in LB broth at 37°C without shaking before administration to BALB/c mice, infection of tissue culture cells, or preparation of extracts containing secreted proteins (see below). Solid LB contained 1.5% Difco agar. Green plates were prepared as described elsewhere (4).
Virulence and Vaccination Assays in BALB/c Mice.
Bacteria were centrifuged at 10,000 × g for 15 min and washed twice in sterile PBS. Serial dilutions were used to infect orally (25 μl) or intraperitoneally (200 μl) groups of 8-wk-old female BALB/c mice. In oral challenge experiments, the acidic pH of the stomach was buffered by suspending bacteria in a 2.5% bicarbonate–0.2% lactose solution before administration. Survival of infected mice was recorded for a minimum of 4 wk. LD50 was calculated by the method of Reed and Muench (17). In vaccination studies 4 wk after infection with the S. typhimurium Dam− mutant SV1610, mice were challenged orally with different doses of the parental virulent strain SL1344. Mouse survival was recorded for 4 additional weeks. In organ colonization studies, lymph nodes, liver, and spleen were removed from two mice at each postinfection time and homogenized in 10 ml of sterile PBS. Dilutions of these homogenates were plated on LB to count the numbers of viable bacteria per organ.
Ligated Loop Model and Electron Microscopy.
Interaction of wild-type and Dam− bacteria with murine intestinal epithelium was examined as described by Jones et al. (18). Briefly, 7-wk-old BALB/c mice were starved for 24 h and anesthetized before surgery by i.p. injection of 2.0 mg of Nembutal. On practicing a small incision, the small bowel was exposed, and a loop was formed by ligating the intestine with silk thread at the ileocaecal junction and at a site 5 cm proximal to the caecum. Two hundred microliters of bacteria (ca. 4 × 108 colony-forming units) were then injected through a 25-gauge needle. The bowel was returned to the abdomen and the incision stapled. Mice were kept alive for 3 h and then sacrificed. The bacteria-exposed intestinal loop was removed, and sections containing Peyer’s patches were fixed in a solution of 2.5% glutaraldehyde/0.8% paraformaldehyde in 0.1 M phosphate buffer. After 15 min, Peyer’s patches were displayed in fresh fixing solution for an additional hour. Postfixation was made with 0.1% osmium tetroxide for 1 h. After three washes with phosphate buffer, samples were dehydrated with ethanol and propylene oxide. Infiltration and embedding was achieved with Epon resin. Resin polymerization was allowed for 48 h at 60°C. Ultrathin sections were contrasted with uranyl acetate and lead and examined in a JEOL1200EX electron microscope.
Eukaryotic Cell Lines.
HeLa epithelial cells (ATCC CCL2) and J774.A1 mouse macrophages were used as prototypes of nonphagocytic and phagocytic cells, respectively. Mouse resident peritoneal macrophages were harvested from BALB/c mice as described by Leung and Finlay (19). Cells were grown in MEM containing 10% fetal bovine serum (HeLa epithelial cells) or in DMEM containing 5% fetal bovine serum and 1 mM glutamine (macrophages).
Bacterial Infection of Eukaryotic Cells.
Eukaryotic cells were seeded the day before the infection in 24-well plates and grown at 37°C, 5% CO2 to obtain 80% confluency. Bacteria were added to reach a multiplicity of infection of 10:1 bacteria/eukaryotic cell. Eukaryotic cells were infected for 15 min (HeLa epithelial cells), 20 min (resident peritoneal macrophages), or 10 min (J774.A1 macrophages) to obtain an infection rate of 70–80% of the cells present. Infected cells were washed three times with PBS and incubated in fresh tissue culture medium containing 100 μg/ml gentamicin. Two hours postinfection, the concentration of gentamicin was lowered to 10 μg/ml. Numbers of viable intracellular bacteria were obtained by lysing infected cells with 1% Triton X-100 and subsequent plating (20). Invasion rates of nonphagocytic cells were determined as the ratio of viable intracellular bacteria at a short postinfection time (2 h) vs. viable bacteria added to infect the eukaryotic cells.
Extracts of Secreted Bacterial Proteins.
Extracts of secreted proteins were prepared by the method of Kaniga et al. (15). Proteins were analyzed by SDS/PAGE in Tris/Tricine buffer by using 8% acrylamide gels (21).
RESULTS
S. typhimurium Dam− Mutants Are Attenuated for Virulence in the BALB/c Mouse Model.
To examine whether Dam methylation is required for virulence in the murine typhoid model, BALB/c mice were infected with various doses of a Dam− strain of S. typhimurium (SV1610) by oral and i.p. routes. In parallel, infections were performed with the virulent strain SL1344 (13). Table 1 shows that LD50 values obtained for the Dam− mutant were over 8.8 × 108 bacteria (oral) and 6.5 × 104 bacteria (i.p.). No orally challenged mice died or showed any symptom of disease during the course of the experiment (4 wk). The LD50 values obtained (Table 1) were at least 103- to 104-fold higher than those of the parental strain SL1344, in agreement with a recent study (6). To confirm that the attenuation of virulence observed in the Dam− mutant was exclusively caused by lack of Dam methylation, we constructed a SV1610 derivative carrying plasmid pTP166, which contains the wild-type dam gene from E. coli (14). The complemented strain was fully virulent (Table 1). Complementation failed when the strain carried high-molecular-weight multimers of pTP166 (data not shown). The latter phenomenon was interpreted as an artifact and was not further investigated. However, the possibility that abnormally high levels of Dam methylase might also cause virulence defects, as suggested by Heithoff et al. (6), can be considered. Complementation of virulence in the in vivo model shows that there is no polar effect of the MudJ element inserted in the dam gene on hypothetical virulence genes located downstream of dam. Altogether, these data show that S. typhimurium requires Dam methylation to cause disease in the murine model. Such a requirement is unrelated to the mismatch repair deficiency of Dam strains, because a MutL− mutant (SV4036) proved to be virulent (Table 1).
Table 1.
Virulence properties of a S. typhimurium Dam− mutant in BALB/c mice
| Bacterial strain | LD50 (oral)* | LD50 (i.p.) |
|---|---|---|
| SL1344 (wild type) | 6 × 104 | <10 |
| SV1610 (Dam−) | >8.8 × 108 | 6.5 × 104 |
| SV1610/pTP166 | 2.75 × 105 | <35 |
| SV4036 (MutL−) | 2.4 × 106 | <17 |
Oral inoculation was done by suspending bacteria in bicarbonate buffer before administration to mice.
Colonization of Target Organs of Infected Mice by the S. typhimurium Dam− Mutant.
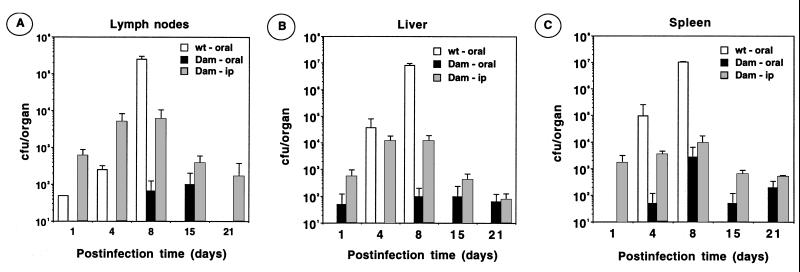
Because there was no sign of disease after oral and i.p. challenge with strain SV1610 (Dam−), we reasoned that Dam methylation could be required for colonization of target organs. To test this possibility, mice were infected orally and intraperitoneally. Lymph nodes, liver, and spleen were harvested periodically from day 1 to 21 after infection, and viable bacteria were counted by plating. Low numbers of Dam− bacteria, approximately 102–103, were found in these organs 3 wk after infection (Fig. 1). In contrast, the parental (virulent) strain SL1344 increased by a factor of 103–104 in number during the first week of infection (Fig. 1). On i.p. challenge, Dam− bacteria increased in number during the first week, dropping afterward to a steady value of 102–103. The complemented strain SV1610/pTP166 (dam-228∷ MudJ/dam+) was likewise able to grow within organs on oral administration (data not shown). Altogether, these data suggest that DNA adenine methylation is essential for S. typhimurium to proliferate within target organs on oral and i.p. administration.
Figure 1.
Colonization of mouse organs by a S. typhimurium Dam− mutant. Groups of 8-wk-old BALB/c mice were infected orally with 8.8 × 108 colony-forming units (cfu) of the strain SV1610 (Dam−) [black histograms] or 7.5 × 105 cfu of strain SL1344 (wild type) [white histograms]. Another group of mice were infected intraperitoneally with 1.5 × 105 cfu of SV1610 (Dam−) [grey histograms]. At the times indicated, lymph nodes (A), liver (B), and spleen (C) were removed from two infected mice, and viable bacteria were determined by plating. At 8 days after infection, all the wild-type infected mice showed clear evidence of disease.
A S. typhimurium Dam− Mutant Elicits Protection to Challenge with the Virulent Strain.
The fact that the Dam− mutant is able to persist, albeit in low numbers, in target organs for 3 wk (Fig. 1) led us to examine whether this attenuated strain could elicit a specific immune response. Mice were infected orally or intraperitoneally with different doses of strain SV1610 (Dam−) and challenged 4 wk after infection with high doses of the virulent strain SL1344. Table 2 shows that infection of BALB/c mice with the Dam− mutant rendered the animal capable of surviving to lethal doses of the virulent strain. Of a total of 42 mice infected with the Dam− mutant, 38 survived the challenge with an amount of virulent bacteria up to 100-fold the LD50 (Table 2). Interestingly, as few as 89 Dam− S. typhimurium cells administered by the i.p. route were capable of eliciting in the animal a specific immune response against the virulent strain (Table 2).
Table 2.
Protection conferred by a S. typhimurium Dam− mutant to challenge with the virulent strain
| Experiment | Dose of SV1610 (Dam−)* | Challenge dose of SL1344 (wt)† | No. dead mice/ inoculated mice |
|---|---|---|---|
| 1 | — | 2.8 × 103 | 1/5 |
| — | 2.8 × 104 | 1/5 | |
| — | 2.8 × 105 | 4/5 | |
| 2 | 2.5 × 106 | 1.2 × 106 | 1/4 |
| 2.5 × 107 | 1.2 × 106 | 1/4 | |
| 2.5 × 108 | 1.2 × 106 | 0/4 | |
| 3 | 7 × 106 | 9.7 × 106 | 0/5 |
| 7 × 107 | 9.7 × 106 | 1/5 | |
| 7 × 108 | 9.7 × 106 | 0/5 | |
| 89 (i.p.) | 9.7 × 106 | 0/5 | |
| 890 (i.p.) | 9.7 × 106 | 0/5 | |
| 8.900 (i.p.) | 9.7 × 106 | 1/5 |
Inoculations were done by the oral route, except when indicated i.p. After 4 wk, all Dam− infected mice survived the doses of bacteria indicated.
All inoculations were done by the oral route. In experiments 2 and 3, challenges with the wild-type strain were done 4 wk after the inoculation with the Dam− mutant. Mouse survival was recorded for 4 additional weeks.
Behavior of the S. typhimurium Dam− Mutant in the Tissue Culture Model.
Dam− mutants of S. typhimurium do not exhibit growth defects in standard media (4). In addition, the Dam− strain used in this study, SV1610, shows a normal serum-resistance response (F.G-P., unpublished work). Therefore, we reasoned that attenuation of the Dam− mutant might reflect an impaired interaction of bacteria with host cells. To test this hypothesis, we analyzed whether the Dam− mutant showed defects in invasion, survival, or growth within nonphagocytic and phagocytic cells. Table 3 shows that the Dam− mutant is partially impaired for invading nonphagocytic cells (30% of wild-type values in HeLa cells). The complemented strain showed wild-type invasion ability (Table 3). Well known invasion mutants such as SipC− and InvJ− yielded values less than 1% (Table 3). The Dam− mutant did not show any proliferation defect within HeLa cells (Table 3). To test whether Dam methylation was required for bacterial survival within macrophages, both the mouse macrophage cell line J774A.1 and mouse resident peritoneal macrophages were used. As previously reported, strain SL1344 was able to proliferate within J774A.1 and to survive within peritoneal macrophages (ref. 22; see also Table 3). Neither the Dam− nor the Dam−/Dam+ (complemented) strain showed any noticeable defect in the interaction with macrophages (Table 3). These data suggest that DNA adenine methylation is not essential for bacteria to proliferate within nonphagocytic cells or to survive within macrophages. In contrast, Dam methylation seems to be required to trigger an efficient bacterial uptake by nonphagocytic cells.
Table 3.
Phenotype of a Dam− mutant in the tissue culture model
| Strain | Invasion, %† | Intracellular proliferation‡ | Survival in macrophages*
|
|
|---|---|---|---|---|
| J774.A1 | Mouse peritoneal | |||
| SL1344 (wt) | 100 ± 23.8 | 25.3 ± 5.05 | 6.85 ± 0.3 | 0.41 ± 0.13 |
| SV1610 (Dam−) | 28.4 ± 9.2 | 16.1 ± 3.4 | 7.7 ± 1.65 | 0.22 ± 0.12 |
| SV1610/pTP166 | 105.4 ± 33.9 | 28.9 ± 0.3 | 10 ± 0.53 | 0.33 ± 0.05 |
| (Dam−/Dam+) | ||||
| SB220 (SipC−) | 0.43 ± 0.05 | ND | ND | ND |
| SB302 (InvJ−) | 0.65 ± 0.23 | ND | ND | ND |
Ratio of viable intracellular bacteria at 20 h vs. 0.5 h after infection. In both types of macrophages, cells were infected for 10 min.
All values are averages of triplicate samples from at least two independent trials. ND, not determined.
Percentage of bacteria that invade HeLa epithelial cells during 15 min and survived a gentamicin protection assay. Plating was done at 2 h after infection. Values were normalized to invasion rate of wild-type bacteria (0.109 ± 0.02).
Ratio of viable intracellular bacteria at 24 h vs. 2 h after infection, obtained after incubating HeLa cells with bacteria for 15 min.
Phenotype of a Dam− Mutant in the Murine Intestinal Epithelium.
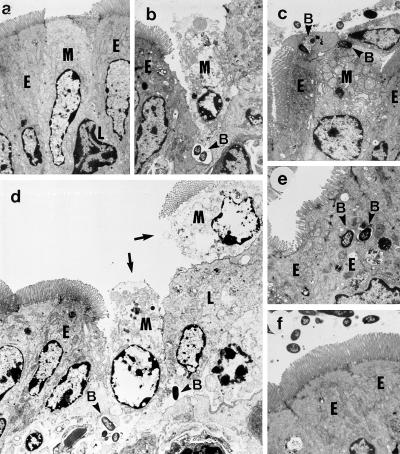
Because Dam− mutants showed a partial defect in invasion of cultured epithelial cells, we addressed the murine ligated loop assay to monitor two critical events linked to Salmonella penetration of the intestinal epithelium: cytotoxicity on M cells of Peyer’s patches and invasion of enterocytes. Elicitation of both processes has been shown to be related to the invasion capacity observed in the in vitro models (23–25). Disruption of M cell integrity was observed on 3 h of infection with the wild type (Fig. 2 a and b). In contrast, very few Dam− (SV1610) bacteria were seen interacting with M cells and, in the rare cases observed, the presence of bacteria did not trigger M cell cytotoxicity (Fig. 2c). Moreover, unlike the wild type, Dam− cells were not seen either causing massive destruction of the intestinal epithelium or moving to deep locations close to the lamina propria (Fig. 2d). Another distinct phenotype of the Dam− mutant was its impaired capacity to invade enterocytes (Fig. 2 e and f). Altogether, these data suggest that Dam methylation modulates the interaction of Salmonella with different cell types in Peyer’s patches.
Figure 2.
Interaction of S. typhimurium Dam− mutants with the murine intestinal epithelium. The ligated loop assay was used to examine the interaction of SL1344 (wild type) and SV1610 (Dam−) with cell types present in Peyer’s patches 3 h after infection. Shown are transmission electron micrographs of: (a) an uninfected M cell; (b) an M cell disrupted by the infection of the wild-type strain; (c) Dam− bacteria interacting with M cells; note the lack of cytotoxicity on the M cell; (d) massive destruction of the intestinal epithelium, which now appears with a clear gap (shown by arrows) as a result of infection by wild-type bacteria. This generalized destruction was never observed in loops infected with Dam− bacteria; (e) wild-type bacteria internalized by enterocytes; (f) Dam− bacteria that do not invade enterocytes. M, M cell; E, enterocyte; L, lymphocyte; B, intracellular bacteria. Note the presence of bacteria in deep locations only in tissues infected with the wild-type strain (b and d). (a–d, ×2,000; e and f, ×3,000.)
Secretion of Proteins in S. typhimurium Dam− Mutants.
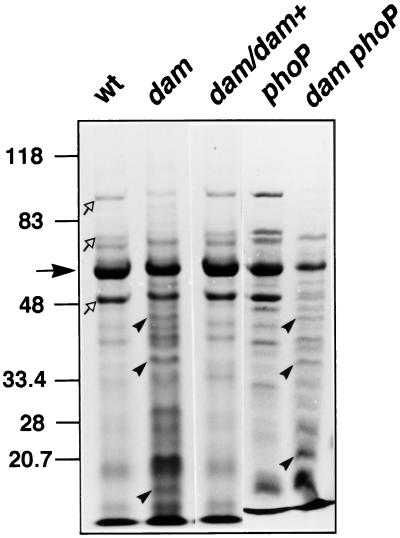
The virulence trait affected in Dam− mutants, invasion of nonphagocytic cells, has been shown to depend on a specialized type III secretion system (reviewed in refs. 26, 27). Type III-secreted proteins have been identified in bacterial culture media supernatants and shown essential for Salmonella invasion (26, 27). We thus analyzed the protein content of supernatants of the wild type, a Dam− mutant, and a Dam+-complemented strain. Extracts containing Salmonella-secreted proteins (ssp) were examined by Coomassie staining. The protein profile obtained for the Dam− mutant was clearly different, and at least three proteins found in the Dam− mutant were not detectable in the wild type (Fig. 3). Interestingly, the dam mutation causes a reduction in the secretion of SPI-1 encoded effector proteins, such as SipA, SipB, and SipC, whereas flagellin secretion is unaffected (Fig. 3). Complementation of the dam mutation fully restored the profile of ssp observed in the wild type (Fig. 3).
Figure 3.
Lack of Dam methylation alters protein secretion. An SDS gel with 8% tricine showing proteins present in culture supernatants from SL1344 (wild type), SV1610 (Dam−), SV1610/pTP166 (Dam−/Dam+), SV4056 (PhoP−), and SV4089 (Dam− PhoP−) strains. Indicated are flagellin (filled arrow), SipA, SipB, and SipC proteins (empty arrows), and three proteins present in supernatants of Dam− strains (arrowheads).
Because an interconnection of regulons under Dam and PhoP control has been recently postulated (6), we examined the effect of dam and phoP mutations on secreted protein profiles. In agreement with previous observations (28), a PhoP− mutant showed a wild-type profile of ssp. In contrast, the ssp profile of a Dam− PhoP− double mutant was similar to that of a Dam− mutant (Fig. 3). Thus, the secretion defect of Dam− mutants is observed in both the presence and the absence of the PhoP product. The observation that the role of Dam methylation in secretion is PhoP independent can be tentatively correlated with invasion assays in HeLa cells: a dam mutation caused a similar invasion reduction in both PhoP+ and PhoP− backgrounds (data not shown). Furthermore, we investigated the involvement of Dam methylase on secretion of SPI1-encoded proteins essential for invasion, such as SipC and InvJ. Western analysis of ssp supernatant extracts by using specific anti-SipC and -InvJ antibodies indicated that secretion of both proteins is reduced by 50% in the Dam− mutant (data not shown). Globally, these assays suggest that Dam methylation might modulate the SPI-1 encoded type III secretion system.
DISCUSSION
In the murine model, attenuation of S. typhimurium virulence in the absence of Dam methylation is observed in both the oral and the intraperitoneal routes. In vivo studies have also shown that, on oral and i.p. administration, the Dam− mutant is able to colonize target organs, but not to proliferate in these locations. The combination of attenuated virulence and organ colonization ability makes Dam− mutants excellent candidates for vaccination. In fact, the Dam− mutant used in this study was able to elicit an immune response sufficient to prevent killing by the virulent strain at doses 100-fold the LD50. Similar observations have recently been reported by Heithoff et al. (6).
Given the high pleiotropy of dam mutations (1, 4), the specific link(s) between Dam methylation and systemic disease cannot be established a priori. However, insights were obtained from the ileal ligated loop assay: the Dam− mutant does not cause cytotoxicity on M cells and is unable to invade enterocytes. These observations explain the low numbers of bacteria recovered from target organs on oral challenge. The M cell has been shown to play a critical role in the intestinal immune response (29). Therefore, lack of cytotoxicity on M cells, together with their proper stimulation by infecting Dam− bacteria, can improve the host immune response and render the Dam− mutant avirulent. In fact, it is known that noninvasive S. typhimurium mutants are avirulent because they do not destroy M cells (30). These results can also explain the optimal vaccine properties of Dam− mutants observed by us and by others (6).
Further insights on the role of Dam methylation in Salmonella virulence were obtained from the tissue culture model: a Dam− mutant showed reduced capacity to invade HeLa cells. However, the invasion impairment of a Dam− mutant was only partial, compared with a SPI1 secretion-deficient InvJ− mutant or a mutant lacking the effector invasion protein SipC. These differences may indicate that a dam mutation causes mild changes in the amount or the activity of these proteins, compared with the effect of knocking out the corresponding genes.
Because secreted proteins are well known effectors in the interaction with eukaryotic cells (26), we considered the possibility that absence of Dam methylation might cause changes in protein secretion mediated by the SPI1 type III system. To test this hypothesis, protein profiles were analyzed in culture supernatants. A distinct pattern of protein secretion was observed in the Dam− mutant. The most relevant differences were: (i) the presence of proteins absent from wild-type extracts and (ii) a reduction in the level of secreted SipA, SipB, and SipC. No defect was observed in flagellin secretion, implying that the effects observed are specific for secretion via the virulence-related type III apparatus. To our knowledge, this is the first description of a DNA modification mechanism that influences secretion of virulence determinants. The effect of Dam methylation on the SPI1 secretion system could be exerted on structural or regulatory genes. Putative target SPI-1-encoded regulators include HilA, InvF (31, 32), and the recently described HilC and HilD (33). Alternatively, Dam methylation could be essential for the activity of other regulators acting on (but not encoded by) SPI1, such as SirA (34, 35) or PhoPQ (28).
Although our protein analyses imply a connection between Dam methylation and the functionality of the SPI1 type III secretion system, our virulence data clearly indicate that the Dam− mutant is also attenuated when administrated intraperitoneally. Because SPI1-defective mutants are still virulent when tested by this route (30, 36, 37), other major virulence genes involved in Salmonella systemic disease might be controlled by Dam methylation. Putative candidates are components of the second type III system encoded in SPI2 (38, 39) or alternative sigma factors as RpoS (σS) or RpoE (σE) (40, 41). However, unlike mutants in these components or regulators, which show clear defects in intracellular proliferation (38, 39) or intracellular survival within macrophages (38–41), our study shows that Dam− mutants are not deficient in these processes. Therefore, further characterization of genes modulated by Dam methylation is required to ascertain which gene products essential for systemic infection are affected in Dam− mutants. A recent report by Heithoff et al. (6) proposes that a large number of Dam-regulated virulence genes exist in S. typhimurium, thus supporting our view that the effects of DNA adenine methylase mutations on Salmonella virulence are pleiotropic.
Acknowledgments
This research was supported by grants from the Ministerio de Educación y Cultura of Spain (PM97-0148-CO2), the Comunidad de Madrid (08.2/0029/97), and the FAIR Program of the European Union (PL96/1743). An institutional grant of the Fundación Ramón Areces to the Centro de Biología Molecular Severo Ochoa is also acknowledged. Strains were kindly provided by J. E. Galán, E. A. Groisman, G. C. Walker, and M. G. Marinus. We thank J. Palacín, M. Guerra, and M. Rejas for their assistance in electron microscopy and animal facilities, D. A. Cano, E. M. Camacho, and J. de la Rosa for help in certain experiments, and C. R. Beuzón and S. Marqués for critical reading of the manuscript.
ABBREVIATION
- Dam
DNA adenine methylation
References
- 1.Marinus M G. Methylation of DNA. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 782–791. [Google Scholar]
- 2.Casadesús J, Torreblanca J. In: Epigenetic Mechanisms of Gene Regulation. Russo V E A, Martienssen R A, Riggs A D, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 141–153. [Google Scholar]
- 3.Noyer-Weidner M, Trautner T A. In: Methylation of DNA in Prokaryotes, in DNA Methylation: Molecular Biology and Biological Significance. Jost J P, Saluz H P, editors. Basel, Switzerland: Birkhäuser; 1993. [Google Scholar]
- 4.Torreblanca J, Casadesús J. Genetics. 1996;144:15–26. doi: 10.1093/genetics/144.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Torreblanca J, Marqués S, Casadesús J. Genetics. 1999;152:31–45. doi: 10.1093/genetics/152.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heithoff D M, Sinsheimer R L, Low D A, Mahan M J. Science. 1999;284:967–970. doi: 10.1126/science.284.5416.967. [DOI] [PubMed] [Google Scholar]
- 7.Charlier D, Gigot D, Huysveld N, Roovers M, Piérad A, Glandsdorff J Mol Biol. 1995;250:383–391. doi: 10.1006/jmbi.1995.0384. [DOI] [PubMed] [Google Scholar]
- 8.Braaten B, Nou X, Kaltenbach L, Low D. Cell. 1994;76:577–588. doi: 10.1016/0092-8674(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 9.Blyn L B, Braaten B A, Low D. EMBO J. 1990;9:4045–4054. doi: 10.1002/j.1460-2075.1990.tb07626.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nou X, Braaten B, Kaltenbach L, Low D. EMBO J. 1995;14:5785–5797. doi: 10.1002/j.1460-2075.1995.tb00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Woude M, Low D. Mol Microbiol. 1994;11:605–618. doi: 10.1111/j.1365-2958.1994.tb00340.x. [DOI] [PubMed] [Google Scholar]
- 12.Huisman T T, de Graaf F K. Mol Microbiol. 1995;16:943–953. doi: 10.1111/j.1365-2958.1995.tb02320.x. [DOI] [PubMed] [Google Scholar]
- 13.Hoiseth S K, Stocker B D A. Nature (London) 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- 14.Marinus M G, Poteete A, Arraj J A. Gene. 1984;28:123–125. doi: 10.1016/0378-1119(84)90095-7. [DOI] [PubMed] [Google Scholar]
- 15.Kaniga K, Tucker S C, Trollinger D, Galán J E. J Bacteriol. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Collazo C M, Zierler M K, Galán J E. Mol Microbiol. 1995;15:25–38. doi: 10.1111/j.1365-2958.1995.tb02218.x. [DOI] [PubMed] [Google Scholar]
- 17.Reed L J, Muench H. Am J Hyg. 1935;27:493–497. [Google Scholar]
- 18.Jones B D, Ghori N, Falkow S. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung K Y, Finlay B B. Proc Natl Acad Sci USA. 1991;88:11470–11474. doi: 10.1073/pnas.88.24.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.García-del Portillo F, Zwick M, Leung K Y, Finlay B B. Proc Natl Acad Sci USA. 1993;90:10544–10548. doi: 10.1073/pnas.90.22.10544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schägger H, von Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 22.Buchmeier N A, Heffron F. Infect Immun. 1989;57:1–7. doi: 10.1128/iai.57.1.1-7.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones B D. Genes Dev. 1997;11:679–687. doi: 10.1101/gad.11.6.679. [DOI] [PubMed] [Google Scholar]
- 24.Jones B D, Falkow S. Annu Rev Immunol. 1996;14:533–561. doi: 10.1146/annurev.immunol.14.1.533. [DOI] [PubMed] [Google Scholar]
- 25.Raupach B, Mecsas J, Heczko U, Falkow S, Finlay B B. Curr Top Microbiol Immunol. 1999;236:137–161. doi: 10.1007/978-3-642-59951-4_8. [DOI] [PubMed] [Google Scholar]
- 26.Galán J E, Collmer A. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 27.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pegues D A, Hantman M J, Behlau I, Miller S I. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 29.Phalipon A, Sansonetti P J. Curr Top Microbiol Immunol. 1999;236:163–189. doi: 10.1007/978-3-642-59951-4_9. [DOI] [PubMed] [Google Scholar]
- 30.Penheiter K L, Mathur N, Giles D, Fahlen T, Jones B D. Mol Microbiol. 1997;24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x. [DOI] [PubMed] [Google Scholar]
- 31.Bajaj V, Lucas R L, Hwang C, Lee C A. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 32.Galán J E. Mol Microbiol. 1996;20:263–271. doi: 10.1111/j.1365-2958.1996.tb02615.x. [DOI] [PubMed] [Google Scholar]
- 33.Schechter L M, Damrauer S M, Lee C A. Mol Microbiol. 1999;32:629–642. doi: 10.1046/j.1365-2958.1999.01381.x. [DOI] [PubMed] [Google Scholar]
- 34.Johnston C, Pegues D A, Hueck C J, Lee A, Miller S I. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 35.Ahmer B M M, van Reeuwijk J, Watson P R, Wallis T S, Heffron F. Mol Microbiol. 1999;31:971–982. doi: 10.1046/j.1365-2958.1999.01244.x. [DOI] [PubMed] [Google Scholar]
- 36.Galán J E, Curtiss R., III Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones B D, Falkow S. Infect Immun. 1994;62:3745–3752. doi: 10.1128/iai.62.9.3745-3752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hensel M, Shea J E, Waterman S R, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang F C, Holden D W. Mol Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- 39.Cirillo D M, Valdivia R H, Monack D M, Falkow S. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- 40.Fang F C, Libby S J, Buchmeier N A, Loewen P C, Switala J, Harwood J, Guiney D G. Proc Natl Acad Sci USA. 1992;89:11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Humphreys S, Stevenson A, Bacon A, Weinhardt A B, Roberts M. Infect Immun. 1999;67:1560–1568. doi: 10.1128/iai.67.4.1560-1568.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]