Abstract
Information theory was used to build a promoter model that accounts for the −10, the −35 and the uncertainty of the gap between them on a common scale. Helical face assignment indicated that base −7, rather than −11, of the −10 may be flipping to initiate transcription. We found that the sequence conservation of σ70 binding sites is 6.5 ± 0.1 bits. Some promoters lack a −35 region, but have a 6.7 ± 0.2 bit extended −10, almost the same information as the bipartite promoter. These results and similarities between the contacts in the extended −10 binding and the −35 suggest that the flexible bipartite σ factor evolved from a simpler polymerase. Binding predicted by the bipartite model is enriched around 35 bases upstream of the translational start. This distance is the smallest 5′ mRNA leader necessary for ribosome binding, suggesting that selective pressure minimizes transcript length. The promoter model was combined with models of the transcription factors Fur and Lrp to locate new promoters, to quantify promoter strengths, and to predict activation and repression. Finally, the DNA-bending proteins Fis, H-NS and IHF frequently have sites within one DNA persistence length from the −35, so bending allows distal activators to reach the polymerase.
INTRODUCTION
Transcriptional regulation is essential to the viability of the cell (1–3). In prokaryotes, many molecules can contribute to or detract from the stability of the initiation complex (4). The minimum requirement for RNA polymerase binding is recognition of the promoter by the σ factor (5–8). In general, prokaryotic RNA polymerases can interchange a number of σ factors which bind and initiate different groups of genes (9). σ70 is the most commonly used σ factor in Escherichia coli and it is responsible for the initiation of most genes (9). This paper will only focus on promoters bound by σ70.
To successfully model initiation, it is necessary to construct a model that unifies multiple components. The conventional model for promoter recognition by σ70 is the binding of two regions upstream of the transcription start point, named the −10 and −35 because of their spacing relative to the first transcribed base (10,11). The initiation complex is also further stabilized by the C-terminal domain of the two α subunits of the core enzyme (αCTD), which can either interact directly with upstream DNA or with regulatory proteins (12). To add to the complexity of the system, recognition of the −10 alone can be sufficient for initiation to occur (13–15). The initiating polymerase can be thought of as moving ship that needs to be anchored down (16). The varying affinities of the binding components for the promoter would correlate to varying weights holding the polymerase in place. The sum of these components must have enough energy to stabilize the polymerase against thermal noise. Therefore, in order to model promoter binding, we need to consider the relative affinity of each molecule affecting the stability of the initiation complex.
In addition, the σ factor is flexible. That is, the distance between the −10 and −35 binding sites is not fixed. This flexibility affects the affinity of the polymerase for the sequence (10). If we treat σ factor bound to core as a simple harmonic oscillator, then expansion or contraction of the polymerase when binding to promoters with varying spacings would strain the molecule and reduce the amount of energy available for stabilization. Since the initiation rate is affected by spacing (10,17–21), our model needs to take into account this internal strain.
Traditionally three possible spacings have been proposed at which the −10 and the −35 bind relative to each other, 17 ± 1 bases, but initiation over a larger range, 15–20 bases, has been shown (10,19,22). These spacings correspond to the number of bases between the 3′ end of the −35 hexamer and the 5′ end of the −10 hexamer. The observed optimal spacing of 17 bases (10) places the centers of the two hexamers on the same face of the DNA 23 bases apart, ∼2 helical twists of B-form DNA (23), suggesting that the polymerase has a DNA-structure-dependent contact. There is also a correlation between the extent of negative supercoiling and the amount of transcription from a promoter (21,22), which demonstrates that the σ factor is sensitive to genomic structure (24,25).
Although neural networks and hidden Markov models (HMMs) have been used to model promoter binding (26–28), constructing these models usually requires the untenable assumption that large stretches of sequence do not contain sites, and the resulting parameters have not been easy to interpret. Other attempts have been made to model promoters using methods not based on HMMs (29–33), but these methods do not uniformly measure the contribution of all components in the initiation complex (−10, −35 and gap). Hertz and Stormo (31) presented a model in which they subtracted the gap penalty for the optimal spacing from each gap value, so that there was no penalty for having the optimal spacing. Their formula to evaluate gap penalties implies that a set of sites that have several equiprobable gap lengths would have no penalties, even though flexibility decreases information (34) (http://www.ccrnp.ncifcrf.gov/~toms/paper/flexrbs/). The method used in this paper, which was previously used to investigate ribosome binding sites (34), does account for gap variability with equiprobable gap lengths. In addition, promoter strengths are not determined purely by the binding of the σ factor. Transcriptional activators and repressors contribute to and detract from the accessibility of DNA by the RNA polymerase. In order to uniformly model the flexible binding of the σ70 in conjunction with transcriptional regulators, we used information theory.
Information theory was developed by Claude Shannon to quantify the transfer of information in communications (35,36) (http://cm.bell-labs.com/cm/ms/what/shannonday/paper.html). It has proven to be useful when applied to a variety of biological systems (37–41) (http://www.ccrnp.ncifcrf.gov/~toms/paper/schneider1986/; http://www.ccrnp.ncifcrf.gov/~toms/paper/rfs/; http://www.ccrnp.ncifcrf.gov/~toms/paper/fisinfo/; http://www.ccrnp.ncifcrf.gov/~toms/paper/lrp/; http://www.ccrnp.ncifcrf.gov/~toms/paper/baseflip/) mainly in quantifying how specific a given DNA-binding protein is, based on the amount of variability within its binding targets. A lower binding site variability corresponds to a higher information content (37). For convenience, information is generally measured in bits, the choice between two equally likely possibilities. A greater information content for a set of binding sites (more bits of information) generally will have more specific binding and a higher binding affinity.
Prokaryotic ribosomes, like the σ factor, have two binding elements separated by a variable distance. In previous work, we used information theory to model 95% of the E. coli ribosome binding sites (34). This flexible model took into account the conservation of both the initiation codon and Shine–Dalgarno regions, and the statistics of the variable spacing between them. Here we applied the same theory to the σ70 promoter components (−35, −10 and their spacing) to create a cohesive model of promoter binding.
An important difference between the ribosome model and the promoter model is that the ribosome model is only composed of two binding elements. The promoter model can be made up of a large number of binding elements with parameters governing how each element behaves, and how the elements interact with each other. This paper shows how an internally consistent multi-part model can be constructed directly from experimentally proven sites, and discusses how this model can be used to predict and identify control systems. We are also interested in understanding the fundamental workings of the RNA polymerase. To this end, we examined the variation in the promoter as a function of spacing, global trends in accessory molecule binding relative to the promoters, and the relationship between ribosome binding sites and promoters. We also propose that the flexible bipartite binding site evolved from a rigid extended −10 binding mode.
MATERIALS AND METHODS
Constructing the promoter model
We built our σ70 binding model by aligning and refining the sequences upstream of 599 experimentally determined transcription starts reported in the RegulonDb database (42) and 85 starts from the PromEC database (43) that were not included in RegulonDb. To align binding sites we used the malign program to maximize the information of either the −10 or −35 by shuffling the sequences (44) (http://www.ccrnp.ncifcrf.gov/~toms/paper/malign). To refine the model, we iteratively removed all sequences with an information content <0 bits of information (45,46) (http://www.ccrnp.ncifcrf.gov/~toms/paper/edmm/; http://www.ccrnp.ncifcrf.gov/~toms/paper/ri/) until we converged on a consistent set of sequences. Since there is both a variable spacing between the −10 and the transcription start point, and between the −35 and the −10, this process was not trivial, and we describe below how we converged on our final model.
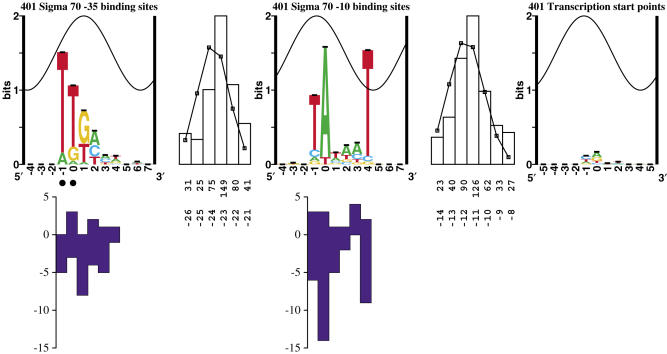
To align the −10, we embedded the DNA sequences −15 to −3 bases upstream of the transcription start site in random DNA, so that our alignment would not be biased by the −35 or by the preference of adenine at the transcription start point. We realigned the −10 region to maximize the information by using the malign program (44) over the range of −12 to −7 bases upstream of the transcription start, allowing for the sequences to shift up to 3 bases in either direction. Since nearby transcription start points could potentially use the same −10, we identified and removed transcription starts from our dataset that were within 15 bases of another site with a lower genomic coordinate (arbitrarily chosen) and had the same orientation. This prevented the same −10 from appearing multiple times in our model, and decreased the size of our dataset from 684 to 620 starts. We then did a cyclic refinement on these sites to remove sequences from our dataset that were not identified as sites by our model. To do this, all sites that had an information content (46) <0 bits were removed, and the model was rebuilt. The zero-bit cutoff was used because it represents a version of the second law of thermodynamics: sites with positive information correspond to negative ΔG of binding (45,46). This approach was successfully used for constructing ribosome (34) and splice site models (47). Removing and rebuilding was continued until no negative sites remained in the set. This reduced our number of sites from 620 to 559. The refined multiple alignment gave us a well conserved −10 region (Figure 1). At each position, the base conservation of the −10 corresponds to the number of mutants found at that position by genetic studies (11).
Figure 1.
Sequence logos of σ70 binding components. From left to right: sequence logo of −35 binding sites, spacing distribution between −35 and −10 binding sites, sequence logo of −10 binding sites, spacing distribution between −10 binding sites and the transcription start point, sequence logo of transcription start points. In a logo, the height of each letter is proportional to the frequency of that base at each position, and the height of the letter stack is the conservation in bits (49). Error bars are shown at the top of the stacks. The total information in the −35 and −10, less the gap uncertainty between them, is (4.02 ± 0.09) + (4.78 ± 0.11) − (2.32 ± 0.04) = 6.48 ± 0.14 bits. The sine wave on each logo represents the 10.6 base helical twist of B-form DNA for the optimal spacing of 23 bases, with the major groove centered at +1 of the −35 (41,51). Black dots indicate the location of important 5-methyl groups on thymine and hence determine the location where the major groove faces the sigma factor (81), along with co-crystal data (65). The top row of numbers in each gap distribution gives the number of cases and the bottom row is the difference between the zero coordinates. A Gaussian curve was fit to each of the two gap distributions (thin black line). Mutational data presented by Hawley and McClure (10,11) are shown under the logos by blue bars. Bars above the abscissa represent the number of observed mutations at each position that have strengthened a promoter, while bars below the abscissa represent the number of mutations that have weakened a promoter. Promoter locations and the information contents of their parts are given in Supplementary Data.
We do not adhere to the conventional numbering system used in describing the distance between the −10 and the −35. The conventional numbering of the spacer is the number of bases between the two hexamers (10). That is, ttgacaNNNtataat would have a spacing of three. Since the convention for position numbering in asymmetric sequence logos is to choose a strongly conserved base, we will refer to the second base in each hexamer as zero, and all spacings will be reported as the difference between those coordinates. Therefore, all values for our spacing are 6 bases greater than the numbering used previously. For example, tTgacannntAtaat would have a spacing of nine (the difference between the capital T in the −35 hexamer and the capital A in the −10 hexamer), rather than three. Therefore, the classical spacing of 17 bases is 23 in our notation. The only way we would be able to adhere to the conventional numbering system would be to assign a base outside one of the hexamers as the zero coordinate. This would be confusing in sequence analysis using sequence walkers (48), (http://www.ccrnp.ncifcrf.gov/~toms/paper/walker/). Furthermore, each sequence walker always has the integer zero in its coordinate system so that one can easily and unambiguously locate a binding site and then specifically identify bases within the binding site.
Aligning the −35 was more difficult than aligning the −10. The traditional model of RNA polymerase binding only allows for three different spacings between the −35 and the −10 (11). Mutational data have shown that this range could be expanded to six positions (10), but that at these expanded spacings the amount of transcription is reduced substantially. The optimal spacing of 23 bp (McClure's spacing of 17 bp) (11) places both hexamers on the same face of the DNA within their respective major grooves (23), indicating that the spacing may be dependent upon DNA structure. Therefore, a new alignment method is needed that takes into account the structure of the DNA.
The algorithm for aligning the −35 is different from our previous approach of aligning upstream sequences in flexible models [i.e. modeling the Shine–Dalgarno relative to the initiation codon in ribosome binding sites (34)]. As with ribosomes, our approach was to create a model de novo from experimental data, so as to avoid biases in previous models. Using a sequence logo (49) (http://www.ccrnp.ncifcrf.gov/~toms/paper/logopaper/), we observed a weak conservation upstream of the aligned −10s in the region expected for the −35, 23 bp upstream. We determined that the conservation of this region was low because a number of sites with a different spacing were overlapping, reducing the total sequence conservation. We performed a cyclic refinement of the region which corresponds to the −35 hexamer at the optimal spacing from the −10 in order to pull out a preliminary −35 model. This cyclic refinement gave a reasonably well conserved −35 sequence logo, which matched the conventional hexamer consensus (11,50) (http://www.ccrnp.ncifcrf.gov/~toms/papers/zen), and this alignment was used for an initial model. After refinement, we used malign to allow for the sites to be moved 1 base in either direction, so as to maximize the information in this refined −35.
Using the program multiscan, the initial −35 model was scanned over the region upstream of the −10 of every promoter in order to find the −35 which most closely matched this model for each site. Of the 559 promoters scanned, 421 had a −35 site >0 bits in the range of 21–26 bases upstream of the −10 [this corresponds to McClure's spacing of 15–20 bases (10)]. The alignment having the strongest site was used, and the total site strength was calculated using the flexible information equation described previously (34):
| 1 |
where Ri(−35) is the individual information (46) of the −35 site, Ri(−10) is the individual information of the −10 site, and GS(d) is the gap surprisal for each spacing d, which is based on the major groove accessibility curve of B-form DNA (41,51) (http://www.ccrnp.ncifcrf.gov/~toms/paper/oxyr/). The equation used to generate a distribution which corresponds to the accessibility of the DNA by the RNA polymerase is:
| 2 |
where w is 10.6 bases (one turn of B-form DNA), center is 23 bases (the optimal spacing between the −35 and the −10), and d is the distance of the −35 from the −10, in bases. This equation describes the major groove accessibility (41,51) in which direct contacts are more accessible and contacts on the opposite face of the DNA are not accessible. The surprisal was then calculated for each position using the gap surprisal equation published previously:
| 3 |
n(d) is the accessibility at spacing d, and n is the sum of all accessibility values over the allowed values of d. e(n) is a small sample correction value (34,37). By using these equations to model the structure of DNA, we gave preference to −35 sequences which were in physically reasonable positions.
Once we had identified the −35, the −10 and the spacing for each promoter, we dispensed with the ‘scaffolding’ equations described above and built a flexible model directly from the sequence data. We did a further cyclic refinement on this set by removing promoters with a flexible information <0 bits, reducing the number of promoters from 421 to 401. Our final model, therefore, contains 59% of the sites that are in our original database. Transcriptional regulators can provide informational contacts through the αCTD and this could account for some of the information used by polymerases (52). As a result, many promoters may have poorly conserved, or highly variable, σ binding sites. The refinement process made the model self-consistent (containing similar sites) and it can therefore be regarded as a basal promoter model. The excluded sites are not consistent with this basal model, and presumably they initiate by some method other than the sole recognition of the −10 and −35 (such as an extended −10 or activation by another protein).
Promoter binding and transcriptional regulation is more complex than our previous flexible modeling system was able to handle because of the contribution of activation proteins (52). Therefore, we created an algorithm that not only considers the strength of the two-part σ70 site, but also can include the information contributed by activating proteins. In order to do this we used the multiscan algorithm.
Multiscan algorithm
Multiscan is an extension of the biscan program that is used to model the flexible prokaryotic ribosome (34). Translational initiation in prokaryotes requires contact at both the P site (or initiation region, IR) and the Shine–Dalgarno (SD). Because of the flexibility of the ribosome, these contacts can occur at different spacings, anywhere between 4 and 18 bases. In order to assess the information present in ribosome binding sites, the contributions of the Shine–Dalgarno, the initiation region and the spacing between them all have to be considered. The equation for calculating the information for a two-part model with variable spacing was given in Equation (1).
The RNA polymerase is similar to the ribosome in that it makes two contacts (the −10 and the−35) with some variable distance between them. Therefore, the flexible information analysis used with ribosomes can also be used to describe the binding of the σ factor to the promoter. The difference between translational and transcriptional initiation is that auxiliary proteins can also bind to either activate or repress transcription. So in order to model the promoter correctly, we need to calculate the information of all the molecules that contribute or interfere.
For activators, as an initial simple model, we assume that their information contributes additively to the total information of the promoter (53), so the new equation is as follows:
| 4 |
where Ri (Act) is the individual information of some activator protein site, and GS(d−35/Act) is the gap surprisal value at spacing d between the activating protein site and the −35. Since the initiation complex is stabilized by the contacts between the αCTD and the regulatory protein (12), the information contribution of an activator is in those contacts, and not in the contacts between the activator and the DNA. Data on these protein–protein contacts are not available through DNA sequences. However, the higher the affinity of the activator for the DNA, the greater the probability of it being bound simultaneously with the polymerase. Therefore, to a first approximation, we can model the indirect contribution of information by the activator as a modulation of the activator information by the protein–protein interaction. We represent this modulation by ρAct, with 0 ≤ ρAct ≤ 1. Since the informatics of interactions with the αCTD are unknown, we will use a ρAct = 1.
This algorithm only includes the activator site and corresponding gap surprisal if they contribute positive information, since that corresponds to favorable binding (46). In addition, the number of potential activators is limited only by the length of the sequence. That is, if multiple activators bind in a range relative to the RNA polymerase that has been observed to be advantageous to transcription initiation, then they are all included into the total information for the site. At present, the algorithm does not account for the possibility of repression of one activator by another (54).
Although it seems reasonable to assume that activator protein information can be scaled by ρAct and added to the total information of the promoter, it is not clear that repressor information should be subtracted. Since repressors block the binding of the polymerase to the DNA, or, in cases such as GalR, cause DNA loops that block binding (55), they do not decrease the strength of the contact but, if present, totally prevent contact from occurring. Therefore, it does not matter what the strength of the repressor is, because a repressor bound to a 1 bit site will prevent initiation as well as a repressor bound to a 10 bit site. The difference between the two is that the 10 bit site will be bound more frequently, so the relative site strengths between the polymerase and the repressor (as well as the concentration of both molecules) can be used to predict the frequency of transcription, but not the ability of the polymerase to bind.
Promoter analysis using the σ70 model
Sequence logos for promoter components were made using the programs delila, alist, encode, rseq, dalvec and makelogo as described previously (49,51). We used the programs diffinst, genhis and genpic to generate the spacing distribution between the binding components.
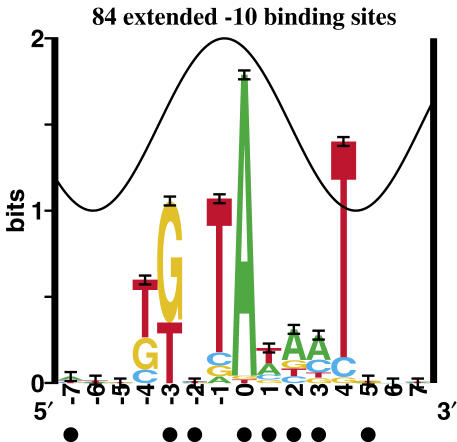
During our refinement process, we identified 138 experimentally verified promoters that did not have an upstream −35. We used this subset to build the extended −10 model (Figure 3). These sites contained a weakly conserved TG two bases upstream of the −10 hexamer. We cyclicly refined (34) these sites over the range −4 to −3 and automatically isolated a subset of 84 sites that turned out to resemble the extended −10 9mer reported previously (13).
Figure 3.
The extended −10 has two additionally conserved bases. This is a sequence logo of −10 regions that have no −35 based on our model, but show conservation at positions −4 and −3. Purines protected from DMS methylation and bromouracil substituted thymines protected by the polymerase are indicated by closed circles (109,110).
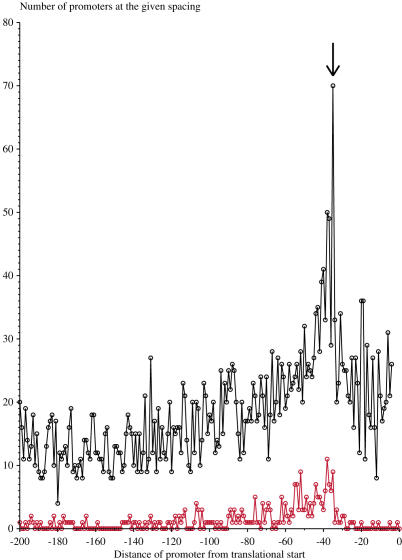
To determine spacings between the −10 and the translational initiation codon (Figure 4), we scanned our promoter model over the 250 bases upstream of all genes in E. coli (56). The site range we used for both the −10 and the −35 was −1 to +4, the range of the hexamer (Figure 1). We plotted the number of occurrences at each distance between the zero position of the −10 and the translational start point of the strongest upstream promoter. To eliminate interference between the −10 model and the initiation codon, we only included sites in our plot that were at least 4 bases upstream of the gene start. We also plotted the number of occurrences at each distance between the −10s of the experimentally determined sites in our model and their respective downstream translational start.
Figure 4.
The optimal spacing of the −10 to the translational initiation codon is ∼35 bases. We plotted the distance between the zero coordinate of the −10 and the translational start point on the abscissa, and the number of promoters at that distance on the ordinate. The upper curve (black) represents data from a scan using our promoter model over the upstream regions of all 4122 genes in E. coli (56). The lower curve (red) represents the location of the −10 relative to translational initiation codons for experimentally verified transcription start points. The arrow pointing to the black curve indicates a peak at −35 bases.
In order to analyze individual sequences using our σ70 model and a transcriptional regulator, we used sequence walker technology (46,48,57). Flexible sites were located using multiscan and displayed as sequence walkers using lister. For Figures 5 and 6, we merely scanned our models over these regions to see how our analysis compared to biochemical data known about these systems. Our Fur model was built from 24 biochemically characterized binding sites (manuscript in preparation) and our Lrp model comes from (34). Once we were confident of the predictive capabilities of our models, we searched for uncharacterized Fur controlled genes in E. coli, two of which are presented in this paper (Figure 7).
Figure 5.
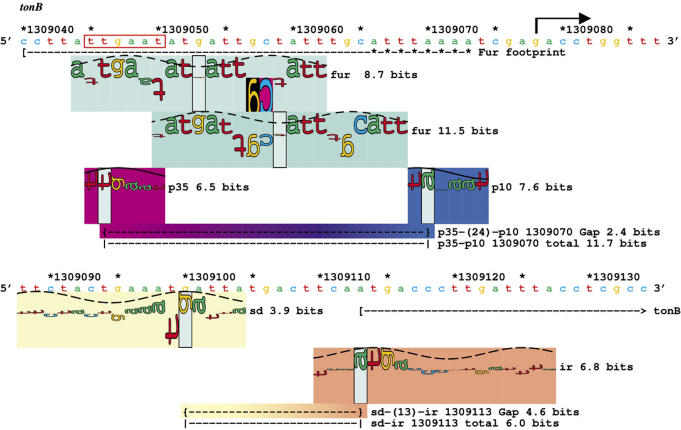
Fur represses transcriptional initiation of tonB. We present here an individual information analysis (46) of the Fur controlled tonB region (71,111) using the sequence walker method (48). Colored rectangles (‘petals’) behind the walkers identify the kind of site (by hue) and the strength of the site (by saturation) (112), (http://www.ccrnp.ncifcrf.gov/~toms/papers/t7island/). The connecting bar between parts of a flexible site transitions linearly between the corresponding colors. The σ70 binding site and the ribosome binding site were both located using flexible binding models (34), in that there are variable distances between binding components. The horizontal dashed line underneath the −10 walker (p10) and the −35 walker (p35) that is labeled ‘Gap’, gives the gap surprisal for whatever distance separates the two components, as well as the coordinate of the downstream component. The dashed line that is labeled ‘total’ gives the total information for the flexible site. Similar lines are underneath the ribosome binding site components (SD, IR). A Fur dimethyl sulfate protection footprint is marked (71,111), and two sequence walkers for Fur fall below it. The downstream side of the Fur protected sequence is somewhere in the region marked by asterisks. The sequence that the polymerase and Fur would both bind is marked with a red box. The transcription start point for tonB is marked with a black arrow, and the translational initiation start point is marked at position 1309113 with a bracket and an arrow. The sequences and coordinates on the map are from GenBank accession number U00096 (113).
Figure 6.
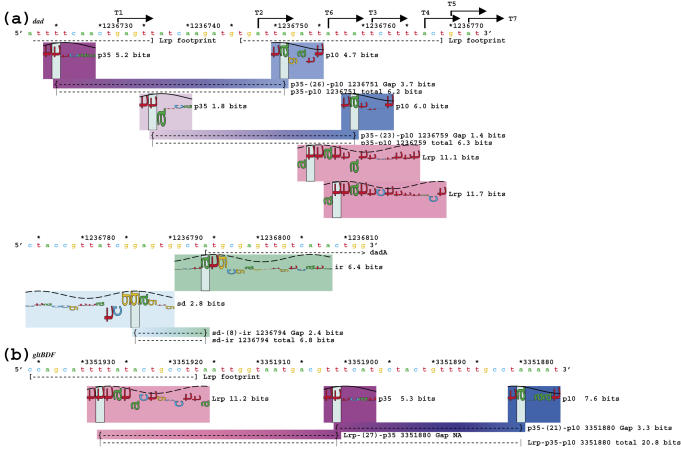
Transcriptional control by Lrp is based on its spacing relative to the polymerase. (a) An individual information analysis of the Lrp repressed dad operon (73–75). (b) An individual information analysis of the Lrp activated gltBDF operon (76). As in Figure 5, the σ70 and ribosome binding sites are each internally connected by lines that report the gap surprisal and total information. Experimentally verified transcription start points are identified with black arrows and named according to Zhi et al. (75), and the dadA gene start is marked with a bracket and arrow at position 1236794. In (b), since Lrp helps to stabilize the initiation complex, its information is added into the total strength of the promoter. Since data on the distance between Lrp sites and the −35 are not available, we did not subtract a gap surprisal and therefore the gap surprisal is marked as NA (not applicable). The sequence and coordinates on the map are from GenBank accession number U00096 (113).
Figure 7.
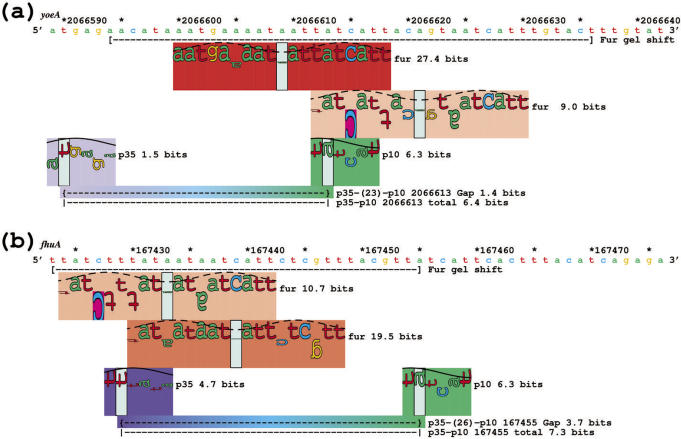
Sequence walkers for σ70 and Fur protein upstream of the (a) yoeA and (b) fhuA genes suggests that these genes are controlled by Fur. Synthetic oligonucleotides that contain sequences marked by brackets under the DNA showed gel mobility shifts by Fur protein (data not shown). The sequence and coordinates on the map are from GenBank accession number U00096 (113).
To identify these novel control elements, we scanned the entire genome for Fur sites that overlapped the σ70 binding sites within 200 bases of translational start points. These two were chosen because of the strength of both the Fur and σ70 sites, and their proximity to each other. Gel shifts confirmed that these sites are bound by Fur (data not shown).
Finally, the relative binding plots (Figure 8) were generated by scanning Fis, H-NS and IHF models over the range −1000 to +1000 bases relative to the transcription start, the −10 and the −35 of all promoters in our model. The frequency of sites was determined at each position relative to a promoter component by dividing the number of predicted sites by the number of promoters. The frequency of sites in the genome was determined by scanning the entire genome with all three regulators, and then dividing by the genome size. We only presented data over the range −400 to +200 because the distributions were flat outside of those ranges and matched the frequency of sites in the entire genome. The Fis and IHF models come from previously published works (39,41,58), while our H-NS model has not been published but resembles another published model (59).
Figure 8.
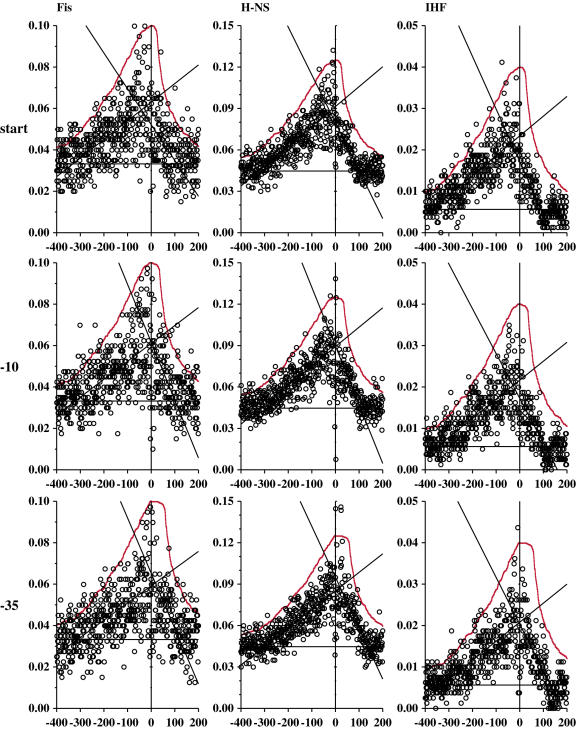
Intergenic binding of DNA bending proteins relative to promoter components. These curves allow one to directly compare the density of non-coding regions to the number of DNA-binding protein sites at each position relative to experimentally determined promoter components. For all graphs, the abscissa is the position of the regulator binding site (either Fis, H-NS or IHF) relative to either the transcription start, the −10, or the −35 in our promoter model. A vertical line marks the zero coordinate of the promoter component. The ordinate is the frequency of sites at that spacing (sites per base). A solid horizontal line marks the frequency of sites per base predicted for the entire genome. Linear regression lines for −400 to 0 and 0 to 200 are shown. A distribution corresponding to the density of intergenic regions surrounding the experimentally verified promoters was fit to the data in each graph, and is shown as a solid red curve (see Materials and Methods).
To compute the intergenic density distributions (Figure 8, red curves), we determined the distance from each promoter part (start, −10 and −35) to the closest upstream and downstream coding region for all sites in our model. We used the EcoGene12 genome annotation to determine coding regions (56). We only used promoters whose zero coordinates are not within coding regions. Out of 401 promoters, 359 transcriptional starts, 356 −10s and 349 −35s were not within a coding region. We then counted the number of sequences that had a non-coding base at each spacing upstream and downstream of the promoter. This curve was then normalized and fit to the relative binding plots in Figure 8, scaled so that the curves are just above the other data points in the graphs, and the zero occurrences level was set equal to the genomic frequency.
RESULTS
The σ70 model
Our dataset for σ70 promoters consisted of both the RegulonDb and the PromEC databases (42,43). Unlike eukaryotic start points, which contain ∼3 bits of information (60), there did not appear to be much information at this prokaryotic transcription start point, only 0.39 ± 0.06 bits (Figure 1). There does appear to be a slight preference for an adenine as the first base, but it is weak. Interestingly, this preference is more conserved in early promoters of bacteriophage T4 (61) where, presumably, it contributes to the phage taking over the cell. Realignment of the start region by allowing shifts of 1 base in either direction (44) gives a pattern of 1.33 bits (data not shown). Since a choice of 1 in 3 requires log2 3 = 1.58 bits, this pattern is not significant.
We were quite easily able to align the −10 relative to the transcription start points (Figure 1). The distance between the −10 and the transcription start point varied between −14 and −8 bases, with the most common spacing of −11 (this is the distance between the transcription start point and the zero position of the −10 logo). The −10 sites contained 4.78 ± 0.11 bits of information over the range of −1 to +4, the range of the hexamer.
The most striking feature of the −10 logo is the strongly conserved T (position +4) where the protein is likely to face the minor groove of the DNA. In other logos for DNA-binding proteins, conservation of bases rarely exceeds 1 bit in the minor groove (62), because in B-form DNA the exposed groups in the minor groove can only be used to distinguish A or T from C or G, but not all four bases individually (63). High conservation in the minor groove suggests that this base is being contacted atypically. Several possibilities are that the helix is distorted when bound, it is interacting with σ70 in the open complex (64), or that it is being flipped out of the helix to initiate open complex formation, as may occur in DNA replication (41).
We had greater difficulty aligning the −35 perhaps because the −35 is often replaced by activators (1,52). Traditionally, the placement of the −35 relative to the −10 is ±1 base relative to the most frequent position of 23 bases [McClure's 17 (11), see Materials and Methods]. Experimental data have shown initiation at a range of −2 to +3 relative to the most frequent position (10,19,22). When we allowed for our model to include sites in this expanded spacing, we identified 107 promoters (>0 bits) that had no possible −35 in the traditional range, suggesting that binding does occur at these peripheral spacings. The amount of conservation in the −35, as in the −10, was low compared with other DNA-binding proteins (37) (4.02 ± 0.09 bits). The conserved region of the −35 appears to fill only one-half of the major groove, as there is an abrupt termination of conservation on the 5′ edge (Figure 1), which is consistent with the abrupt termination of contacts in the σA/−35 co-crystal and the 5′ most edge of the polymerase (65). [σA is the primary sigma factor in Thermus aquaticus, corresponding to σ70 in E. coli (8)]. This suggests that there is space for other binding components to come in and to help stabilize the complex. This is supported by the observation that positive control mutants are immediately adjacent to the portion of σ which binds into the −35 groove (65). As shown by the sine waves in Figure 1, when we place the G at +1 of the −35 close to the center of the major groove [as observed in the co-crystal (65)], at the optimal spacing of 23 bases, the T at +4 of the −10 is exactly positioned in the minor groove. This agrees with the proposal that position +4 (−7 in conventional numbering) is contacted atypically, for example by base flipping to initiate transcription (41).
We allowed for the spacing range between the two hexamers to be between 21 and 26 bases. The optimal spacing between the zero coordinates of the −35 and −10 was 23 bases. The spacing distribution appeared to be approximately Gaussian with an uncertainty of 2.32 ± 0.04 bits.
The total sequence conservation (Rsequence) for the σ70 model (−35, gap, −10) is 6.48 ± 0.14 bits (Figure 1). It contains 401 of the 684 sites in our combined RegulonDb–PromEC database. As discussed in Materials and Methods, the number of sites in our dataset was reduced through a series of refinements in order to identify a consistent subset of promoters that presumably can initiate without accessory molecules.
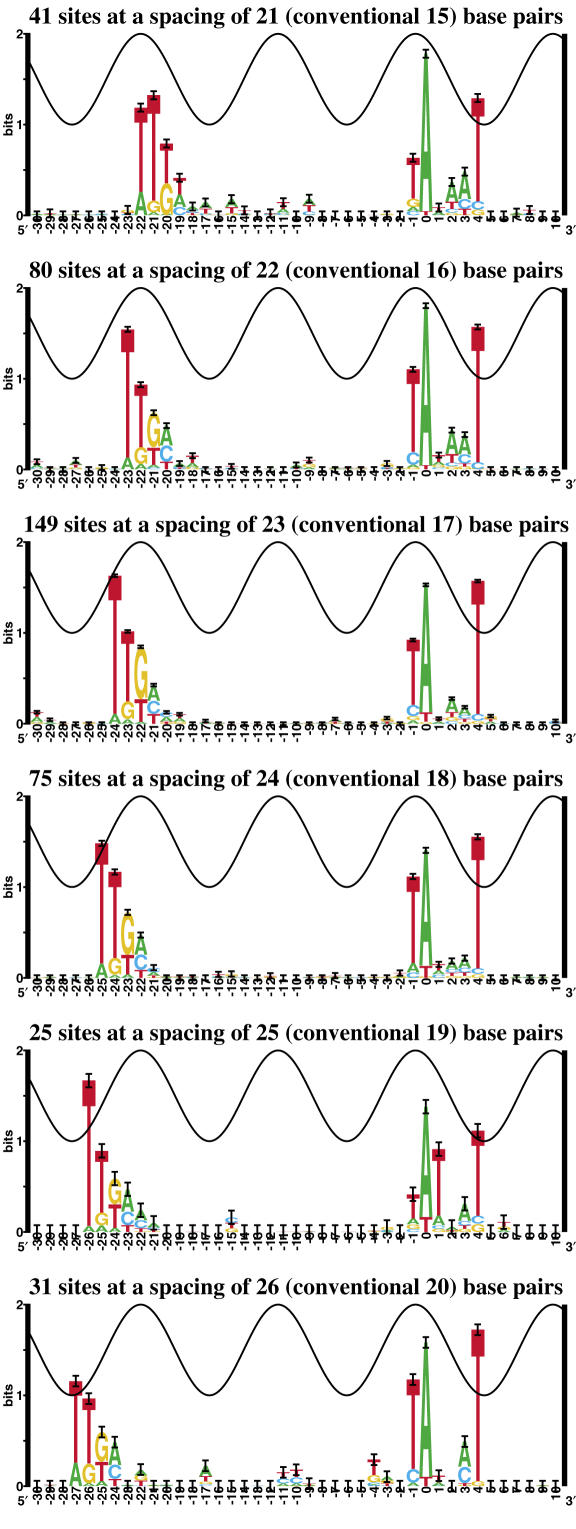
To see how the σ70 promoter varies with spacing, logos were made for each of the spacing classes (Figure 2). The logos for the three most frequently used spacings (22,23,24) look fairly similar. In these three cases, the bulk of the −35 logo falls in the major groove on the same DNA face as the −10, two helical turns away. The further spacings do look a little different, but this could be from smaller sample sizes. The logo for spacing 25 is the most unique, having what appears to be a distorted −10 (prominent T at position +1 and poorly conserved Ts at −1 and at +4). Also, at spacing 21 the logo is a little different with perhaps a slightly more conserved T (22 cases) instead of G (18 cases) in the third position of the −35 hexamer (position −20).
Figure 2.
Sequence logos for σ70 promoters as a function of spacing. The spacings correspond to the −35 to −10 gap distribution in Figure 1. Conventional spacings are given in parentheses.
The conservation of the −35 at each spacing in Figure 2 appears to follow the sine wave. That is, the conserved bases do not go above the wave, except at position −26 of spacing 25. This suggests that the polymerase preferentially contacts the two components when they are on the same face of the DNA (23), but that the σ 4.2 region (which contacts the −35) can rotate relative to the σ 2.4 region (which contacts the −10) or that the DNA twist can change. It has been suggested that models be made for each spacing class (32); this may be useful for sequences outside of the central three spacings. Differences in logos outside of the three similar central spacings (22–24) could be caused by awkward contacts with the polymerase at the extremes of rotation of the −35 relative to the −10.
We looked at the −10 as a function of spacing relative to the transcription start point (data not shown). There was a slight increase in the conservation of the 5′ T of the −10 hexamer at greater spacings. Besides that, there was little variability in the logos of the different spacing classes. The amount of information at the transcription start point was small (∼0.4 bits) for all spacings, and the slight variability between them could be accounted for by noise.
To avoid duplicate sites, we had excluded 64 promoters from our dataset which were within 15 bases of another transcript start (see Materials and Methods). Having built the model, we went back and scanned it over these regions to see if we could predict promoters upstream of more complex overlapping transcripts (data not shown). In 25 cases, for every experimentally determined transcriptional start point there were one or more distinct predicted promoters. In 17 of the 64 cases there was only one predicted promoter and both transcripts fell within the known distance distribution of 8–14 bases downstream from the −10 (Figure 1). In 10 cases there was only one predicted promoter and only one of the transcripts fell within 8–14 bases downstream. Twelve of the starts had no predicted promoter upstream, suggesting that these transcripts are regulated. These results confirm that the basal model functions reasonably well on sequences from which it was not constructed.
To further verify our promoter model, we scanned it over the starts of 36 small RNAs presented by Hershberg et al. (66) (data not shown). The model identified promoters upstream of 23 of the 36 starts. That is, ∼64% of the sites had a promoter with a total information >0 bits from 8 to 14 bases (Figure 1) upstream of the small RNA transcription start. This percentage is similar to that of empirically determined promoters that formed a coherent ‘basal’ set in our refinement process (59%), suggesting that initiation by the basal machinery may only occur ∼60% of the time both in general and at small RNAs. None of these sites has been used to build our model. Therefore, this result again shows that our model can identify the promoters of transcriptional starts that were not included in the model.
The conservation of bases that we observed in our model resembled previous non-information theory based alignments (11), mutation data (10,11,20), an in vivo selection assay (67), and the −10 sequence logo published previously for a smaller dataset (41). The mutation data of Moyle et al. (68) had a 0.6 correlation coefficient to the predicted individual information for our complete promoter model (data not shown). These results are consistent with observations by Mirny and Gelfand (69) who demonstrated a good correlation between sequence conservation and the number of base contacts a protein makes with DNA.
Creating a model for the −35 was difficult, presumably because many promoters are activated and the activator could take over the sequence conservation from the −35, as proposed by Raibaud and Schwartz (52). To test this hypothesis we scanned the 14 sequences in the E. coli genome reported to be positively activated by Raibaud and Schwartz and determined which −35 was strongest in the 10 bp window they allowed. In contrast to the 4.0 ± 0.1 bits in our −35 model, these −35 sequences in activated promoters were no more than 1 ± 4 bits. The weak conservation of positively activated sites probably does explain why creating a −35 model is difficult. To our knowledge this is the only published dataset of confirmed positively activated E. coli promoters.
The extended minus 10
Initiation has been shown to occur in the absence of a −35 in conjunction with an extension to the −10 region (13,14). During our refinement process, a subset of 138 promoters did not have a predicted −35 binding site, and these were subsequently removed from the flexible basal model. A sequence logo revealed that the removed subset of promoters contained a weakly conserved TG upstream of the −10 hexamer, in the region identified as the extended −10. We therefore did a cyclic refinement of the two bases containing the weakly conserved TG, and a well-conserved extended −10 emerged (Figure 3). There was no conservation of bases observed outside of the range −4 to +4. Interestingly, the new bases in the −10 appear to follow the sine wave (41), and this region is protected, suggesting that it is bound in the major groove. The Rsequence for the extended −10 over the range of −4 to +4 bases is 6.74 ± 0.25 bits, almost the same value as the total Rsequence for the flexible σ70 model, which includes the −35 and the gap surprisal. There was also a slight increase in the strength of the −10 hexamer (−1 to +4) from 4.78 ± 0.11 bits in the flexible model to 5.05 ± 0.23 bits in the extended −10 model.
The relationship between the promoter and the ribosome binding site
In order to determine if there are any spacing preferences between the zero coordinate of the −10 and the translational initiation codon, we scanned our σ70 model upstream of all 4122 annotated genes in E. coli (56). We saw one substantial peak in the spacing histogram of predicted promoters, ∼30–40 bases upstream of the ATG (Figure 4). We also plotted the distance between the −10s of the experimentally verified transcription starts and their corresponding translational start codons, which gave a similar peak around 35 bases (Figure 4). In both plots, promoters were predicted as far as 200 bases upstream of the translational start. Similar results were obtained by Huerta and Collado-Vides (70).
For all 401 promoters in our model, we did not see any correlation between promoter strength and the strength of its downstream ribosome binding site as measured by the individual information contents of each flexible model (data not shown). We did find that the lowest combined individual information of a promoter and an RBS was 3.49 bits.
Transcriptional regulation
We used the σ70 model in conjunction with transcriptional regulator models to study promoter structures in non-basal conditions. As an example, we show the experimentally verified Fur-controlled gene tonB (71) and the degree to which Fur represses it (Figure 5, Z. Chen, K.A. Lewis, R.K. Shultzaberger, I.G. Lyakhov, M. Zheng, B. Doan, G. Storz and T.D. Schneider, manuscript in preparation). The results are conveniently displayed using sequence walkers, which show the individual contribution of each base to a binding site as the height of a letter, with the scale being in bits (46,48,57). Multiscan found a strong 11.7 bit RNA polymerase site, which was in our model, 8 bases upstream of the experimentally proven transcription start point (72). This is displayed as two sequence walkers, one for the −35 and one for the −10. To show that they are part of the same promoter, a dashed line is shown connecting the zero coordinates of each walker. Over this site there were two Fur binding sites (8.7 and 11.5 bits), one of which shows clear sequence competition with the −35 sequence walker (red box). That is, both sequence walkers show positive contributions of bases in the same major groove, so binding by σ70 and the dimeric Fur protein cannot occur simultaneously. This is a good example of how these models can show not only the affinity of the promoter and its competing repressor, but also the mechanism of transcriptional regulation. The translational start is 43 bases downstream of the −10, close to the optimal spacing indicated by our data (Figure 4).
As a second example, we used the transcriptional regulator Lrp (40), which can both activate and repress transcription in E. coli (Figure 6). As with Fur at tonB (Figure 5), the sequence walkers readily show that Lrp repression of the dad operon (73–75) is probably caused by an occlusion of the promoter by the repressor in the −10 region (Figure 6a).
Since there are at least seven identified transcription start points for the dad operon (74,75), we used our model to see if we could identify the corresponding promoters. The three most downstream starts marked in Figure 6a (transcripts 4, 5 and 7) presumably use the same promoter (total information 6.3 bits). These starts are 8, 11 and 13 bases from the −10, respectively, which are all reasonable distances between the −10 and transcription start (Figure 1). The next upstream start (transcript 3) is clearly the result of the binding of a 6.2 bit promoter 10 bases upstream. Transcript 6 may be initiated by a 3.0 bit promoter only 6 bases upstream, and transcript 2 is probably initiated by a 5.6 bit promoter 8 bases upstream (data not shown). Our model did not identify a potential promoter upstream of the start at transcript 1, suggesting that initiation at this point is stabilized by other accessory molecules; likely candidates are CRP (there is a 15.3 bit site 44 bases upstream of transcript 1) and Lrp (75).
Interestingly, the computed strength of the dad promoters increases as they get closer to the gene start point but this effect is not observed in arcA (data not shown), which also has seven verified transcript starts (70). Two bound Lrp molecules (11.1 and 11.7 bits) could block the binding and initiation of the two downstream dad promoters and four subsequent downstream transcripts (3, 4, 5, 7), and possibly prevent the opening of transcript 6. The most downstream promoter (transcript 7) is 38 bases away from the translational start point, which produces a transcript only a few bases longer than needed to contain the conserved Shine–Dalgarno region (34), showing an optimization of cellular resources by minimizing the length of mRNAs. We predicted Lrp binding in the region protected by the upstream footprint, but the site was relatively weak at 1.5 bits (data not shown).
Lrp activation of the gltBDF (76) operon can also be predicted using sequence walkers (Figure 6b). In this instance, a strong Lrp-binding site (11.2 bits) is just upstream of a strong promoter (5.3 + 7.6 − 3.3 = 9.6 bits). Since bits are additive, we suggest that the upstream Lrp site increases the overall affinity of the initiation complex for the promoter, giving a total information that could be as high as 20.8 bits. As mentioned in Materials and Methods, this value is an upper bound on the contribution to promoter binding by Lrp through the αCTD.
Based on the two examples in Figure 6, we propose that Lrp is stabilizing the αCTD (12) and promoting initiation when bound upstream of the σ70 binding site, but repressing initiation by occlusion when overlapping the σ70 binding site.
Besides being able to dissect well-understood genetic control systems, we would like to be able to predict new ones for testing. Using a Fur model and the flexible sigma model, we identified a number of potential Fur repressed genes in the E. coli genome. We report two of these cases here (Figure 7). For yoeA (Figure 7a), two strong Fur binding sites (27.4 and 9.0 bits) overlap an average promoter (6.4 bits). The −10 is located 48 bases upstream of the translational start point. A similar result is seen with the fhuA gene (Figure 7b), where two strong Fur sites (10.7 and 19.5 bits, confirmed by gel shift experiments, data not shown) overlap an above-average promoter (7.3 bits) (77,78). The −10 is located 29 bases upstream of the fhuA coding region.
Where DNA bending proteins bind relative to promoter components
We searched for the relative placement of transcriptional regulators near the starts of all of the promoters in our model (Figure 8). We present data for models of Fis (39), H-NS (59) and IHF (41,58). All three proteins are transcriptional activators that are involved in chromosomal compaction (79). We plotted the number of sites predicted at each position relative to the transcription start point, the −10 and the −35 in order to determine which component the regulators were primarily interacting with. We observed in these graphs that there tends to be a peak in the range of −300 to +100, maximizing at the −35 and the −10 alignments, but not at the transcription start. In all cases, fewer sites were predicted downstream than upstream of the promoter. Although these curves are not necessarily linear, to quantify this we did a linear regression for the regions −400 to 0 and from 0 to +200. The slope of the −400 to 0 line is always smaller than that for 0 to +200. The intersection of these two lines is consistently to the left of the alignment point for alignment by the start base, but close for the −10 and the −35. This is reasonably consistent with the idea that the transcriptional factors tend to cluster around the −35, as might be expected from the αCTD contact (53). Other proteins (Fnr, Fur, LexA, ArgR, CRP, TrpR and LacI) were also analyzed, but because they have higher information the models predicted fewer binding sites, so their graphs were too noisy to interpret.
We determined the range of non-coding sequences surrounding each promoter component in our dataset of 401 promoters (Figure 1). We counted the number of intergenic regions at various distances from each promoter component. These curves were then normalized to the graphs in Figure 8 so that they fit just above the data, and so that zero occurrences of intergenic regions was matched to the genomic baseline frequency for each protein (Figure 8, red curves). The curves matched fairly well, in that the curves downstream of the components are consistently steeper than the curves upstream of the promoter components.
Previous analysis by Robison et al. had also identified a preference for genetic control elements to bind in intergenic regions (59), but that analysis was not done in reference to the alignment of promoter components.
DISCUSSION
Genetic control systems often consist of multiple binding components with variable distances between them. These variable distances can affect the stability of the binding complex. Our approach is to use experimentally demonstrated binding sites to construct models. Unlike neural networks, this approach avoids the assumption that untested stretches of nucleotide sequence do not contain binding sites, and it sets the model upon a firm foundation. Our model construction uses information theory, which not only allows measurements of the patterns at the binding sites, but can also account for distance preferences on the same quantitative and universal scale of bits (37). We previously used a flexible modeling method for ribosome binding sites (34). That successful application suggested that information theory can be applied to any multi-part binding system where binding is affected by the spacing between components. In this paper we show that the same approach works well to quantify prokaryotic promoters, which have two binding components at approximately −10 and −35 bases from the start of transcription (11).
A σ70 model based on information theory
The amount of sequence conservation in the −10 and in the −35 is fairly low, ∼5 and 4 bits, respectively. As is found for most DNA-binding proteins (41,62), both sequence logos appear to follow a sine wave, which represents the 10.6 base helical twist of B-form DNA (Figure 1). There are unique characteristics to each logo though.
We used the T. aquaticus σA/−35 co-crystal (65) to determine the location of where the σ protein faces the major groove with respect to the−35. Using the average gap distance, this assignment places the major groove of the T–A base pair at position +4 of the −10 on exactly the opposite face of the DNA as the −35 (Figure 1). With this assignment, the well-conserved T at position +4 in the −10 logo exceeds the sine wave. In instances where conservation exceeds 1 bit in the minor groove, DNA distortion or base flipping was proposed (41). DNA breathes, opening base pairs on a millisecond time scale (80). By stabilizing this specific flipped-out base, the polymerase could initiate promoter opening. An enzymatic mechanism to initiate this process was proposed by Dubendorff et al. (81).
In addition, with the exception of +4, the pattern of sequence conservation of the extended −10 follows the sine wave (Figure 3). This effect has been observed in numerous other sequence logos (51,62), and it can be used to precisely assign the location of protein contacts (41,82) (http://www.ccrnp.ncifcrf.gov/~toms/paper/repan3/), so the extended −10 pattern further suggests that the T at +4 faces the polymerase through the minor groove. Because position −1 has predominantly T instead of equiprobable A and T (Figure 1), it is unlikely to be bound by a minor groove contact in B-form DNA (62). This is consistent with our assignment that the major groove side of base −1 faces the polymerase.
Sclavi et al. used hydroxy radical footprinting to look at intermediates in open complex formation (83). They observed that protection at position 0 (−11 in conventional numbering) occurs after protection in the region of +3 to +5 (−8 to −6 in conventional numbering). This is consistent with the T at +4 (−7 in conventional numbering) initiating DNA melting through a base flipping mechanism (41), which would explain why this position appears anomalous in the sequence logo. In contrast, it has been proposed that flipping of the A at position −11 in the minus ten (our number 0) initiates DNA melting to form the open complex (84–90). If this is the case, why is our groove assignment 5 bases (180°) different? One possibility is that the DNA helix could be distorted between the −35 and the −10, which our model does not account for. However, Young et al. showed that a small part of the σ factor and the first 314 amino acids of the β subunit are sufficient to initiate promoter melting, which probably excludes DNA bending or twisting (91). Furthermore, a co-crystal structure of a fork-junction DNA bound to a holoenzyme (7) shows smoothly bent B-form DNA from the −35 to just before the −10. In this structure the extended −10 is contacted in the major groove, consistent with Figure 3. We conclude that DNA distortions are not sufficient to explain the discrepancy. If base −11 is flipping first, then it may be that −7 is bound specifically after promoter opening, but this mechanism is apparently inconsistent with the hydroxyl radical footprinting data (83). We have not found a satisfactory explanation for why our clear groove assignment implies that the flipped base is −7 while previous reports suggest −11.
The amount of conservation in the −35 is fairly weak. The clear absence of sequence conservation in the major groove immediately upstream of the −35 could leave room for activating proteins to bind and to stabilize the polymerase [this is supported by the σA/−35 co-crystal structure (65)]. By interacting with the polymerase near the −35 contact, accessory molecules could make σ70 a much more discriminate binder.
Penotti found that the distance between the human TATA sites and the transcriptional start is variable, with an uncertainty of ∼3 bits (60). He also observed that there are ∼3 bits of information at the start point itself. In other words, the information of the start (Rsequence) is just sufficient for it to be located with respect to the TATA (Rfrequency), which is the smallest known example of this evolutionary principle (37,92) (http://www.ccrnp.ncifcrf.gov/~toms/paper/ev/). Unlike eukaryotic transcription starts, there is a low conservation of bases at the transcription start point of E. coli (0.39 ± 0.06 bits). Because the average gap surprisal between the −10 and the transcriptional start (2.56 ± 0.04 bits) exceeds the information at the start point, we propose that the determination of which base to begin polymerization is influenced more by the detailed path of the RNA through the open complex (7), than by the actual base at the start.
The conventional spacing allowed between the −10 and the −35 only varies by 3 bases (11), but to account for experimental data (10,17,18), we allowed six bases. Most promoters do fall into the traditional three spacing classes, but binding at further spacings seems experimentally and statistically (Figure 2) reasonable since 24% of the promoters have their strongest −35 outside of the three central spacings. The sequence logos for the most common spacings of 22, 23 and 24 are fairly similar, while those for the outside spacings are a little different. This suggests that at extreme spacings the polymerase may be contacting the promoter differently. At a spacing of 25, the least frequently bound spacing class, the most peculiar −10 logo is seen. The −10 at spacing 25 is unique, with a prominent T at position +1 and poorly conserved Ts at positions +4 and −1. Although this anomaly may simply be an artifact of the small sample (n = 25), this spacing could be conformationally awkward for the polymerase, and this may in itself account for the rarity of promoters with a spacing of 25 bases. It has been shown that at higher superhelical densities, the rate of transcription significantly increased for promoters with this spacing (22), suggesting that activity from this spacing may require helical distortions.
The overall low information content of the entire σ70 binding site suggests that the RNA polymerase binds frequently along the genome (70,93). With a total information of 6.48 ± 0.14 bits, σ70 would bind approximately once in every 90 bases in random equiprobable DNA. This would lead to 10 times more transcripts than genes in E. coli. The promiscuous nature of the polymerase may be necessary to allow transcription of many different genes in the genome. The polymerase must bind independently of gene function, so it must be indiscriminate enough to bind to a variety of control regions. This suggests that transcription is frequently influenced not only by the strength of the sigma binding site, but also by regulatory molecules. It is also possible that many small RNAs are generated, as has been discovered recently [66,93,94].
In fragments from E. coli with lengths of 163 ± 24 bp, Kawano et al. found 0.76 promoters in one orientation (93). From this we compute Rfrequency = −log2(2 × 0.76/163 ± 24) = 6.7 ± 0.2 bits per site. This is remarkably close to the value for our model, Rsequence = 6.48 ± 0.14 bits per site, and it shows that, as with other genetic systems, the information in the binding sites is sufficient to locate the sites in the genome (37,92). This quantitatively demonstrates that information theory provides a reasonable basal model of polymerase binding.
Evolutionary implications of the extended minus 10
Surprisingly, we were easily able to isolate 84 promoters that lack a −35 and exhibit an extended −10 (13,14). The information content of extended −10 promoters is almost identical to the information content of the entire σ70 model (6.7 and 6.5 bits, respectively). That is, the information contribution of the −35 hexamer in the flexible promoter (Figure 1) is approximately compensated by a short extension of the −10 (Figure 3). The additional information of the larger conserved region in the −35 is restricted by the information penalty of the gap surprisal. [The gap surprisal accounts for how the variable spacing between the −10 and −35 affects transcription (10,17,18)].
There is a correlation between the amount of conservation within binding sites (the average information content or Rsequence) and the amount of information needed to locate binding sites in the genome (37,92). Also, it appears that the information of a site (or group of sites) relates to the energetics of the system (45). Therefore, since these two promoter classes have a similar information content, we assume that they are equally able to be identified in the genome and to stabilize the polymerase. A single binding element, such as the extended −10, is a much simpler machine to evolve than a two-part flexible binder. The bacteriophage T7 RNA polymerase (95) has only one binding element (96), so having two widely separated parts is not essential for transcription. Therefore, we suggest that in prokaryotes the extended −10 may be an evolutionary predecessor to the modern bipartite promoter. Another possibility is that the bipartite promoter is the evolutionary predecessor of the extended −10, but this does not explain the origin of bipartite promoters.
Although they have a similar amount of information, the single-element promoter (Figure 3) and the more complicated bipartite promoter (Figure 1) have differences in their sequence conservation. Not only is the conservation of the two upstream bases of the extended −10 lost in the bipartite promoter, but there is also a slight decrease in strength of the part of the extended −10 which corresponds to the bipartite −10 (5.05 bits to 4.78 bits, respectively, over the range −1 to +4). The information in the extended −10 that was lost from the bipartite promoter is important for the polymerase to function correctly. That information was apparently reallocated to the −35, so as to produce a promoter that was functionally equivalent to its predecessor. The energy lost by the internal strain of the flexing polymerase is compensated by the additional information of the−35. This is reminiscent of the apparent evolutionary information flow from the exon to the intron sides of both donor and acceptor splice junctions (97), (http://www.ccrnp.ncifcrf.gov/~toms/paper/splice/).
An advantage of having two widely separated binding components may be to increase promoter strength disparities through interactions with transcriptional regulators. By having a larger binding region, there are more spatial opportunities for accessory proteins to affect the initiation complex. As shown in Figures 5 and 6, both the −10 and −35 are targeted by transcriptional repressors, so having two binding elements provides a larger target region within which to evolve repression.
Why would the cell evolve a flexible bipartite binding mechanism? A possible explanation could be that this mechanism allows a polymerase bound to the promoter to sense genomic structure. Indeed, transcriptional initiation has been observed to vary with the superhelicity of the DNA (22,25,98,99). These differences in the rate of transcription could be from differences in the meltability of the promoter or the stability of the closed complex (11,22). Also, the spacing between the −10 and −35 is large, two helical turns of DNA, which increases polymerase sensitivity to the overall structure, since twist or bending effects are amplified over larger distances. Twist and bending strain could affect polymerase contacts at both the −10 and −35, as shown in Figure 2. Therefore, a large flexible bipartite polymerase may have the advantage of higher sensitivity for superhelical regulation, whereas a rigid single-groove binder could be much less sensitive.
The two extra bases of the extended −10 are similar to positions 0 and +1 of the −35 (Figures 1 and 3). In both cases there is a T > G next to a G > T. Both T–A and G–C base pairs have an exposed hydrogen acceptor and donor in the major groove (O4–N6 contact for T–A, O6–N4 contact for G–C) (51), suggesting that the polymerase could contact either base pair at these moieties. Preferences for T > G or vice versa should depend on the exact positioning of amino acid contacts between the polymerase and the base, since these contacts are in slightly different positions on the base. Indeed, according to models constructed by Barne et al. (14), glutamic acids E458 and E585 contact the extended −10 G at −3 and the −35 G at +1, respectively, suggesting a correspondence between these two regions. Furthermore, there is a gap in sequence conservation at position −2 between the −10 hexamer and the additionally conserved bases of the extended −10 (Figure 3). Correspondingly, the amino acids that contact the two bases at −1 and −3, T440 and E458, are separated by 17 amino acids (14). These observations suggest that there are two separable binding elements contacting the −10 region. The similarities between the −35 and the extension of the −10 region suggest that the protein element recognizing the −10 extension was duplicated and then the duplicate merely drifted away from the −10 to form the−35. This is consistent with the structure of the σ factor, which has two parts separated by an extended polypeptide (23).
In comparison to the E. coli σ70 and σ32, both of which appear to contain two helix–turn–helix DNA-binding domains, the σ55 factor produced by bacteriophage T4 for late transcription, contains only one helix–turn–helix motif (1). Correspondingly, the T4 σ55 only recognizes a −10 region which contains about 16.2 bits of information (61). This is close to the 17.6 bits required to locate the 50 known late promoters in the E. coli genome (37). A pared-down RNA polymerase is able to recognize and open an extended −10 (91). These observations are consistent with the hypothesis that −10 recognition evolved first, followed by appearance of the −35.
The relationship between the promoter and the ribosome binding site
The information content of the flexible ribosome binding site (34) is greater than the σ70 model, 9.28 ± 0.06 versus 6.48 ± 0.14 bits, respectively. This most simply suggests that there is often more than one promoter per coding region in the cell as, for example, shown in Figure 6 and suggested by Huerta and Collado-Vides (70). In addition, the information in the promoters is lower than that in ribosome binding sites because many promoters rely on activation. A quantitative estimation of these contributions would require detailed knowledge of the number of transcripts and activator binding sites throughout the genome.
When we scanned our promoter model upstream of all annotated genes in E. coli (56), our model frequently identified sites at a spacing of ∼35 bases between the zero coordinate of the −10 and the first base of the start codon (Figure 4). The peak represents transcripts starting 11 bases downstream from the −10 (Figure 1) to produce an ∼24 bp mRNA leader. Accounting for the 12–14 bases of mRNA inside the polymerase (23,100) this leader leaves ∼10–12 bases exposed. This is just sufficient space to encode for a Shine–Dalgarno (3 bases on the 5′ side), a typical spacer (most commonly 9 bases) and an initiation codon (2 bases on the 3′ side, for a total of 14 bases) (34), and for the ribosome to dock as soon as 0–2 more bases have been synthesized (101). The 35 base spacing allows the earliest possible loading of ribosomes onto the mRNA. There is a gradual decrease in the number of sites upstream of the peak around −35, which suggests a preference for the polymerase to bind close to the translational start.
Transcriptional regulation
Our original dataset of experimentally verified transcription start points was larger than the number of sites in our final model (684 versus 401). The cyclic refinement that removed sites focused the original group down by selecting a subset that is coherent. The excluded sites were weak (information content is <0 bits) compared to the retained subset and are therefore presumably activated or use a different sigma factor. Also, as previously noted, the average information of unregulated promoters is fairly low (6.48 ± 0.14), implying that the polymerase binds frequently throughout the genome. These observations are consistent with the role of regulatory proteins to help stabilize weak promoters.
Intergenic regions have a composition that is different from coding regions, and protein-binding domains have evolved to bind the intergenic regions (59). As shown in Figure 4, the polymerase has a tendency to bind close to the translational start. Regulatory proteins have the same tendency (Figure 8, circles), but this appears to be because the density of non-coding regions varies relative to the promoter (Figure 8, red curves). Interestingly, the steepness of the density curve is greater downstream of the promoter than upstream. This suggests that in each intergenic region, RNA polymerase binding sites tend to be located where they will maximize the amount of upstream regulatory DNA, while minimizing the length of transcribed mRNA. These effects would preserve cellular resources.
The −35 is the most upstream component of the promoter, and it is closest to the αCTD, to which activator proteins bind (12,53). This explains why the alignments shown in Figure 8 are best matched by the −35 and −10 in contrast to the start point of the flexible promoter.
Without DNA bending, activators more than 20 bases upstream would have difficulty binding to the αCTD (53), because at distances shorter than the persistence length (150–200 bp) DNA is like a rigid rod (102). Furthermore, pairs of DNA sites come together most easily when they are ∼300 bp apart (103,104). These physical limits mean that activators would be restricted to be either immediately upstream of the polymerase, as noted previously (105), or at least 300 bases away. DNA bending proteins and curved DNA, which is in the intergenic regions (79,106), loosen these restrictions and allow activators bound within 300 bases upstream of the promoter to function. This explains why Fis, H-NS and IHF are often found within 300 bases of the promoter (Figure 8), as noted previously by Ussery et al. (79). Given that DNA bending proteins exist, intergenic regions could evolve to be <300 bases and still allow activation by proteins further than a few bases upstream of the promoter. This leads to another cellular evolutionary conservation principle in which the cell maximizes potential activator access while minimizing total genome length. Thus the intergenic region distribution follows from the distributions of the DNA bending proteins, and the DNA bending protein locations are in turn a consequence of the persistence length of DNA.
The number of Fis dimers in the cell has been shown to drastically increase in response to nutritional upshifts (107). If a major role of Fis in the cell is to facilitate activation through DNA-bending within the persistence length, then these results show how Fis can act as a powerful global regulator, linking transcription to cellular nutrition (39,79,108).
Our individual information analysis (Figures 5–7) confirms that the mechanism of transcriptional control can be predicted by the spatial position of transcription factors (105). Sequence walkers reveal that proteins whose positions share information with the promoter will probably bind antagonistically, while those which do not interfere with the −35 or −10 may be activators. Quantification of promoter strengths, however, is a difficult task because of the number of components that can be involved in the stabilization, or occlusion, of the initiation complex. Our approach is simple, clear, powerful, and useful in the design and analysis of experiments aimed at understanding genetic control mechanisms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We would like to thank Dmitry Vassylyev for providing atomic coordinates for the closed promoter and Heladia Salgado for proving us with the RegulonDb database. We would also like to thank Brent Jewett, Danielle Needle, Michael Levashov, Aidan Ryan and Pete Rogan for their comments. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Funding to pay the Open Access publication charges for this article was provided by NCI.
Conflict of interest statement. None declared.
REFERENCES
- 1.Horwitz M.S., Loeb L.A. Structure–function relationships in Escherichia coli promoter DNA. Prog. Nucleic Acid Res. Mol. Biol. 1990;38:137–164. doi: 10.1016/s0079-6603(08)60710-2. [DOI] [PubMed] [Google Scholar]
- 2.Gralla J.D. Promoter recognition and mRNA initiation by Escherichia coli E σ70. Methods Enzymol. 1990;185:37–54. doi: 10.1016/0076-6879(90)85006-a. [DOI] [PubMed] [Google Scholar]
- 3.deHaseth P.L., Zupancic M.L., Record M.T., Jr RNA polymerase–promoter interactions: the comings and goings of RNA polymerase. J. Bacteriol. 1998;180:3019–3025. doi: 10.1128/jb.180.12.3019-3025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Browning D.F., Busby S.J. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 5.Burgess R.R., Travers A.A., Dunn J.J., Bautz E.K. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- 6.Gross C.A., Chan C., Dombroski A., Gruber T., Sharp M., Tupy J., Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harb. Symp. Quant. Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- 7.Murakami K.S., Masuda S., Campbell E.A., Muzzin O., Darst S.A. Structural basis of transcription initiation: an RNA polymerase holoenzyme–DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 8.Young B.A., Gruber T.M., Gross C.A. Views of transcription initiation. Cell. 2002;109:417–420. doi: 10.1016/s0092-8674(02)00752-3. [DOI] [PubMed] [Google Scholar]
- 9.Gross C., Lonetto M., Losick R. Bacterial sigma factors. In: McKnight S.L., Yamamoto K.R., editors. Transcriptional Regulation. New York: Cold Spring Harbor Laboratory Press; 1992. pp. 129–176. [Google Scholar]
- 10.Hawley D.K., McClure W.R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McClure W.R. Mechanism and control of transcription initiation in prokaryotes. Annu. Rev. Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 12.Benoff B., Yang H., Lawson C.L., Parkinson G., Liu J., Blatter E., Ebright Y.W., Berman H.M., Ebright R.H. Structural basis of transcription activation: the CAP–αCTD–DNA complex. Science. 2002;297:1562–1566. doi: 10.1126/science.1076376. [DOI] [PubMed] [Google Scholar]
- 13.Keilty S., Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J. Biol. Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- 14.Barne K.A., Bown J.A., Busby S.J., Minchin S.D. Region 2.5 of the Escherichia coli RNA polymerase σ70 subunit is responsible for the recognition of the ‘extended -10’ motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar A., Malloch R.A., Fujita N., Smillie D.A., Ishihama A., Hayward R.S. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an ‘extended minus 10’ promoter. J. Mol. Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 16.Eichenberger P., Dethiollaz S., Buc H., Geiselmann J. Structural kinetics of transcription activation at the malT promoter of Escherichia coli by UV laser footprinting. Proc. Natl Acad. Sci. USA. 1997;94:9022–9027. doi: 10.1073/pnas.94.17.9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandecki W., Reznikoff W.S. A lac promotor with a changed distance between -10 and -35 regions. Nucleic Acids Res. 1982;10:903–912. doi: 10.1093/nar/10.3.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aoyama T., Takanami M., Ohtsuka E., Taniyama Y., Marumoto R., Sato H., Ikehara M. Essential structure of E. coli promoter: effect of spacer length between the two consensus sequences on promoter function. Nucleic Acids Res. 1983;11:5855–5864. doi: 10.1093/nar/11.17.5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dombroski A.J., Johnson B.D., Lonetto M., Gross C.A. The sigma subunit of Escherichia coli RNA polymerase senses promoter spacing. Proc. Natl Acad. Sci. USA. 1996;93:8858–8862. doi: 10.1073/pnas.93.17.8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stefano J.E., Gralla J.D. Mutation-induced changes in RNA polymerase–lac ps promoter interactions. J. Biol. Chem. 1982;257:13924–13929. [PubMed] [Google Scholar]
- 21.Borowiec J.A., Gralla J.D. All three elements of the lac pS promoter mediate its transcriptional response to DNA supercoiling. J. Mol. Biol. 1987;195:89–97. doi: 10.1016/0022-2836(87)90329-9. [DOI] [PubMed] [Google Scholar]
- 22.Aoyama T., Takanami M. Supercoiling response of E. coli promoters with different spacer lengths. Biochim. Biophys. Acta. 1988;949:311–317. doi: 10.1016/0167-4781(88)90157-1. [DOI] [PubMed] [Google Scholar]
- 23.Vassylyev D.G., Sekine S., Laptenko O., Lee J., Vassylyeva M.N., Borukhov S., Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 24.Lim H.M., Lewis D.E., Lee H.J., Liu M., Adhya S. Effect of varying the supercoiling of DNA on transcription and its regulation. Biochemistry. 2003;42:10718–10725. doi: 10.1021/bi030110t. [DOI] [PubMed] [Google Scholar]
- 25.Peter B.J., Arsuaga J., Breier A.M., Khodursky A.B., Brown P.O., Cozzarelli N.R. Genomic transcriptional response to loss of chromosomal supercoiling in Escherichia coli. Genome Biol. 2004;5:R87. doi: 10.1186/gb-2004-5-11-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukashin A.V., Anshelevich V.V., Amirikyan B.R., Gragerov A.I., Frank-Kamenetskii M.D. Neural network models for promoter recognition. J. Biomol. Struct. Dyn. 1989;6:1123–1133. doi: 10.1080/07391102.1989.10506540. [DOI] [PubMed] [Google Scholar]
- 27.Weller K., Recknagel R.D. Promoter strength prediction based on occurrence frequencies of consensus patterns. J. Theor. Biol. 1994;171:355–359. doi: 10.1006/jtbi.1994.1239. [DOI] [PubMed] [Google Scholar]
- 28.GuhaThakurta D., Stormo G.D. Identifying target sites for cooperatively binding factors. Bioinformatics. 2001;17:608–621. doi: 10.1093/bioinformatics/17.7.608. [DOI] [PubMed] [Google Scholar]
- 29.Galas D.J., Eggert M., Waterman M.S. Rigorous pattern-recognition methods for DNA sequences. Analysis of promoter sequences from Escherichia coli. J. Mol. Biol. 1985;186:117–128. doi: 10.1016/0022-2836(85)90262-1. [DOI] [PubMed] [Google Scholar]
- 30.Mulligan M.E., Hawley D.K., Entriken R., McClure W.R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984;12:789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hertz G.Z., Stormo G.D. Escherichia coli promoter sequences: analysis and prediction. Methods Enzymol. 1996;273:30–42. doi: 10.1016/s0076-6879(96)73004-5. [DOI] [PubMed] [Google Scholar]
- 32.O'Neill M.C. Consensus methods for finding and ranking DNA binding sites: application to Escherichia coli promoters. J. Mol. Biol. 1989;207:301–310. doi: 10.1016/0022-2836(89)90256-8. [DOI] [PubMed] [Google Scholar]
- 33.Harley C.B., Reynolds R.P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987;15:2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shultzaberger R.K., Bucheimer R.E., Rudd K.E., Schneider T.D. Anatomy of Escherichia coli ribosome binding sites. J. Mol. Biol. 2001;313:215–228. doi: 10.1006/jmbi.2001.5040. [DOI] [PubMed] [Google Scholar]
- 35.Shannon C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948;27:379–423. 623–656. [Google Scholar]
- 36.Pierce J.R. An Introduction to Information Theory: Symbols, Signals and Noise. NY: Dover Publications, Inc.; 1980. [Google Scholar]
- 37.Schneider T.D., Stormo G.D., Gold L., Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J. Mol. Biol. 1986;188:415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- 38.Rogan P.K., Faux B.M., Schneider T.D. Information analysis of human splice site mutations. Hum. Mutat. 1998;12:153–171. doi: 10.1002/(SICI)1098-1004(1998)12:3<153::AID-HUMU3>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 39.Hengen P.N., Bartram S.L., Stewart L.E., Schneider T.D. Information analysis of Fis binding sites. Nucleic Acids Res. 1997;25:4994–5002. doi: 10.1093/nar/25.24.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shultzaberger R.K., Schneider T.D. Using sequence logos and information analysis of Lrp DNA binding sites to investigate discrepancies between natural selection and SELEX. Nucleic Acids Res. 1999;27:882–887. doi: 10.1093/nar/27.3.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider T.D. Strong minor groove base conservation in sequence logos implies DNA distortion or base flipping during replication and transcription initiation. Nucleic Acids Res. 2001;29:4881–4891. doi: 10.1093/nar/29.23.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salgado H., Santos-Zavaleta A., Gama-Castro S., Millan-Zarate D., Diaz-Peredo E., Sanchez-Solano F., Perez-Rueda E., Bonavides-Martinez C., Collado-Vides J. RegulonDB (version 3.2): transcriptional regulation and operon organization in Escherichia coli K-12. Nucleic Acids Res. 2001;29:72–74. doi: 10.1093/nar/29.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hershberg R., Bejerano G., Santos-Zavaleta A., Margalit H. PromEC: an updated database of Escherichia coli mRNA promoters with experimentally identified transcriptional start sites. Nucleic Acids Res. 2001;29:277. doi: 10.1093/nar/29.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider T.D., Mastronarde D. Fast multiple alignment of ungapped DNA sequences using information theory and a relaxation method. Discrete Appl. Math. 1996;71:259–268. doi: 10.1016/S0166-218X(96)00068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schneider T.D. Theory of molecular machines II. Energy dissipation from molecular machines. J. Theor. Biol. 1991;148:125–137. doi: 10.1016/s0022-5193(05)80467-9. [DOI] [PubMed] [Google Scholar]
- 46.Schneider T.D. Information content of individual genetic sequences. J. Theor. Biol. 1997;189:427–441. doi: 10.1006/jtbi.1997.0540. [DOI] [PubMed] [Google Scholar]
- 47.Rogan P.K., Svojanovsky S., Leeder J.S. Information theory-based analysis of CYP2C19, CYP2D6 and CYP3A5 splicing mutations. Pharmacogenetics. 2003;13:207–218. doi: 10.1097/00008571-200304000-00005. [DOI] [PubMed] [Google Scholar]
- 48.Schneider T.D. Sequence walkers: a graphical method to display how binding proteins interact with DNA or RNA sequences [Erratum (1998) Nucleic Acids Res., 26, 1135.] Nucleic Acids Res. 1997;25:4408–4415. doi: 10.1093/nar/25.21.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider T.D., Stephens R.M. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider T.D. Consensus sequence zen. Appl. Bioinformatics. 2002;1:111–119. [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider T.D. Reading of DNA sequence logos: prediction of major groove binding by information theory. Methods Enzymol. 1996;274:445–455. doi: 10.1016/s0076-6879(96)74036-3. [DOI] [PubMed] [Google Scholar]
- 52.Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu. Rev. Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- 53.Busby S., Ebright R.H. Transcription activation by catabolite activator protein (CAP) J. Mol. Biol. 1999;293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 54.Hengen P.N., Lyakhov I.G., Stewart L.E., Schneider T.D. Molecular flip-flops formed by overlapping Fis sites. Nucleic Acids Res. 2003;31:6663–6673. doi: 10.1093/nar/gkg877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Semsey S., Virnik K., Adhya S. Three-stage regulation of the amphibolic gal operon: from repressosome to GalR-free DNA. J. Mol. Biol. 2006;358:355–363. doi: 10.1016/j.jmb.2006.02.022. [DOI] [PubMed] [Google Scholar]
- 56.Rudd K.E. EcoGene: a genome sequence database for Escherichia coli K-12. Nucleic Acids Res. 2000;28:60–64. doi: 10.1093/nar/28.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider T.D., Rogan P.K. 1999. Computational analysis of nucleic acid information defines binding sites. United States Patent 5867402. [Google Scholar]
- 58.Goodrich J.A., Schwartz M.L., McClure W.R. Searching for and predicting the activity of sites for DNA binding proteins: compilation and analysis of the binding sites for Escherichia coli integration host factor (IHF) Nucleic Acids Res. 1990;18:4993–5000. doi: 10.1093/nar/18.17.4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robison K., McGuire A.M., Church G.M. A comprehensive library of DNA-binding site matrices for 55 proteins applied to the complete Escherichia coli K-12 genome. J. Mol. Biol. 1998;284:241–254. doi: 10.1006/jmbi.1998.2160. [DOI] [PubMed] [Google Scholar]
- 60.Penotti F.E. Human DNA TATA boxes and transcription initiation sites: A statistical study. J. Mol. Biol. 1990;213:37–52. doi: 10.1016/S0022-2836(05)80120-2. [DOI] [PubMed] [Google Scholar]
- 61.Miller E.S., Kutter E., Mosig G., Arisaka F., Kunisawa T., Ruger W. Bacteriophage T4 genome. Microbiol. Mol. Biol. Rev. 2003;67:86–156. doi: 10.1128/MMBR.67.1.86-156.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Papp P.P., Chattoraj D.K., Schneider T.D. Information analysis of sequences that bind the replication initiator RepA. J. Mol. Biol. 1993;233:219–230. doi: 10.1006/jmbi.1993.1501. [DOI] [PubMed] [Google Scholar]
- 63.Seeman N.C., Rosenberg J.M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc. Natl Acad. Sci. USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brodolin K., Zenkin N., Severinov K. Remodeling of the sigma70 subunit non-template DNA strand contacts during the final step of transcription initiation. J. Mol. Biol. 2005;350:930–937. doi: 10.1016/j.jmb.2005.05.048. [DOI] [PubMed] [Google Scholar]
- 65.Campbell E.A., Muzzin O., Chlenov M., Sun J.L., Olson C.A., Weinman O., Trester-Zedlitz M.L., Darst S.A. Structure of the bacterial RNA polymerase promoter specificity σ subunit. Mol. Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- 66.Hershberg R., Altuvia S., Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oliphant A.R., Struhl K. Defining the consensus sequences of E. coli promoter elements by random selection. Nucleic Acids Res. 1988;16:7673–7683. doi: 10.1093/nar/16.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Moyle H., Waldburger C., Susskind M.M. Hierarchies of base pair preferences in the P22 ant promoter. J. Bacteriol. 1991;173:1944–1950. doi: 10.1128/jb.173.6.1944-1950.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mirny L.A., Gelfand M.S. Structural analysis of conserved base pairs in protein–DNA complexes. Nucleic Acids Res. 2002;30:1704–1711. doi: 10.1093/nar/30.7.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huerta A.M., Collado-Vides J. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 2003;333:261–278. doi: 10.1016/j.jmb.2003.07.017. [DOI] [PubMed] [Google Scholar]
- 71.Young G.M., Postle K. Repression of tonB transcription during anaerobic growth requires Fur binding at the promoter and a second factor binding upstream. Mol. Microbiol. 1994;11:943–954. doi: 10.1111/j.1365-2958.1994.tb00373.x. [DOI] [PubMed] [Google Scholar]
- 72.Postle K., Good R.F. DNA sequence of the Escherichia coli tonB gene. Proc. Natl Acad. Sci. USA. 1983;80:5235–5239. doi: 10.1073/pnas.80.17.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhi J., Mathew E., Freundlich M. Lrp binds to two regions in the dadAX promoter region of Escherichia coli to repress and activate transcription directly. Mol. Microbiol. 1999;32:29–40. doi: 10.1046/j.1365-2958.1999.01314.x. [DOI] [PubMed] [Google Scholar]
- 74.Mathew E., Zhi J., Freundlich M. Lrp is a direct repressor of the dad operon in Escherichia coli. J. Bacteriol. 1996;178:7234–7240. doi: 10.1128/jb.178.24.7234-7240.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhi J., Mathew E., Freundlich M. In vitro and in vivo characterization of three major dadAX promoters in Escherichia coli that are regulated by cyclic AMP-CRP and Lrp. Mol. Gen. Genet. 1998;258:442–447. doi: 10.1007/s004380050754. [DOI] [PubMed] [Google Scholar]
- 76.Wiese D.E.,II, Ernsting B.R., Blumenthal R.M., Matthews R.G. A nucleoprotein activation complex between the leucine-responsive regulatory protein and DNA upstream of the gltBDF operon in Escherichia coli. J. Mol. Biol. 1997;270:152–168. doi: 10.1006/jmbi.1997.1057. [DOI] [PubMed] [Google Scholar]
- 77.Tsolis R.M., Baumler A.J., Stojiljkovic I., Heffron F. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J. Bacteriol. 1995;177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Masse E., Vanderpool C.K., Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ussery D., Larsen T.S., Wilkes K.T., Friis C., Worning P., Krogh A., Brunak S. Genome organisation and chromatin structure in Escherichia coli. Biochimie. 2001;83:201–212. doi: 10.1016/s0300-9084(00)01225-6. [DOI] [PubMed] [Google Scholar]
- 80.Leroy J.L., Kochoyan M., Huynh-Dinh T., Guéron M. Characterization of base-pair opening in deoxynucleotide duplexes using catalyzed exchange of the imino proton. J. Mol. Biol. 1988;200:223–238. doi: 10.1016/0022-2836(88)90236-7. [DOI] [PubMed] [Google Scholar]
- 81.Dubendorff J.W., deHaseth P.L., Rosendahl M.S., Caruthers M.H. DNA functional groups required for formation of open complexes between Escherichia coli RNA polymerase and the λ PR promoter. Identification via base analog substitutions. J. Biol. Chem. 1987;262:892–898. [PubMed] [Google Scholar]
- 82.Lyakhov I.G., Hengen P.N., Rubens D., Schneider T.D. The P1 phage replication protein RepA contacts an otherwise inaccessible thymine N3 proton by DNA distortion or base flipping. Nucleic Acids Res. 2001;29:4892–4900. doi: 10.1093/nar/29.23.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sclavi B., Zaychikov E., Rogozina A., Walther F., Buckle M., Heumann H. Real-time characterization of intermediates in the pathway to open complex formation by Escherichia coli RNA polymerase at the T7A1 promoter. Proc. Natl Acad. Sci. USA. 2005;102:4706–4711. doi: 10.1073/pnas.0408218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Helmann J.D., deHaseth P.L. Protein–nucleic acid interactions during open complex formation investigated by systematic alteration of the protein and DNA binding partners. Biochemistry. 1999;38:5959–5967. doi: 10.1021/bi990206g. [DOI] [PubMed] [Google Scholar]
- 85.Fenton M.S., Lee S.J., Gralla J.D. Escherichia coli promoter opening and -10 recognition: mutational analysis of σ70. EMBO J. 2000;19:1130–1137. doi: 10.1093/emboj/19.5.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lim H.M., Lee H.J., Roy S., Adhya S. A ‘master’ in base unpairing during isomerization of a promoter upon RNA polymerase binding. Proc. Natl. Acad. Sci. USA. 2001;98:14849–14852. doi: 10.1073/pnas.261517398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.deHaseth P.L., Tsujikawa L. Probing the role of region 2 of Escherichia coli σ70 in nucleation and maintenance of the single-stranded DNA bubble in RNA polymerase–promoter open complexes. Methods Enzymol. 2003;370:553–567. doi: 10.1016/S0076-6879(03)70047-0. [DOI] [PubMed] [Google Scholar]
- 88.Roy S., Lim H.M., Liu M., Adhya S. Asynchronous basepair openings in transcription initiation: CRP enhances the rate-limiting step. EMBO J. 2004;23:869–875. doi: 10.1038/sj.emboj.7600098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee H.J., Lim H.M., Adhya S. An unsubstituted C2 hydrogen of adenine is critical and sufficient at the −11 position of a promoter to signal base pair deformation. J. Biol. Chem. 2004;279:16899–16902. doi: 10.1074/jbc.C400054200. [DOI] [PubMed] [Google Scholar]
- 90.Heyduk E., Kuznedelov K., Severinov K., Heyduk T. A consensus adenine at position −11 of the nontemplate strand of bacterial promoter is important for nucleation of promoter melting. J. Biol. Chem. 2006;281:12362–12369. doi: 10.1074/jbc.M601364200. [DOI] [PubMed] [Google Scholar]
- 91.Young B.A., Gruber T.M., Gross C.A. Minimal machinery of RNA polymerase holoenzyme sufficient for promoter melting. Science. 2004;303:1382–1384. doi: 10.1126/science.1092462. [DOI] [PubMed] [Google Scholar]
- 92.Schneider T.D. Evolution of biological information. Nucleic Acids Res. 2000;28:2794–2799. doi: 10.1093/nar/28.14.2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kawano M., Storz G., Rao B.S., Rosner J.L., Martin R.G. Detection of low-level promoter activity within open reading frame sequences of Escherichia coli. Nucleic Acids Res. 2005;33:6268–6276. doi: 10.1093/nar/gki928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wassarman K.M., Zhang A., Storz G. Small RNAs in Escherichia coli. Trends Microbiol. 1999;7:37–45. doi: 10.1016/s0966-842x(98)01379-1. [DOI] [PubMed] [Google Scholar]
- 95.Yin Y.W., Steitz T.A. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 96.Schneider T.D., Stormo G.D. Excess information at bacteriophage T7 genomic promoters detected by a random cloning technique. Nucleic Acids Res. 1989;17:659–674. doi: 10.1093/nar/17.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stephens R.M., Schneider T.D. Features of spliceosome evolution and function inferred from an analysis of the information at human splice sites. J. Mol. Biol. 1992;228:1124–1136. doi: 10.1016/0022-2836(92)90320-j. [DOI] [PubMed] [Google Scholar]
- 98.Travers A., Muskhelishvili G. DNA supercoiling—a global transcriptional regulator for enterobacterial growth? Nature Rev. Microbiol. 2005;3:157–169. doi: 10.1038/nrmicro1088. [DOI] [PubMed] [Google Scholar]
- 99.Chen Y.C., Jeng S.T. Binding affinity of T7 RNA polymerase to its promoter in the supercoiled and linearized DNA templates. Biosci. Biotechnol. Biochem. 2000;64:1126–1132. doi: 10.1271/bbb.64.1126. [DOI] [PubMed] [Google Scholar]
- 100.Darst S.A. Bacterial RNA polymerase. Curr. Opin. Struct. Biol. 2001;11:155–162. doi: 10.1016/s0959-440x(00)00185-8. [DOI] [PubMed] [Google Scholar]
- 101.Rudd K.E., Schneider T.D. Compilation of E. coli ribosome binding sites. In: Miller J.H., editor. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 17.19–17.45. [Google Scholar]
- 102.Hagerman P.J. Flexibility of DNA. Annu. Rev. Biophys. Biophys. Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- 103.Halford S.E., Szczelkun M.D. How to get from A to B: strategies for analysing protein motion on DNA. Eur. Biophys. J. 2002;31:257–267. doi: 10.1007/s00249-002-0224-4. [DOI] [PubMed] [Google Scholar]
- 104.Ringrose L., Chabanis S., Angrand P.O., Woodroofe C., Stewart A.F. Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances. EMBO J. 1999;18:6630–6641. doi: 10.1093/emboj/18.23.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Collado-Vides J., Magasanik B., Gralla J.D. Control site location and transcriptional regulation in Escherichia coli. Microbiol. Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bolshoy A., Nevo E. Ecologic genomics of DNA: upstream bending in prokaryotic promoters. Genome Res. 2000;10:1185–1193. doi: 10.1101/gr.10.8.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ball C.A., Osuna R., Ferguson K.C., Johnson R.C. Dramatic changes in Fis levels upon nutrient upshift in Escherichia coli. J. Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Travers A., Schneider R., Muskhelishvili G. DNA supercoiling and transcription in Escherichia coli: the FIS connection. Biochimie. 2001;83:213–217. doi: 10.1016/s0300-9084(00)01217-7. [DOI] [PubMed] [Google Scholar]
- 109.Siebenlist U., Simpson R.B., Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980;20:269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- 110.Roberts C.W., Roberts J.W. Base-specific recognition of the nontemplate strand of promoter DNA by E. coli RNA polymerase. Cell. 1996;86:495–501. doi: 10.1016/s0092-8674(00)80122-1. [DOI] [PubMed] [Google Scholar]
- 111.Althaus E.W., Outten C.E., Olson K.E., Cao H., O'Halloran T.V. The ferric uptake regulation (Fur) repressor is a zinc metalloprotein. Biochemistry. 1999;38:6559–6569. doi: 10.1021/bi982788s. [DOI] [PubMed] [Google Scholar]
- 112.Chen Z., Schneider T.D. Comparative analysis of tandem T7-like promoter containing regions in enterobacterial genomes reveals a novel group of genetic islands. Nucleic Acids Res. 2006;34:1133–1147. doi: 10.1093/nar/gkj511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Blattner F.R., Plunkett G., III, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.