Abstract
Deinococcus radiodurans RNA ligase (DraRnl) is a template-directed ligase that seals nicked duplexes in which the 3′-OH strand is RNA. DraRnl is a 342 amino acid polypeptide composed of a C-terminal adenylyltransferase domain fused to a distinctive 126 amino acid N-terminal module (a putative OB-fold). An alanine scan of the C domain identified 9 amino acids essential for nick ligation, which are located within nucleotidyltransferase motifs I, Ia, III, IIIa, IV and V. Seven mutants were dysfunctional by virtue of defects in ligase adenylylation: T163A, H167A, G168A, K186A, E230A, F281A and E305A. Four of these were also defective in phosphodiester formation at a preadenylylated nick: G168A, E230A, F281A and E305A. Two nick sealing-defective mutants were active in ligase adenylylation and sealing a preadenylylated nick, thereby implicating Ser185 and Lys326 in transfer of AMP from the enzyme to the nick 5′-PO4. Whereas deletion of the N-terminal domain suppressed overall nick ligation and ligase adenylylation, it did not compromise sealing at a preadenylylated nick. Mutational analysis of 15 residues of the N domain identified Lys26, Gln31 and Arg79 as key constituents. Structure–activity relationships at the essential residues were determined via conservative substitutions. We propose that DraRnl typifies a new clade of polynucleotide ligases. DraRnl homologs are detected in several eukaryal proteomes.
INTRODUCTION
The recent identification and characterization of an RNA ligase (DraRnl) from the radiation-resistant bacterium Deinococcus radiodurans raised the prospect that RNA end sealing might be pertinent to bacterial physiology (1). DraRnl is a 342 amino acid polypeptide encoded by the D.radiodurans DRB0094 ORF, which is one of several genes transiently up-regulated during recovery from radiation exposure (2). DraRnl, like all other ATP-dependent RNA/DNA ligases, joins 3′-OH and 5′-PO4 termini via a series of three nucleotidyl transfer steps: (i) the enzyme reacts with ATP to form a covalent ligase-(lysyl-N)-AMP intermediate plus pyrophosphate, (ii) AMP is transferred from ligase-adenylate to the 5′-PO4 end to form a polynucleotide-adenylate intermediate and (iii) an RNA 3′-OH attacks the 5′-PO4 of the polynucleotide-adenylate to seal the two strands via a 3′–5′ phosphodiester bond and release AMP (3–5).
There are two distinct branches of the RNA ligase family, exemplified by bacteriophage T4 RNA ligase 1 (Rnl1) and RNA ligase 2 (Rnl2), respectively (6–8). Whereas Rnl1-like ligases prefer to join single-stranded RNA termini emanating from RNA stems (9), the Rnl2-like ligases display optimal activity in sealing nicks embedded within duplex RNAs or RNA–DNA hybrids (10–12). Initial biochemical characterization of DraRnl highlighted functional similarities to Rnl2, e.g. activity in sealing duplex RNA nicks, but no activity in circularizing short single-stranded RNA substrates (this being the preferred reaction for Rnl1-type ligases) (1). Yet, the primary structure of DraRnl reveals novel features and domain arrangements that confound its classification as either Rnl1-like or Rnl2-like.
T4 Rnl1 (374 amino acids) and T4 Rnl2 (334 amino acids) are composed of two structural domains (7,13). They have an N-terminal nucleotidyltransferase domain, shared with DNA ligases and mRNA capping enzymes, that includes the six peptide motifs (I, Ia, III, IIIa, IV and V) that define the covalent nucleotidyltransferase superfamily (14). Rnl1 and Rnl2 are distinguished from one another by their C-terminal domains, which adopt unique α-helical folds that are unrelated to the OB-fold modules appended to the C-termini of the nucleotidyltransferase domains of DNA ligases and RNA capping enzymes. Available evidence suggests that the biological specificity of polynucleotide ligases is dictated in part by their carboxyl domains. In the case of ATP-dependent DNA ligases and RNA capping enzymes, the C-terminal OB domain is required for the initial step of covalent enzyme nucleotidylation at the lysine of motif I (KxDG) (15–18). T4 Rnl2 can form ligase-AMP in the absence of its C domain (19), but is then unable to execute the composite RNA nick sealing reaction. The Rnl2 C domain is required for nick recognition and catalysis of nick adenylylation, yet it is not required for phosphodiester formation at a preadenylylated nick (19). The remarkable feature of DraRnl is that is has no C domain at all. Rather, it has a distinctive N-terminal module, which is important for strand joining activity, but has no primary structure similarity to any known polynucleotide ligase (Figure 1). The DraRnl N-terminal module is a putative homolog of the OB-fold of phenylalanyl-tRNA synthetases (1).
Figure 1.
DraRnl subfamily of RNA ligases. The amino acid sequence of DraRnl is aligned to the sequences of homologous proteins from S.avermitilis (Sav) and bacteriophage 44RR.8t (44RR). Putative nucleotidyltransferase motifs I, Ia, III, IIIa, IV and V are highlighted in shaded boxes. The essential motif I lysine nucleophile is denoted by |. Residues subjected to mutational analysis in the present study are denoted by •. The translation start site of the N-terminal deletion mutant NΔ126 is indicated by an arrow above the sequence.
DraRnl is a template-directed RNA ligase capable of sealing nicks in which the 3′-OH strand is RNA. DraRnl can join a RNAOH end to a 5′-pRNA or 5′-pDNA-strand, but it is unable to join when the 3′-OH strand is DNA (1). In light of this specificity, it is conceivable that DraRnl contributes to radiation resistance by either repairing broken RNAs or by sealing broken DNAs that have acquired 3′-OH RNA termini by ribonucleotide addition (perhaps as a stop-gap measure during double-strand break repair). Unlike other polynucleotide ligases, DraRnl relies on manganese (not magnesium) as a cofactor for strand sealing (though either Mg or Mn can support the formation of the DraRnl-AMP intermediate). Manganese exerts unique effects on Deinococcus growth, metabolism and radiosensitivity (20,21). Manganese is associated with the Deinococcus genome (22) and extracellular manganese concentration affects genome condensation (23).
DraRnl homologs are found in the proteomes of Streptomyces avermitilis, Hahella chejuensis and the Aeromonas phage 44RR2.8t. A primary structure alignment indicates that they recapitulate the distinctive features of DraRnl. They have a conserved N-terminal module not found in other ligase clades and they terminate ∼30 amino acids downstream of the presumptive counterpart of nucleotidyltransferase motif IV (305EGVVV309 in DraRnl; see Figure 1). DraRnl and its cousins have an obvious counterpart of motif I that contains the lysine to which AMP becomes covalently attached. The motif I Lys165 side chain of DraRnl is essential, insofar as its replacement by alanine abolished RNA nick sealing and ligase adenylylation activities (1). The sequence alignment in Figure 1 highlights the location of candidate DraRnl equivalents of several other nucleotidyltransferase motifs.
Here, we address the following questions. What other residues of the DraRnl nucleotidyltransferase domain are essential for sealing? At which step of the ligation reaction do the essential residues act? How does the distinctive N domain of DraRnl promote RNA sealing? Are specific residues in the N domain critical for activity? Answering these questions provides new insights to the evolution of polynucleotide ligases.
MATERIALS AND METHODS
Recombinant DraRnl
pET plasmids encoding His10-tagged versions of wild-type (WT) DraRnl and the NΔ126 deletion variant were described previously (1). Alanine and conservative amino acid substitution mutations were introduced by site-directed mutagenesis. Oligodeoxynucleotide primers encoding the desired mutation were annealed to plasmid pET-DraRNL and extended by PfuTurbo DNA polymerase. The template DNA was digested with DpnI and the nicked circular plasmids were transformed into Escherichia coli XL1 Blue. The DraRNL ORFs of all mutant plasmids were sequenced completely to exclude the acquisition of unwanted changes. The plasmids were then transformed into E.coli BL21(DE3). Cultures (0.5 l) of E.coli BL21(DE3)/pET-DraRNL were grown at 37°C in Luria–Bertani (LB) medium containing 0.1 mg/ml ampicillin until the A600 reached ∼0.6. The cultures were chilled on ice for 30 min, adjusted to 0.1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) and 2% ethanol, and then incubated at 17°C for 16 h with continuous shaking. Cells were harvested by centrifugation and the cell pellet was stored at −80°C. All subsequent procedures were performed at 4°C. Thawed bacteria were resuspended in 30 ml of buffer A [50 mM Tris–HCl (pH 7.5), 0.5 M NaCl and 10% sucrose]. Lysozyme, phenylmethylsulfonyl fluoride (PMSF) and Triton X-100 were added to final concentrations of 1 mg/ml, 1 mM and 0.1%, respectively. The lysates were sonicated to reduce viscosity and insoluble material was removed by centrifugation. The soluble extracts were applied to 1 ml columns of Ni-nitrilotriacetic acid-agarose (Qiagen) that had been equilibrated with buffer A. The columns were washed with 8 ml of the same buffer and then eluted stepwise with 4 ml aliquots of 25, 50 and 200 mM imidazole in buffer B [50 mM Tris–HCl (pH 8.0), 0.5 M NaCl and 10% glycerol]. The polypeptide compositions of the column fractions were monitored by SDS–PAGE. The His10-DraRnl proteins adsorbed to the column and were recovered in the 200 mM imidazole eluates. The eluates were dialyzed against buffer containing 50 mM Tris–HCl (pH 8.0), 200 mM NaCl, 2 mM DTT, 2 mM EDTA, 10% glycerol, 0.1% Triton X-100 and then stored at −80°C. Protein concentrations were determined by SDS–PAGE analysis of serial dilutions of the DraRnl preparations in parallel with serial dilutions of a BSA standard. The gels were stained with Coomassie blue, and the staining intensities of the DraRnl and BSA polypeptides were quantified using a digital imaging and analysis system from Alpha Innotech Corporation.
Nicked duplex ligase substrate
A 12mer DNA-strand (5′-CACTATCGGAAT) was 5′ 32P-labeled using T4 polynucleotide kinase and [γ-32P]ATP, then purified by electrophoresis through a nondenaturing 17% polyacrylamide gel. To form the nicked substrate, a mixture of the radiolabeled 5′-PO4 DNA-strand, a 12mer 3′-OH RNA-strand (5′-CAAUUGCGACCC), and complementary 24mer template DNA-strand (3′-GTTAACGCTGGGGTGATAGCCTTA) was annealed at a molar ratio of 1:5:2 in 150 mM NaCl, 10 mM Tris–HCl (pH 8.0), 1 mM EDTA by incubation for 10 min at 65°C, followed by incubation for 15 min at 37°C and then 30 min at 22°C. The ligase substrate was stored at −20°C and thawed on ice immediately prior to use.
Adenylylated nicked duplex substrate
A preadenylylated 32P-labeled DNA-strand (AppDNA) was prepared by enzymatic adenylylation of 5′ 32P-labeled nicked duplex and gel-purification of the adenylylated strand. A nicked duplex was formed by annealing 0.2 nmol of 5′ 32P-labeled 12mer DNA, 1 nmol of a 3′-deoxy-substituted 18mer RNA [5′-r(UUUAAUCAAUUGCGACC)(dC)], and 0.4 nmol of a complementary 24mer DNA (3′-GTTAACGCTGGGGTGATAGCCTTA) in 120 μl of the annealing buffer described above. The nicked duplex was reacted with 1 nmol of T4 Rnl2-H37A (24) in a 150 μl mixture that had been adjusted to 50 mM Tris–acetate (pH 6.5), 2 mM MgCl2 and 5 mM DTT. The reaction was quenched after 90 s at 22°C by adding 75 μl of 95% formamide, 20 mM EDTA. The mixture was heated at 95°C for 5 min and the products were resolved by electrophoresis through a 40 cm 17% native polyacrylamide gel. The separated 32P-labeled AppDNA and pDNA strands were located by autoradiography. A gel slice containing the AppDNA was excised, and the DNA was eluted by incubating the slice overnight in 250 μl of 10 mM Tris–HCl (pH 8.0) and 1 mM EDTA. The preadenylylated nicked substrate was formed by annealing the radiolabeled AppDNA-strand and the 12mer RNAOH strand to the 24mer DNA template as described above.
Ligation at a 5′-PO4 nick
Reaction mixtures (10 μl) containing 50 mM Tris–acetate (pH 6.5), 5 mM DTT, 10 mM MnCl2, 200 μM ATP, 0.2 pmol 32P-labeled nicked duplex substrate and 15 ng of WT or mutant DraRnl were incubated for 15 min at 37°C. The reactions were quenched by adding 10 μl of 20 mM EDTA, 95% formamide. The samples were heated for 5 min at 95°C and then analyzed by electrophoresis through a 14 cm 17% polyacrylamide gel containing 7 M urea in 45 mM Tris–borate and 1 mM EDTA. The extent of ligation, calculated as 24mer/(12 + 24mer), was quantified with a Fuji BAS-2500 imaging apparatus. WT DraRnl sealed 55% of the input pDNA-strand under these conditions. The extents of ligation by the mutant proteins were normalized to the WT level (redefined as 100%).
Adenylyltransferase assay
Reaction mixtures (10 μl) containing 50 mM Tris–HCl (pH 9.0), 10 mM MgCl2, 5 mM DTT, 50 μM [α-32P]ATP and 1 μg of WT or mutant DraRnl were incubated for 15 min at 37°C. The reactions were quenched with SDS, and the products were analyzed by SDS–PAGE. The DraRnl-32P AMP adduct was quantified with a Fuji BAS-2500 imaging apparatus. As reported previously (1), ∼8% of the input WT DraRnl was labeled in vitro with 32P AMP. The extents of adenylylation of the mutant proteins were normalized to the WT level (redefined as 100%).
Ligation at a preadenylylated nick
Reaction mixtures (10 μl) containing 50 mM Tris–acetate (pH 6.5), 5 mM DTT, 10 mM MnCl2, 0.2 pmol 32P-labeled preadenylylated nicked duplex substrate and 30 ng DraRnl were incubated at 22°C for 20 s. The reactions were quenched by adding 10 μl of 20 mM EDTA in 95% formamide. The samples were heated for 5 min at 95°C and then analyzed by electrophoresis through a 14 cm 8% polyacrylamide gel containing 7 M urea in 45 mM Tris–borate, 1 mM EDTA. The extent of ligation, calculated as 24mer/(AppDNA + 24mer), was quantified with a Fuji BAS-2500 imaging apparatus. The extents of ligation by the mutant proteins were normalized to the WT level (redefined as 100%).
RESULTS AND DISCUSSION
Effects of alanine substitutions in the nucleotidyltransferase domain
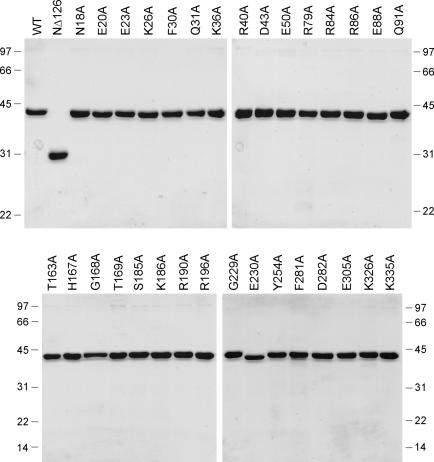
Inspection of the C-terminal domain of DraRnl revealed putative counterparts of nucleotidyl transferase motifs I, III and IV (Figure 1) and two plausible candidates each for motifs Ia, IIIa and V. In order to identify, which residues are critical for DraRnl activity, and thereby distinguish the real catalytic motifs from the ‘impostors’, we performed an alanine scan of the 16 residues indicated by ‘•’ in Figure 1. The DraRnl-Ala proteins were produced in E.coli as His10-tagged fusions and purified by Ni-agarose chromatography (Figure 2).
Figure 2.
DraRnl-Ala and NΔ126 mutants. Aliquots (3 μg) of the nickel-agarose preparations of WT DraRnl, the NΔ126 mutant, and the indicated alanine mutants of DraRnl were analyzed by SDS–PAGE. The Coomassie blue stained gels are shown. The positions and sizes (kDa) of marker polypeptide are indicated on the left and right.
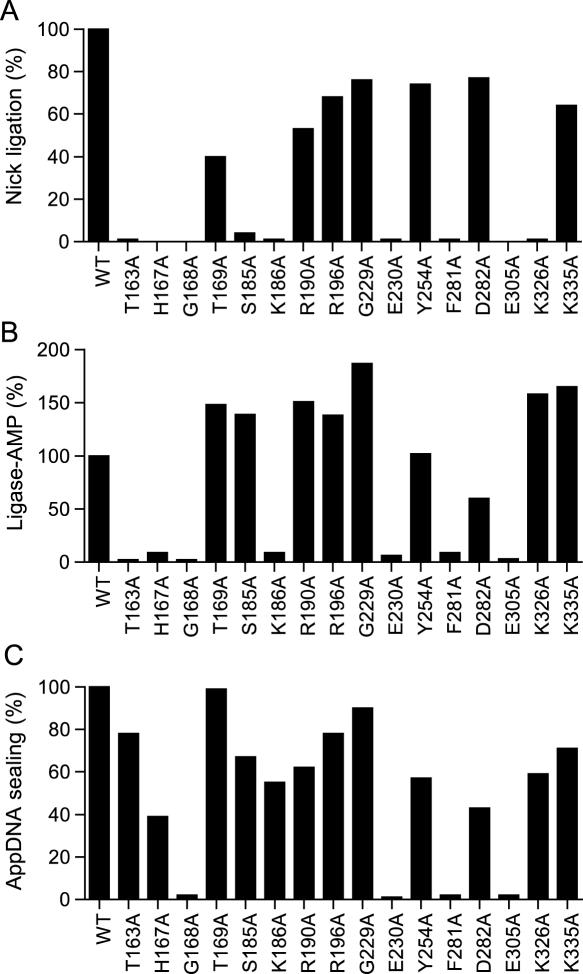
The WT and mutant DraRnl proteins were assayed in parallel for their ability to seal a singly nicked duplex substrate composed of an unlabeled 12mer RNA 3′-OH strand and a 5′ 32P-labeled 12mer DNA-strand annealed to a complementary 24mer DNA template strand. Ligation was evinced by conversion of the input labeled 12mer substrate oligonucleotide to a 24mer product (1). The extents of ligation by the Ala mutants were normalized to the WT value (Figure 3A). We deemed an amino acid to be important when its replacement by alanine resulted in at least a 10-fold decrement in activity relative to WT DraRnl. By this criterion, nine residues were important for strand sealing: Thr163 (1% of WT), His167 (<1%), Gly168 (<1%), Ser185 (4%), Lys186 (1%), Glu230 (1%), Phe281 (1%), Glu305 (<1%) and Lys326 (1%). Six other residues were judged to be non-essential (Thr169, Arg190, Arg196, Gly229, Tyr254, Asp282 and Lys335) because the respective Ala mutants retained between 40 and 80% of WT nick sealing function.
Figure 3.
Effect of alanine mutations in the nucleotidyltransferase domain. (A) Nick sealing. The extents of RNAOH/pDNA ligation by DraRnl mutants were normalized to the WT value. (B) Adenylyltransferase activity. The extents of ligase-AMP formation by the DraRnl mutants were normalized to the WT value. (C) Phosphodiester formation at a preadenylylated nick. The extents of RNAOH/AppDNA sealing by DraRnl mutants were normalized to the WT value (120 fmol sealed).
The DraRnl-Ala mutants were also assayed for adenylyltransferase activity (step 1 of the ligation pathway) by incubating the enzyme in the presence of [α32P]-ATP and a divalent cation and monitoring the transfer of label to the DraRnl polypeptide to form a covalent ligase-AMP adduct. The extents of adenylylation by the Ala mutants were normalized to the WT value (Figure 3B). Seven of the mutants that were dysfunctional in nick ligation were defective in the ligase adenylylation reaction: T163A (2% of WT ligase-AMP formation), H167A (9%), G168A (2%), K186A (9%), E230A (6%), F281A (9%) and E305A (3%). Thus, the defects in nick sealing by these proteins can be attributed to their inability to react with ATP to form the ligase-AMP intermediate. Two of the mutants defective for overall nick sealing retained full-activity in ligase adenylylation: S185A and K326A (Figure 3). We surmise that Ser185 and Lys326 make essential contributions during steps 2 or 3 of the ligation pathway. As expected, all seven of the Ala mutants that retained overall nick sealing activity also retained adenylyltransferase activity: T169A, R190A, R196A, G229A, Y254A, D282A and K355A.
The third step of the ligation reaction (phosphodiester formation) can be studied in isolation by assaying the sealing of a preadenylylated nicked duplex (18,19). The preadenylylated substrate was composed of an unlabeled 12mer RNA 3′-OH strand and a 5′ 32P-labeled 12mer AppDNA-strand annealed to a complementary 24mer DNA template. WT DraRnl reacted with this substrate in the absence of ATP to catalyze phosphodiester bond formation, as evinced by the conversion of the 5′ 32P-labeled AppDNA-strand into a 24mer product. The extents of AppDNA sealing by the Ala mutants were normalized to the WT value (Figure 3C). As expected, the seven Ala mutants that retained 5′-PO4 nick sealing activity also were able to seal the preadenylylated nick. Four residues were required for step 3 catalysis: Gly168 (motif I), Glu230 (motif III), Phe281 (candidate motif IIIa) and Glu305 (motif IV). As these four were also required for step 1, we surmise that Gly168, Glu230, Phe281 and Glu305 are critical globally for ligase activity. The instructive findings were that three of the mutations (T136A, H167A and K186A) that severely impaired overall sealing and ligase adenylylation had little effect on sealing at a preadenylylated nick. Furthermore, we see that the S185A and K326A proteins, which were defective overall but did catalyze step 1, were also able to catalyze step 3 in isolation (Figure 3), implying that Ser185 and Lys326 are essential for step 2 (nick recognition and adenylate transfer to the nick 5′-phosphate). These results underscore that different constellations of amino acids are required for the three component steps of the ligation pathway.
Alanine scanning identifies the nucleotidyltransferase motifs
We have now identified 10 essential DraRnl residues (9 presently and Lys165 previously) that are located within what we surmise are the counterparts of conserved nucleotidyltransferase motifs I (163TEKLHG168), Ia (185SK186), III (225VQIFGEVV232), IIIa (277YEGPFDEAT285), IV (230EGVVV234) and V (324SVK326). The alanine scan appears to have distinguished the ‘real’ motifs Ia, IIIa and V from nearby ‘imposters’ of similar sequence composition. For example, motif Ia is defined by a Ser-Lys or Ser-Arg dipeptide located at the tip of a surface loop. The basic Lys/Arg side chain of motif Ia contacts the γ phosphate of the NTP during step 1 and/or the 5′ phosphate of the polynucleotide during step 2 of the reaction pathway (7,13,15,17,19). The DraRnl segment separating motifs I and III has several candidate motifs Ia: 185SK186, 189SR190 and 195LR196. Our findings that Arg190 and Arg196 are non-essential, while Ser185 and Lys186 are both essential, support the designation of 185SK186 as the genuine motif Ia. Motif V is located just distal to motif IV and contains one or two lysines that contact the α phosphate of the NTP substrate and/or the lysyl-NMP intermediate (7,13,15,17,19,25,26). Of the two candidate conserved lysines situated downstream of DraRnl motif IV (Lys326 and Lys335), only Lys326 is essential for activity. Motif IIIa typically includes an essential aromatic side chain (Phe or Tyr) flanked by an aspartate. The aromatic side chain stacks on the purine ring of the NTP substrate. Of two candidate motif IIIa-like dipeptides in DraRnl—254YD255 and 281FD282—only the latter has a functionally important aromatic side chain.
Structure–activity relationships at essential residues of DraRnl
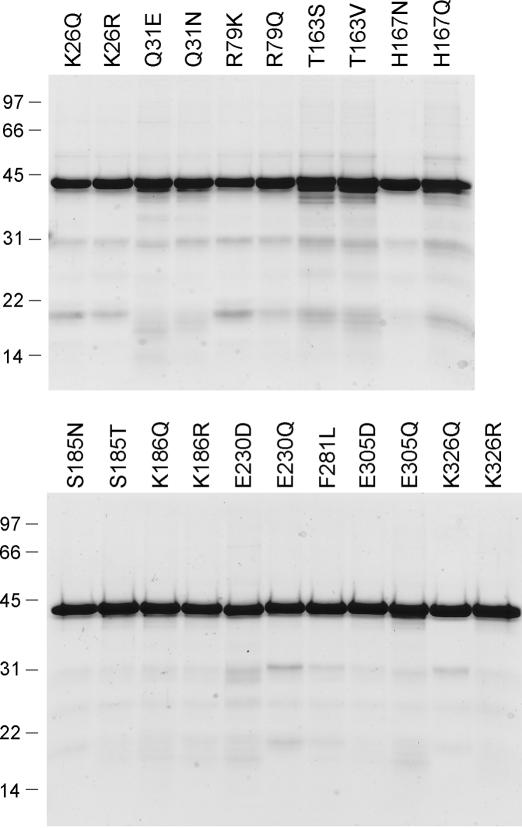
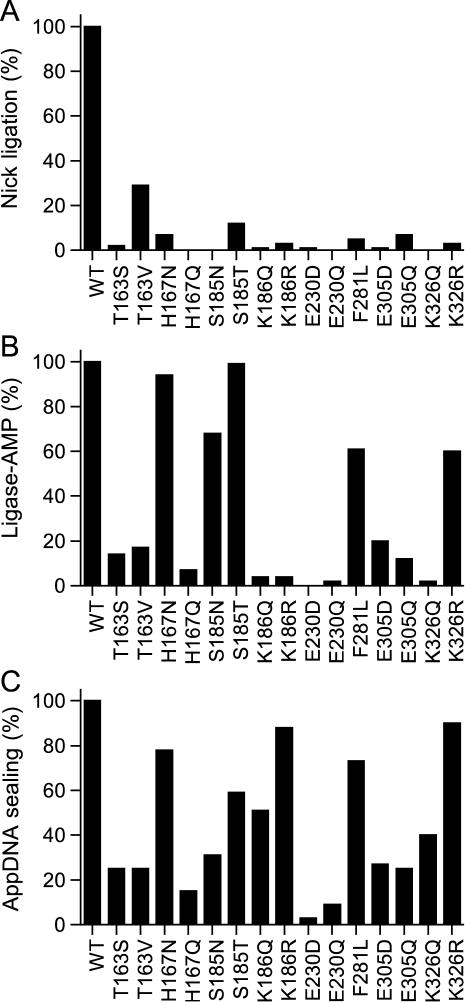
The eight non-glycine residues in the nucleotidyltransferase domain identified presently as essential for DraRnl activity were subjected to conservative substitutions. Lysine was replaced by arginine and glutamine, glutamate by aspartate and glutamine, threonine by serine and valine, histidine by asparagine and glutamine, serine by threonine and asparagine and phenylalanine by leucine. Fifteen conservative mutants were produced in E.coli as His10-tagged fusions and purified by Ni-agarose chromatography (Figure 4). Fourteen of the conservative substitutions reduced nick sealing activity to <10% of the WT level (Figure 5A): T163S (2%); H167N (7%), H167Q (<1%), S185N (<1%), K186Q (1%), K186R (3%), E230D (1%), E230Q (<1%), F281L (5%), E305D (1%), E305Q (7%), K326Q (<1%) and K326R (3%). S185T and T163V displayed 12 and 30% of WT nick ligation, respectively.
Figure 4.
Conservative mutants of DraRnl. Aliquots (3 μg) of the nickel-agarose preparations of the indicated mutants of DraRnl were analyzed by SDS–PAGE. The Coomassie blue stained gels are shown. The positions and sizes (kDa) of marker polypeptides are indicated on the left.
Figure 5.
Effects of conservative mutations in the nucleotidyltransferase domain. (A) Nick sealing. The extents of RNAOH/pDNA ligation by DraRnl mutants were normalized to the WT value. (B) Adenylyltransferase activity. The extents of ligase-AMP formation by the DraRnl mutants were normalized to the WT value. (C) Phosphodiester formation at a preadenylylated nick. The extents of RNAOH/AppDNA sealing by DraRnl mutants were normalized to the WT value (90 fmol sealed).
Effects of the conservative changes on the adenylyltransferase (step 1) and phosphodiester formation (step 3) reactions are shown in Figure 5B and C, respectively. Several classes of mutational effects were noted, whereby the conservative changes either failed to restore activity compared to the Ala mutant, or they selectively restored activity in a subset of the component steps of the pathway. For example, we find that Glu230 (in motif III) is strictly essential for both steps 1 and 3, insofar as the E230Q and E230D mutants were severely defective and phenocopied E230A. These results indicate that the carboxylate is required (i.e. the isosteric amide of glutamine is ineffective) but not sufficient. We infer a strict distance requirement from the main chain to the carboxylate group that is satisfied by glutamate, but not by aspartate. Based on structural and mutational data for other covalent nucleotidyltransferases (7,16,27,28), it is likely that Glu230 of DraRnl coordinates a ribose hydroxyl of ATP during step 1 and of the adenylylated nick during step 3. Replacing Glu305 (in motif IV) with Asp or Gln failed to revive the composite nick sealing activity, but did partially restore function in step 1 (to 17% for E305D and 14% for E305Q) and step 3 (27% for E305D and 25% for E305Q). Thus, the overall defects of the E305D and E305Q proteins could either reflect additive effects on steps 1 and 3, or a stringent requirement for Glu305 during step 2. Structural and mutational studies of other nucleotidyltransferases implicate the motif IV glutamate in binding the divalent cation cofactor, either directly or via water (7,13,17).
Whereas Lys186 (in motif Ia) is strictly essential for ligase adenylylation (i.e. K186R and K186Q phenocopied K186A at this step), it is not required for step 3. The K186Q and K186R mutants resembled the Ala mutant in their ability to seal the preadenylylated nicked substrate. We ascribe the requirement for motif Ia Lys186 in ligase adenylylation to a likely interaction of this side chain with the γ phosphate of ATP (13,15). The neighboring motif Ia residue Ser185 is also required for nick sealing; S185T and S185N were 12 and 1% as active as WT, respectively. S185T retained step 1 and step 3 activities similar to S185A, whereas S185N was slightly less active. We surmise that: (i) the serine hydroxyl is essential during step 2, (ii) it makes a critical hydrogen-bonding interaction either to substrate or another constituent of the enzyme and (iii) its function in step 2 is hindered sterically by the extra methyl group of threonine.
Lys326 (in motif V) is strictly essential for nick sealing; neither Gln nor Arg could replace this lysine. Whereas K326R, like K326A, was active in the ligase-AMP formation, the K326Q mutant was defective. Thus, glutamine interferes with adenylylation. The K326R mutant displays WT activity in sealing at a preadenylylated nick (while K326Q is about half as active as K326R). These results suggest that the stringent requirement for lysine is imposed at step 2 of the ligation pathway. Based on structural and mutational studies of other nucleotidyltransferases, we infer that the motif V Lys326 contacts the AMP phosphate during the polynucleotide 5′-adenylylation reaction (7,13,15,25,26).
Replacing the putative motif IIIa Phe281 side chain with leucine (a partial isostere of phenylalanine) caused a severe decrement in nick joining comparable to that of the F281A mutation. Yet, in contrast to alanine, the leucine mutation revived the step 1 and step 3 activities. The results suggest that the aromatic quality of this residue is critical during step 2, whereas an aliphatic γ-branched side chain suffices during steps 1 and 3. The motif IIIa Phe is predicted to comprise part of the adenosine binding pocket, by stacking on the adenine base (14).
The DraRnl motif I (163TEKLHG168) contains three essential residues (Thr163, His167 and Gly168) in addition to the lysine nucleophile for the adenylyltransferase reaction. DraRnl resembles the Rnl2 subfamily in having a histidine 2 amino acids downstream of the motif I lysine. Their signature histidine deviates from the ‘classical’ motif I sequence found in DNA ligases, T4 Rnl1 and RNA capping enzymes, which have an aspartate at the equivalent position. This motif I His/Asp side chain is implicated in binding a divalent cation cofactor (7,13,25). The His167 side chain is required for overall nick sealing by DraRnl. The essentiality of His167 for the DraRnl adenylylation reaction that we observe here is at odds with mutational data for most other covalent nucleotidyltransferases. The equivalent motif I Asp or His side chain is not required for step 1 adenylylation by Chlorella virus DNA ligase (17,29), human DNA ligase 1 (30), Methanobacterium DNA ligase (31), T4 Rnl1 (32), T4 Rnl2 (6), Thermus thermophilus DNA ligase (33), or E.coli LigA (28), but is instead essential during downstream steps of the ligation reactions catalyzed by those enzymes. In the case of RNA capping enzymes, the motif I aspartate, though conserved, is not required for enzyme-guanylylation in vitro or the composite RNA capping reaction in vivo (16,34,35). To our knowledge, DraRnl is only the second instance in which the motif I His/Asp plays an essential role in the attack of lysine on the NTP substrate, the first example being Mycobacterium DNA ligase D (25).
DraRnl adenylyltransferase activity was restored by replacing His167 with asparagine, but not glutamine. Asparagine and glutamine are both partially isosteric with histidine, such that the Asn-Nδ can superimpose on His-Nδ, while Gln-Nɛ can mimic His-Nɛ. We surmise from the conservative mutational effects that His167 Nδ engages in a critical contact, most likely with the divalent cation cofactor for step 1. The H167N mutant is proficient in sealing a preadenylylated nick, which suggests that the same atomic contacts apply during step 3. The fact that H167N displays feeble composite nick sealing activity implies a more stringent requirement for the native histidine during step 2 of the ligation pathway.
The serine change at Thr163 was as deleterious as alanine with respect to overall nick ligation, yet T163S was less active than T163A in the isolated step 3 reaction. Introduction of valine, which is isosteric to threonine but unable to engage in hydrogen-bonding, partially restored overall nick joining (to 30% of WT). Yet, T163V was no better than T163S in its adenylyltransferase and step 3 sealing activities. The structural basis for the requirement for this motif I residue is not clear based on studies of other covalent nucleotidyltransferases. The equivalent residue in T4 Rnl2 is an arginine (Arg33), which is essential for the composite RNA nick joining reaction of Rnl2 and for the adenylyltransferase step (10). Rnl2 Arg33 is proposed to stabilize the fold of the nucleotidyltransferase domain via intramolecular contacts to other amino acids (10).
The N-terminal domain is dispensable for sealing at a preadenylylated nick
Initial characterization of a series of N-terminal deletions of DraRnl established a role for the N domain in overall nick sealing and ligase-AMP formation. Whereas a 126 amino acid N-terminal deletion reduced DraRnl activity by a factor of 10, removal of 136 amino acids abolished its sealing and adenylylation activity (1). Here we queried the impact of the NΔ126 truncation on phosphodiester formation at a preadenylylated nick. The His-tagged NΔ126 version of DraRnl was produced in bacteria and purified by Ni-agarose chromatography (Figure 2). NΔ126 was 8% as active as full-length DraRnl in overall nick ligation (Figure 6A) and generated 4% as much ligase-AMP intermediate (Figure 6B). The instructive finding was that NΔ126 was twice as active as WT DraRnl in sealing at a preadenylylated nick (Figure 6C). Thus, we conclude that the adenylyltransferase domain suffices for phosphodiester synthesis. The increase in step 3 activity accompanying the deletion of the N-terminal domain of DraRnl agrees with previous findings that deleting the C-terminal domain of T4 Rnl2 enhances its step 3 activities (19).
Figure 6.
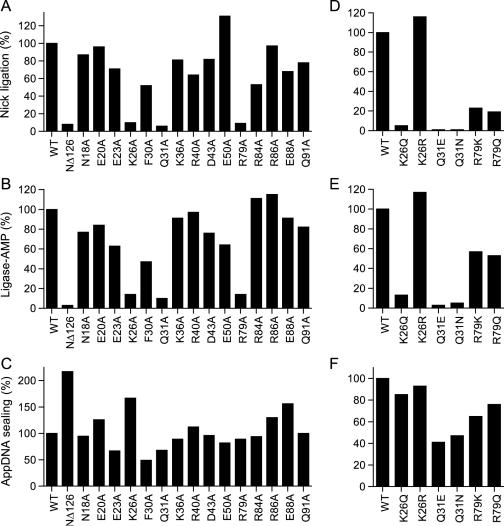
Mutational analysis of the N-terminal domain of DraRnl. (A and D) Nick sealing. The extents of RNAOH/pDNA ligation by DraRnl mutants were normalized to the WT value. (B and E) Adenylyltransferase activity. The extents of ligase-AMP formation by the DraRnl mutants were normalized to the WT value. (C and F) Phosphodiester formation at a preadenylylated nick. The extents of RNAOH/AppDNA sealing by DraRnl mutants were normalized to the WT values (120 and 90 fmol sealed in C and F, respectively).
Alanine scanning of the N-terminal domain
We performed an alanine scan of 15 residues of the N-terminal domain that are conserved in at least one other DraRnl-like protein, focusing on charged and polar residues as plausible participants in the ligase adenylylation step. The targeted residues (Asn18, Glu20, Glu23, Lys26, Phe30, Gln31, Lys36, Arg40, Asp43, Glu50, Arg79, Arg84, Arg86, Glu88 and Gln91) are denoted by ‘•’ in Figure 1. The 15 DraRnl-Ala proteins were produced in E.coli as His10-tagged fusions and purified by Ni-agarose chromatography (Figure 2). Three of the mutations—K26A, Q31A and R79A—reduced overall nick sealing activity to ∼10% of the WT level, thereby phenocopying the NΔ126 mutant. The twelve other Ala mutants retained nick sealing activity well above the threshold of significance for deeming any of the targeted residues as important for function (Figure 6A). The K26A, Q31A and R79A proteins were also defective in ligase adenylylation (10–14% of WT activity), whereas the 12 other Ala mutations had little effect, or only a modest effect, on the yield of ligase-adenylate (Figure 6B). None of the single alanine mutations resulted in a significant defect in sealing at a preadenylylated nick (Figure 6C). We surmise that Lys26, Gln31 and Arg79 are important constituents of the N-terminal domain of DraRnl.
Structure–activity relationships at Lys26, Gln31 and Arg79
Lys26 was substituted conservatively with arginine and glutamine; Gln31 was replaced with glutamate and asparagine; Arg79 was changed to lysine and glutamine. The purified mutant proteins (Figure 4) were tested for nick sealing activity (Figure 6D), ligase adenylylation (Figure 6E) and sealing at a preadenylylated nick (Figure 6F). Arginine restored full nick sealing and adenylyltransferase function in lieu of Lys26, whereas glutamine phenocopied the alanine mutant. We conclude that a positive charge at position 26 suffices for DraRnl adenylyltransferase activity. Gln31 was strictly essential for function, insofar as Q31E and Q31N were severely defective for nick sealing and ligase-AMP formation, though they remained capable of sealing an adenylylated nick. We observed a partial gain of ligase and adenylyltransferase function when Arg79 was replaced by lysine or glutamine, suggesting that hydrogen bond donation is a key property of this residue. Yet, the finding that arginine is needed for optimal activity hints that Arg79 makes a bidentate hydrogen bond. Of the three functionally relevant side chains in the N domain, only Arg79 is conserved in other DraRnl-like proteins (Figure 1). It is conceivable that Arg79 facilitates step 1 by interactions with the β or γ phosphates of the ATP substrate that assist in positioning the pyrophosphate leaving group apical to the attacking Lys165 nucleophile. This is analogous to the roles in leaving group orientation postulated for the C-terminal OB domains of ATP-dependent DNA ligases and GTP-dependent mRNA capping enzymes (15,17) and the N-terminal Ia domain of NAD+-dependent DNA ligases (26,36). Alternatively, Arg79 might stabilize the fold of the N domain. Lys26 and Gln31 are not conserved, making it less plausible that they play a direct role in substrate binding or catalysis. We speculate that Lys26 and Gln31 are important for the tertiary structure of the N domain.
A new variant in ligase evolution
DraRnl can now be confidently assigned membership in the covalent nucleotidyltransferase superfamily, within which DraRnl and its homologs comprise a novel clade by dint of their unique domain organization. DraRnl has no structural module fused to the C-terminus of the core nucleotidyltransferase domain; instead it has a putative OB-like domain fused to the N-terminus. Although definitive classification of the DraRnl N-terminal domain awaits a crystal structure, an analysis of its primary structure highlights similarity to the B2 domain of the β subunit of bacterial phenylalanyl-tRNA synthetase (1), which does adopt an OB-fold (37). Of the three important residues in the DraRnl N domain, only Arg79 is conserved in the OB domain of phenylalanyl-tRNA synthetases (as a lysine). There is no apparent primary structure similarity between the N domain of DraRnl and the OB domains of DNA ligases and RNA capping enzymes. The novel domain composition and order of DraRnl provides added support for the proposal that ligases evolved from an undifferentiated ‘stand-alone’ adenylyltransferase by incorporating distinct protein modules that conferred biochemical specificity and participation in a particular biological transaction (10).
Candidate DraRnl-like ligases in eukarya
One of the dividends of structure-function analysis is the ability to distinguish the wheat from the chaff when inspecting the large number of low scoring hits that emerge from a Blast search. In particular, segmental similarities within otherwise dissimilar polypeptides take on meaning when they include the functional groups required for enzyme activity. Also, even one or two segments of conservation, if they include active site residues, can be expanded by manual alignments to identify a likely family member. It was in just this manner (mutagenesis and manual sequence realignments) that the nucleotidyltransferase motifs were first identified and the evolutionary connections between DNA ligases, RNA ligases, RNA capping enzymes were established (38). Our presumption is that the DraRnl clade is not limited to bacteria and bacteriophage. Thus, it is noteworthy that we have identified putative DraRnl-type ligases is several eukaryal proteomes, including the fungi Magnaporthe grisea (a 428 amino acid polypeptide; GenBank accession no. XP_367846), Neurospora crassa (408 amino acids; accession no. CAE76396) and Gibberella zeae (338 amino acids, accession no. XP_380758) and the social amoeba Dictyostelium discoideum (434 amino acids; accession no. EAL61744). Each of these hypothetical eukaryal proteins contains an N-terminal module similar to DraRnl (a putative OB-fold) fused to a C-terminal nucleotidyltransferase-like domain. An alignment of DraRnl to the M.grisea polypeptide highlights the conservation of nearly all of the side chains defined presently as essential for the ligase activity of DraRnl (Figure 7). Thus, we posit the existence of a novel family of eukaryal ligases.
Figure 7.
Eukaryal DraRnl homolog. The amino acid sequence of DraRnl (Dra) is aligned to that of a homologous 428 amino acid polypeptide from the fungus M.grisea (Mgr). Positions of side chain identity/similarity are denoted by • above the sequence. The putative equivalents of motifs I, Ia, III, IV and V are highlighted in shaded boxes, as is the aromatic residue in putative motif IIIa.
Acknowledgments
This work was supported by NIH grant GM42498 to S.S. and NIH Fellowship GM076887 to A.R. S.S. is an American Cancer Society Research Professor. Funding to pay the Open Access publication charges for this article was provided by NIH grant GM42498.
Conflict of interest statement. None declared.
REFERENCES
- 1.Martins A., Shuman S. An RNA ligase from Deinococcus radiodurans. J. Biol. Chem. 2004;279:50654–50661. doi: 10.1074/jbc.M407657200. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y., Zhou J., Omelchenko M.V., Beliaev A.S., Venkateswaran A., Stair J., Wu L., Thompson D.K., Xu D., Rogozin I.B., et al. Transcriptome dynamics of Deinococcus radiodurans recovering from ionizing radiation. Proc. Natl Acad. Sci. USA. 2003;100:4191–4196. doi: 10.1073/pnas.0630387100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cranston J.W., Silber R., Malathi V.G., Hurwitz J. Studies on ribonucleic acid ligase. Characterization of an adenosine triphosphate-inorganic pyrophosphate exchange reaction and demonstration of an enzyme-adenylate complex with T4 bacteriophage-induced enzyme. J. Biol. Chem. 1974;249:7447–7456. [PubMed] [Google Scholar]
- 4.Silber R., Malathi V.G., Hurwitz J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc. Natl Acad. Sci. USA. 1972;69:3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugino A., Snoper T.J., Cozzarelli N.R. Bacteriophage T4 RNA ligase. Reaction intermediates and interaction of substrates. J. Biol. Chem. 1977;252:1732–1738. [PubMed] [Google Scholar]
- 6.Ho C.K., Shuman S. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc. Natl Acad. Sci. USA. 2002;99:12709–12714. doi: 10.1073/pnas.192184699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nandakumar J., Shuman S., Lima C.D. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell. 2006;127:71–84. doi: 10.1016/j.cell.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 8.Wang L.K., Ho C.K., Pei Y., Shuman S. Mutational analysis of bacteriophage T4 RNA ligase 1. Different functional groups are required for the nucleotidyl transfer and phosphodiester bond formation steps of the ligation reaction. J. Biol. Chem. 2003;278:29454–29462. doi: 10.1074/jbc.M304320200. [DOI] [PubMed] [Google Scholar]
- 9.Amitsur M., Levitz R., Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nandakumar J., Ho C.K., Lima C.D., Shuman S. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J. Biol. Chem. 2004;279:31337–31347. doi: 10.1074/jbc.M402394200. [DOI] [PubMed] [Google Scholar]
- 11.Nandakumar J., Shuman S. How an RNA ligase discriminates RNA versus DNA damage. Mol. Cell. 2004;16:211–221. doi: 10.1016/j.molcel.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 12.Blanc V., Alfonzo J.D., Aphasizhev R., Simpson L. The mitochondrial RNA ligase from Leishmania tarentolae can join RNA molecules bridged by a complementary RNA. J. Biol. Chem. 1999;274:24289–24296. doi: 10.1074/jbc.274.34.24289. [DOI] [PubMed] [Google Scholar]
- 13.El Omari K., Ren J., Bird L.E., Bona M.K., Klarmann G., LeGrice S.F., Stammers D.K. Molecular architecture and ligand recognition determinants for T4 RNA ligase. J. Biol. Chem. 2006;281:1573–1579. doi: 10.1074/jbc.M509658200. [DOI] [PubMed] [Google Scholar]
- 14.Shuman S., Lima C.D. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Hakansson K., Doherty A.J., Shuman S., Wigley D.B. X-ray crystallography reveals a large conformational change during guanyl transfer by mRNA capping enzymes. Cell. 1997;89:545–553. doi: 10.1016/s0092-8674(00)80236-6. [DOI] [PubMed] [Google Scholar]
- 16.Sawaya R., Shuman S. Mutational analysis of the guanylyltransferase component of mammalian mRNA capping enzyme. Biochemistry. 2003;42:8240–8249. doi: 10.1021/bi034396d. [DOI] [PubMed] [Google Scholar]
- 17.Odell M., Sriskanda V., Shuman S., Nikolov D.B. Crystal structure of eukaryotic DNA ligase-adenylate illuminates the mechanism of nick sensing and strand joining. Mol. Cell. 2000;6:1183–1193. doi: 10.1016/s1097-2765(00)00115-5. [DOI] [PubMed] [Google Scholar]
- 18.Sriskanda V., Shuman S. Mutational analysis of Chlorella virus DNA ligase: catalytic roles of domain I and motif VI. Nucleic Acids Res. 1998;26:4618–4625. doi: 10.1093/nar/26.20.4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho C.K., Wang L.K., Lima C.D., Shuman S. Structure and mechanism of RNA ligase. Structure. 2004;12:327–339. doi: 10.1016/j.str.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 20.Chou F.I., Tan S.T. Manganese(II) induces cell division and increases in superoxide dismutase and catalase activities in an aging deinococcal culture. J. Bacteriol. 1990;172:2029–2035. doi: 10.1128/jb.172.4.2029-2035.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y.M., Wong T.Y., Chen L.Y., Lin C.S., Liu J.K. Induction of a futile embden-meyerhof-parnas pathway in Deinococcus radiodurans by Mn: possible role of the pentose phosphate pathway in cell survival. Appl. Environ. Microbiol. 2000;66:105–112. doi: 10.1128/aem.66.1.105-112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leibowitz P.J., Schwartzberg L.S., Bruce A.K. The in vivo association of manganese with the chromosome of Micrococcus radiodurans. Photochem. Photobiol. 1976;23:45–50. doi: 10.1111/j.1751-1097.1976.tb06769.x. [DOI] [PubMed] [Google Scholar]
- 23.Levin-Zaidman S., Englander J., Shimoni E., Sharma A.K., Minton K.W., Minsky A. Ringlike structure of the Deinococcus radiodurans genome: a key to radioresistance? Science. 2003;299:254–256. doi: 10.1126/science.1077865. [DOI] [PubMed] [Google Scholar]
- 24.Nandakumar J., Shuman S. Dual mechanisms whereby a broken RNA end assists the catalysis of its repair by T4 RNA ligase 2. J. Biol. Chem. 2005;280:23484–23489. doi: 10.1074/jbc.M500831200. [DOI] [PubMed] [Google Scholar]
- 25.Akey D., Martins A., Aniukwu J., Glickman M.S., Shuman S., Berger J.M. Crystal structure and nonhomologous end-joining function of the ligase component of Mycobacterium DNA ligase D. J. Biol. Chem. 2006;281:13412–13423. doi: 10.1074/jbc.M513550200. [DOI] [PubMed] [Google Scholar]
- 26.Gajiwala K.S., Pinko C. Structural rearrangement accompanying NAD+ synthesis within a bacterial DNA ligase crystal. Structure. 2004;12:1449–1459. doi: 10.1016/j.str.2004.05.017. [DOI] [PubMed] [Google Scholar]
- 27.Sriskanda V., Shuman S. Role of nucleotidyltransferase motifs I, III and IV in the catalysis of phosphodiester bond formation by Chlorella virus DNA ligase. Nucleic Acids Res. 2002;30:903–911. doi: 10.1093/nar/30.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu H., Shuman S. Structure-guided mutational analysis of the nucleotidyltransferase domain of Escherichia coli NAD+-dependent DNA ligase (LigA) J. Biol. Chem. 2005;280:12137–12144. doi: 10.1074/jbc.M413685200. [DOI] [PubMed] [Google Scholar]
- 29.Sriskanda V., Shuman S. Chlorella virus DNA ligase: nick recognition and mutational analysis. Nucleic Acids Res. 1998;26:525–531. doi: 10.1093/nar/26.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kodama K., Barnes D.E., Lindahl T. In vitro mutagenesis and functional expression in Escherichia coli of a cDNA encoding the catalytic domain of human DNA ligase I. Nucleic Acids Res. 1991;19:6093–6099. doi: 10.1093/nar/19.22.6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sriskanda V., Kelman Z., Hurwitz J., Shuman S. Characterization of an ATP-dependent DNA ligase from the thermophilic archaeon Methanobacterium thermoautotrophicum. Nucleic Acids Res. 2000;28:2221–2228. doi: 10.1093/nar/28.11.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heaphy S., Singh M., Gait M.J. Effect of single amino acid changes in the region of the adenylylation site of T4 RNA ligase. Biochemistry. 1987;26:1688–1696. doi: 10.1021/bi00380a030. [DOI] [PubMed] [Google Scholar]
- 33.Luo J., Barany F. Identification of essential residues in Thermus thermophilus DNA ligase. Nucleic Acids Res. 1996;24:3079–3085. doi: 10.1093/nar/24.15.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwer B., Shuman S. Mutational analysis of yeast mRNA capping enzyme. Proc. Natl Acad. Sci. USA. 1994;91:4328–4332. doi: 10.1073/pnas.91.10.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cong P., Shuman S. Covalent catalysis in nucleotidyl transfer. A KTDG motif essential for enzyme–GMP complex formation by mRNA capping enzyme is conserved at the active sites of RNA and DNA ligases. J. Biol. Chem. 1993;268:7256–7260. [PubMed] [Google Scholar]
- 36.Sriskanda V., Shuman S. Conserved residues in domain Ia are required for the reaction of Escherichia coli DNA ligase with NAD+ J. Biol. Chem. 2002;277:9695–9700. doi: 10.1074/jbc.M111164200. [DOI] [PubMed] [Google Scholar]
- 37.Goldgur Y., Mosyak L., Reshetnikova L., Ankilova V., Lavrik O., Khodyreva S., Safro M. The crystal structure of phenylalanyl-tRNA synthetase from thermus thermophilus complexed with cognate tRNAPhe. Structure. 1997;5:59–68. doi: 10.1016/s0969-2126(97)00166-4. [DOI] [PubMed] [Google Scholar]
- 38.Shuman S., Schwer B. RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol. Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]