Abstract
GEK1, an Arabidopsis thaliana gene product, was recently identified through its involvement in ethanol tolerance. Later, this protein was shown to display 26% strict identity with archaeal d-Tyr-tRNATyr deacylases. To determine whether it actually possessed deacylase activity, the product of the GEK1 open reading frame was expressed in Escherichia coli from a multi-copy plasmid. Purified GEK1 protein contains two zinc ions and proves to be a broad-specific, markedly active d-aminoacyl-tRNA deacylase in vitro. Moreover, GEK1 expression is capable of functionally compensating in E. coli for the absence of endogeneous d-Tyr- tRNATyr deacylase. Possible connections between exposure of plants to ethanol/acetaldehyde and misaminoacylation of tRNA by d-amino acids are considered.
INTRODUCTION
Several aminoacyl-tRNA synthetases have been reported to have the ability to transfer the d-isomer of their amino acid onto their cognate tRNA. Depending on the synthetase, the rates of such misacylations are 15–2000-fold lower than the rates obtained with the cognate l-amino acids (1,2). However, these rates are high enough to cause accumulation in the cell of metabolically inactive d-aminoacyl-tRNAs (3). To recycle such misacylated tRNA molecules, cells display enzyme activities capable of hydrolyzing the ester bond between the polynucleotide and the d-amino acid. These activities were detected for the first time in Escherichia coli, yeast, rabbit reticulocytes and rat liver by Calendar and Berg (1967) (4). Much later, the E. coli gene encoding deacylase activity was identified (5). Disruption of this gene, called dtd, causes reduced growth of the bacterium on minimal medium containing either d-tyrosine, d-aspartate or d-tryptophan (2,5). Further studies showed that, in a Δdtd context and in the presence of d-tyrosine, nearly half of the pool of cellular tRNA specific of l-tyrosine was esterified with this d-amino acid, thus explaining the impairment of growth (3).
The product of E. coli dtd (DTD1) is called d-aminoacyl-tRNA deacylase or d-Tyr-tRNATyr deacylase. It has been studied in depth (3,6). The reaction catalyzed by DTD1 is highly specific for the d-isomer of the amino acid esterified to tRNA. The enzyme is active with different substrates such as d-Tyr-tRNATyr, d-Asp-tRNAAsp or d-Trp-tRNATrp. Under optimal in vitro conditions, the hydrolysis of d-Tyr-tRNATyr proceeds at a maximal rate of 6 s−1 with a Km value of 1 μM (5).
Homologs of the E. coli dtd gene are recognizable in the genomes of many bacteria and eukaryotes, not in those of archaea. Another type of d-Tyr-tRNATyr deacylase (DTD2) has been discovered in archaea (7). This enzyme displays the fold of a bacterial peptidyl-tRNA hydrolase and carries two firmly bound zinc ions, crucial to the activity (7). Despite marked differences from its bacterial counterpart, the structural gene of the archaeal hydrolase (dtd2) can complement an E. coli Δdtd mutant for resistance to d-tyrosine. Homologs of dtd2 are found in most archaea and in plants.
In Arabidopsis thaliana, the dtd2 homolog, previously named GEK1, has been shown to be involved in ethanol resistance (8,9). Mutants in which GEK1 expression is impaired no longer germinate on a culture medium containing 0.04% ethanol. Mutant seedlings are 10–100-fold more sensitive to ethanol than wild-type plants while overexpression of GEK1 improves the tolerance to ethanol (9). The ethanol hypersensitivity of the gek1 mutants is attributed to enhanced sensitivity to acetaldehyde, a metabolite of ethanol (9). Remarkably, the gek1 mutants do not display any particular behavior in response to anoxic, heat shock or salt stresses (9).
The GEK1 product (GEK1) displays ∼26% strict identity with available archaeal d-Tyr-tRNATyr deacylase sequences (7). Moreover, residues essential to the activity of archaeal deacylases are conserved in GEK1. This strongly suggests that GEK1 carries the activity of a d-aminoacyl-tRNA deacylase. To test this idea, we expressed the GEK1 product in an E. coli context. The purified recombinant protein proves to be a very active d-aminoacyl-tRNA deacylase in vitro. We also establish that expression of GEK1 functionally complements an E. coli Δdtd mutant. Possible metabolic pathways linking the deacylase activity of GEK1 to exposure of plants to acetaldehyde are discussed.
MATERIALS AND METHODS
d-[methylene-3H]tyrosine (211 GBq/mmol) was custom-prepared by Amersham Biosciences. d-[2,3-3H]aspartic acid (1.33 TBq/mmol) was directly purchased at Amersham Biosciences. l-[14C]tyrosine (16 GBq/mmol) was from NEN Life Science Products and unlabeled d-tyrosine was from Sigma.
Strains and plasmids used in this study are listed in Table 1.
Table 1.
Strains and plasmids
| Description | Reference | |
|---|---|---|
| Strains | ||
| E. coli strains | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F′ proAB lacIq lacZΔM15 Tn10) | Stratagene |
| K37 | galK rpsL | (10) |
| K37ΔrecAλDE3 | K37 ΔrecA938::cat (λDE3) | (7) |
| K37ΔrecAΔtyrHλDE3 | K37 ΔrecA938::cat Δdtd::kan (λDE3) | (7) |
| S. cerevisiae strains | ||
| DBY2057 | MATa ura3-52 | (11) |
| DBY2057ΔDTD1 | DBY2057 Δdtd1::kan | (12) |
| Plasmids | ||
| pET3alpa | ApR | (13) |
| pET15blpa | ApR | (13) |
| pET3alpa::GEK1 | ApR GEK1 derivative of pET3alpa | This work |
| pET15blpa::GEK1 | ApR GEK1 derivative of pET15blpa | This work |
Cloning of the GEK1 gene in the pET3alpa and in the pET15blpa vectors
To introduce the coding sequence of GEK1 into the expression vectors pET3alpa and pET15blpa (13), the plasmid harbored by clone S81170 (provided by the Arabidopsis Biological Resource Center, (14)) was used as template in PCR amplifications. In view of the insertion into pET3alpa, the following two primers were chosen for amplification: NdeIGEK1-for (5′-GGGAATTCCATATGGTAACACTAATCGTGGCCACCGCCGATCCAGCGTCGATCAACCCTGC-3′) and NotIGEK1-rev (5′-TTTTCCTTTTGCGGCCGCTCATGTGAAATCGTTTGGCTTCCCA-3′). For the insertion into pET15blpa, primers were: SW1 (5′-CTAGTCTAGACTAGTTTAAGGAGATATACATATGGTAACACTAATCGTGG-3′) and NotIGEK1-rev. The two resulting amplified fragments were purified with the help of the Qiagen PCR purification Kit 50 and digested by either NdeI plus NotI or by XbaI plus NotI. Digestion products were inserted into plasmid pET3alpa or pET15blpa to give plasmids pET3alpa::GEK1 and pET15blpa::GEK1, respectively. In each plasmid, the cloned gene was verified by DNA sequencing.
Preparation of crude extracts
Bacteria were grown at 37°C in 75 ml of 2xTY medium containing 100 μg/ml of ampicillin until the stationary phase of growth. Then, isopropyl-1-thio-β-d-galactopyranoside (IPTG) was added at a final concentration of 1 mM and the culture was further incubated at room temperature for 5 h with agitation. After centrifugation at 9800 g for 15 min at 4°C, bacteria were resuspended in 5 mM potassium phosphate (pH 7.2) containing 160 μM zinc acetate and 10 mM 2-mercaptoethanol (PZM buffer). The volume of this buffer was adjusted to obtain an optical density (OD) at 650 nm of ∼100. Cells were disrupted by sonication (3 min, 0°C) and debris were removed by centrifugation (10 min, 20 600 g, 4°C). Deacylase activity was measured in the supernatant. The total amount of proteins in the extract was determined by using the Bradford protein assay (Biorad), with bovine serum albumin (BSA) as the standard.
Preparation of substrates
E. coli tRNATyr and tRNAAsp were prepared as described previously (2,5,15). tRNATyr was aminoacylated with d-[3H]tyrosine (500 Ci/mol) or l-[14C]tyrosine (434 Ci/mol) as described (2,5) with the following modifications. To synthesize d-Tyr-tRNATyr, the reaction mixture (1 ml) contained 20 mM Tris-HCl (pH 7.8), 7 mM MgCl2, 2 mM ATP, 3.5 μM d-[3H]tyrosine, 3.5 μM tRNATyr and 1.2 μM purified E. coli tyrosyl-tRNA synthetase. l-[14C]Tyr-tRNATyr was prepared with a similar procedure, with the exception that l-[14C]tyrosine, tRNATyr and tyrosyl-tRNA synthetase were added to the reaction mixture at final concentrations of 10, 7 and 0.5 μM, respectively. d-Asp-tRNAAsp was synthetized in a reaction mixture containing 20 mM Tris-HCl (pH 7.8), 7 mM MgCl2, 2 mM ATP, 0.1 mM EDTA, 60 μM d-[3H]aspartic acid (500 Ci/mol), 5 μM tRNAAsp and 6.5 μM purified E. coli aspartyl-tRNA synthetase.
Reaction conditions (10 min, 28°C) allowed nearly full esterification of tRNATyr by d- or l-tyrosine, and 20% esterification of tRNAAsp by d-aspartic acid. Nucleic acids were precipitated with the addition of 2.5 volumes of ethanol and 300 mM sodium acetate (pH 4.8), and recovered by centrifugation (30 min, 20 600 g, 4°C). The pellets were resuspended in 50 μl of 20 mM sodium actetate (pH 4.8) containing 100 mM KCl and 0.1 mM EDTA. Finally, the samples were chromatographed on a Trisacryl GF05 column (0.25 × 16 cm) equilibrated in the same buffer. Recovered d-aminoacyl tRNAs were stored at −20°C.
Diacetyl-l-[14C]Lys-tRNALys (310 Ci/mol) was prepared as described previously (15).
Measurement of deacylation rates
Unless otherwise stated, d-Tyr-tRNATyr hydrolysis by GEK1 was followed under initial rate conditions for 5 min at 28°C in 100 μl assays containing 50 nM [3H]d-Tyr-tRNATyr, 20 mM Tris-HCl (pH 7.8), 5 mM MgCl2, 40 μM zinc acetate, 50 μg/ml BSA and 2.5 mM 2-mercaptoethanol. Prior to its addition to the assay, the sample containing enzyme activity was diluted in 20 mM Tris-HCl (pH 7.8) containing 160 μM zinc acetate, 200 μg/ml BSA and 10 mM 2-mercaptoethanol. The reaction was quenched by the addition of 340 μl ethanol, 14 μl sodium acetate 3 M (pH 4.8) and 20 μl carrier RNA from yeast at 4 mg/ml. Samples were centrifuged (20 min, 20 600 g, 4°C) and the radioactivity in the supernatant was measured by scintillation counting (5). Initial rates of l-[14C]Tyr-tRNATyr, d-[3H]Asp-tRNAAsp or diacetyl-l-[14C]-Lys-tRNALys hydrolysis were measured under the same conditions.
To measure the catalytic constants of purified GEK1 in the reaction of d-Tyr-tRNATyr hydrolysis, initial rates were followed as above in miniaturized (20 μl) assays containing increasing concentrations of the substrate (100–4000 nM) and GEK1 concentrations ranging from 3 to 6 pM. Km and kcat values were derived from iterative non-linear fits of the theoretical Michaelis equation to the experimental values using the Levenberg–Marquardt algorithm (16).
E. coli DTD1 deacylase activity was assayed at 28°C for 5 min in 20 mM Tris-HCl (pH 7.8) containing 5 mM MgCl2, 0.025 mM EDTA, 50 μg/ml BSA and 2.5 mM 2-mercaptoethanol. Prior to its addition to the assay, the enzyme was diluted in 20 mM Tris-HCl (pH 7.8), 0.1 mM EDTA, 200 μg/ml BSA and 10 mM 2-mercaptoethanol.
Pyrococcus abyssi DTD2 deacylase activity was measured at 37°C for 5 min in 20 mM Tris-HCl (pH 7.8) containing 4 mM MgCl2, 40 μM zinc acetate, 50 μg/ml BSA and 2.5 mM 2-mercaptoethanol. Prior to its addition to the assay, the enzyme was diluted in 20 mM Tris-HCl (pH 7.8) containing 160 μM zinc acetate, 200 μg/mL BSA and 10 mM 2-mercaptoethanol.
Purification of GEK1
E. coli strain K37ΔrecAΔtyrHλDE3 was transformed by plasmid pET3alpa::GEK1. Transformed cells were grown at 37°C in 1 l of 2 × TY medium containing 100 μg/ml ampicillin. When the OD650 of the culture reached 1.2, IPTG was added at a final concentration of 1 mM and growth was continued for 5 h at room temperature. Cells were harvested by centrifugation (15 min, 11 300 g, 4°C) and resuspended in PZM buffer containing 0.1 mM phenylmethylsulfonyl fluoride (PMSF). Cells (OD650 = 50) were disrupted by sonication (10 min, 0°C) and debris removed by centrifugation (40 min, 7700 g, 4°C). Nucleic acids were precipitated by the addition of streptomycin (30 mg/ml) to the supernatant. Then, the sample was centrifuged for 20 min at 17 400 g (4°C). The resulting supernatant was brought to 70% ammonium sulfate saturation and centrifuged for 15 min at 17 400 g (4°C). The protein pellet was dissolved in 20 ml of PZM buffer and dialyzed against 3 l of the same buffer. The resulting solution was applied on a column of Q-Sepharose Fast Flow (3.2 × 11.2 cm, Pharmacia) equilibrated in PZM buffer. Elution was performed at a flow rate of 1.3 ml/min with a 1.5 l linear NaCl gradient (0–0.5 M NaCl in PZM buffer). Enzyme activity was recovered between 208 and 272 min of the gradient. Active fractions were pooled and immediately applied on a hydroxylapatite column (1.1 × 10 cm, Biorad) equilibrated in PZM buffer. This column was eluted at a flow of 0.2 ml/min with 100 ml of a linear potassium phosphate buffer gradient (5–500 mM potassium phosphate, pH 7.2, in PZM buffer). Enzyme activity recovered between 165 and 260 min of the gradient was pooled. According to SDS-PAGE analysis, the purified protein was at least 95% homogeneous. The protein was then concentrated by ammonium sulfate precipitation (70% saturation). After centrifugation at 17 400 g for 30 min (4°C), the pellet was dialyzed twice, first against PZM buffer to eliminate ammonium sulfate and second against PZM buffer containing 60% glycerol. The purification procedure yielded 6.6 mg of protein from 1.7 g (wet weight) cells. Storage was at −20°C.
Concentrations of GEK1 were calculated using a molecular weight of 34 726 Da and an extinction coefficient of 1.737 at 280 nm.
Zinc concentration measurements
Zinc content of GEK1 was estimated from mass spectrometry experiments. An aliquot of GEK1 in 70% ammonium sulfate was dialyzed overnight against 10 mM ammonium acetate (pH 7.5) containing 10 μM zinc acetate. Then, zinc measurements were performed on a Bruker APEX III FT-ICR mass spectrometer equipped with a 7.0 T actively shielded magnet and an unmodified Apollo electrospray source, as previously described (7). The mass was determined under native or denaturing (50% acetonitrile and 0.2% formic acid) conditions.
Zinc in GEK1 was also assessed by flame atomic absorption spectroscopy. In this case, prior to zinc analysis, GEK1 was submitted to a size-exclusion chromatography on a TSK SWXL 3000 column equilibrated in a zinc-depleted buffer (20 mM Tris-HCl, pH 7.8, 300 mM KCl), as described previously in the case of an archaeal DTD2 protein (7). The eluted protein was followed through its absorbancy at 280 nm and through measurement of its deacylase activity. Active fractions were submitted to zinc analysis at 213.9 nm, in the peak height mode, with 90 μl injections, using a Varian AA220 spectrophotometer equipped with an air–acetylene burner. Various zinc solutions (0–40 μM) in the above buffer were used as standards.
Measurement of d-tyrosine or d-aspartic acid toxicity
E. coli cells were grown overnight at 37°C in M9-glucose minimal medium containing 100 μg/ml of ampicillin, plus either 2.4 mM d-tyrosine or 11 mM d-aspartic acid. Control experiments without d-amino acid were performed in parallel. Then, bacteria were diluted in the same media containing or not a d-amino acid, to a final OD650 of 0.05 and left to grow at 37°C. The OD650 of the cultures were measured every 90 min, during 540 min, to calculate generation times.
Effect of ethanol and acetaldehyde on the growth of various E. coli and Saccharomyces cerevisiae strains
E. coli strains K37ΔrecAλDE3 and K37ΔrecAΔtyrHλDE3 were plated on M9 minimal medium supplemented with 0.2% glucose and 0–16% ethanol or 0–12% acetaldehyde. Then, cells were left to grow for a week at 37°C. Possible effects of ethanol or acetaldehyde on the growth at 30°C of S. cerevisiae strains DBY2057 and DBY2057ΔDTD1 were searched for in the same manner, except that a yeast nitrogen base supplemented with 50 μg/ml uracil and 2% glucose was used as the growth medium.
RESULTS
Expression of GEK1 in E. coli
To express GEK1 in E. coli, the coding sequence of the GEK1 gene was inserted into either the pET3alpa or the pET15blpa expression vector, under control of the T7 promoter. After introduction of the resulting plasmids in the Δdtd E. coli strain K37ΔrecAΔtyrHλDE3, GEK1 expression was induced by addition of IPTG. Cells grown in a rich culture medium were harvested after 5 h of induction, and deacylase activity in crude cell extracts was measured. Extracts from strains K37ΔrecAλDE3 (dtd+) and K37ΔrecAΔtyrHλDE3 each containing either plasmid pET3alpa or plasmid pET15blpa were also examined (Table 2). In the presence of plasmids pET3alpa::GEK1 or pET15blpa::GEK1, activities in cell extracts against d-Tyr-tRNATyr as substrate were increased by a factor of at least 40 000, compared to the activity of the crude extract from the Δdtd mutant, or by a factor of 4000, compared to the activity of the crude extract from the strain containing the endogeneous E. coli deacylase.
Table 2.
Deacylase activity in crude extracts
| Strain | Deacylase activity (U/mg) |
|---|---|
| K37ΔrecAλDE3 + pET3alpa | 0.9 |
| K37ΔrecAΔtyrHλDE3 + pET3alpa | <0.1 |
| K37ΔrecAΔtyrHλDE3 + pET3alpa::GEK1 | 7700 |
| K37ΔrecAλDE3 + pET15blpa | 1.0 |
| K37ΔrecAΔtyrHλDE3 + pET15blpa | <0.1 |
| K37ΔrecAΔtyrHλDE3 + pET15blpa::GEK1 | 4000 |
Cells were grown overnight at 37°C in 2xTY medium with 100 μg/ml ampicillin. Then, IPTG was added at a final concentration of 1 mM, and growth was continued for 5 h at room temperature. Specific deacylase activities were measured in crude extracts obtained by sonication. Final total protein concentration in the extracts was 10–20 mg/ml. Rates of hydrolysis of d-[3H]Tyr-tRNATyr in the presence of 20 mM Tris-HCl (pH 7.8), 100 nM d-[3H]Tyr-tRNATyr, 5 mM MgCl2, 40 μM zinc acetate, 50 μg/ml BSA and 2.5 mM 2-mercaptoethanol were measured as described in Materials and Methods. One unit corresponds to the enzyme activity capable of hydrolyzing 1 pmol of d-[3H]Tyr-tRNATyr per min under the above assay conditions.
These results establish that, in an E. coli context, a protein with d-Tyr-tRNA hydrolase activity can be produced from the open reading frame of GEK1.
Activity of purified GEK1 protein
To precisely measure the deacylase activity of GEK1, this protein was purified to homogeneity. Under initial rate conditions (28°C, 20 mM Tris-HCl, pH 7.8, 5 mM MgCl2, 50 nM d-[3H]Tyr-tRNATyr, 40 μM zinc acetate, 50 μg/ml BSA, 2.5 mM 2-mercaptoethanol and 3–6 pM of enzyme), the activity of the purified GEK1 protein was 12.7 s−1.
Previous experiments showed that ionic strength stimulated the activities of E. coli and P. abyssi d-Tyr-tRNATyr deacylases, probably through the folding of the tRNA 3D structure (5,7). In order to determine the effect of ionic strength on the activity of GEK1, initial rates of d-Tyr-tRNATyr hydrolysis were measured in the presence of various MgCl2 and KCl concentrations. Addition of one or the other of these salts strongly stimulated the deacylase activity (Table 3). At optimal concentrations of MgCl2 or KCl (5 and 50 mM, respectively), the deacylase activity was increased by factors of ∼10 and ∼8, respectively, with respect to assay conditions without any added salt. The simultaneous addition of MgCl2 and KCl did not increase further the reaction rate. On the contrary, addition of KCl in the presence of the optimal MgCl2 concentration (5 mM) led to a diminution of the deacylase activity. Such experiments suggest that ionic strength improves the rate of the reaction through formation of a native tRNA structure. At excessive ionic strength, the enzyme activity becomes impaired probably through inhibition of the binding of the substrate.
Table 3.
Activity of GEK1 under various assay conditions
| Added component | Initial rate (s−1) |
|---|---|
| None | 1.3 |
| 3 mM MgCl2 | 12.1 |
| 5 mM MgCl2 | 12.7 |
| 7 mM MgCl2 | 11.3 |
| 10 mM MgCl2 | 8.3 |
| 15 mM MgCl2 | 3.7 |
| 20 mM MgCl2 | 3.4 |
| 20 mM KCl | 5.6 |
| 50 mM KCl | 10.4 |
| 100 mM KCl | 9.1 |
| 200 mM KCl | 2.7 |
| 400 mM KCl | 0.3 |
| 5 mM MgCl2 + 20 mM KCl | 9.6 |
| 5 mM MgCl2 + 50 mM KCl | 6.1 |
| 5 mM MgCl2 + 100 mM KCl | 3.1 |
| 5 mM MgCl2 + 200 mM KCl | 1.2 |
| 5 mM MgCl2 + 400 mM KCl | 0.1 |
| 40 μM zinc acetate | 1.7 |
| 250 μM EDTA | 1.3 |
| 5 mM MgCl2 + 40 μM zinc acetate | 11.9 |
| 5 mM MgCl2 + 250 μM EDTA | 13.7 |
| 5 mM MgCl2 + 40 μM zinc acetate + 250 μM EDTA | 11.9 |
| 50 mM KCl + 40 μM zinc acetate | 9.5 |
| 50 mM KCl + 250 μM EDTA | 11.7 |
| 50 mM KCl + 40 μM zinc acetate + 250 μM EDTA | 10.1 |
Initial rates of hydrolysis of d-[3H]Tyr-tRNATyr catalyzed by purified GEK1 were measured at 28°C for 5 min in the presence of 20 mM Tris-HCl (pH 7.8), 50 nM d-[3H]Tyr-tRNATyr and the indicated components. Prior to the assay, GEK1 was diluted in 20 mM Tris-HCl buffer (pH 7.8) containing 200 μg/ml BSA and 10 mM 2-mercaptoethanol. Shown values are within ± 15%.
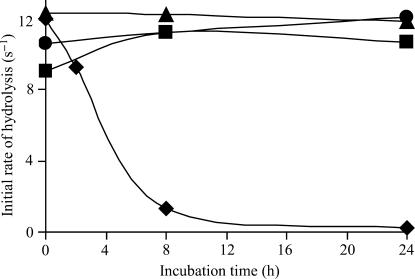
Because the archaeal deacylase carries strongly bound zinc ions, we examined whether GEK1 needed these metal ions to display full activity. The initial rate of d-Tyr-tRNATyr hydrolysis by the plant hydrolase was not sensitive to the presence in a 5-min assay of either 40-μM zinc acetate or 250-μM EDTA, an efficient chelator of zinc ions. In another set of experiments, GEK1 was left to incubate in the presence of 1 mM EDTA prior to the assay (Figure 1). At various times, aliquots were withdrawn and assayed (5 min) for deacylase activity. Measured activity progressively decreased, with a half-life time of ∼3 h. After 24 h, almost 100% of the initial activity was lost. However, if the inactivated enzyme was assayed in the presence of 40 μM zinc acetate, nearly full activity could be recovered. If GEK1 was incubated with 160 μM zinc acetate instead of 1 mM EDTA, enzyme activity remained constant for at least 24 h. This behavior markedly resembles that of an archaeal DTD2 deacylase (7). It strongly suggests that zinc ions are essential to the activity of the protein.
Figure 1.
Incubation of GEK1 in the presence of EDTA or of zinc acetate: effect on the initial rate of d-Tyr-tRNATyr hydrolysis. GEK1 (2 µM) was incubated in 20 mM Tris-HCl buffer (pH 7.8) containing 200 µg/ml BSA, 10 mM 2-mercaptoethanol and either 1 mM EDTA or 160 μM zinc acetate. At the times indicated, an aliquot of GEK1 was withdrawn and assayed for 5 min in the presence of 20 mM Tris-HCl (pH 7.8), 50 nM d-[3H]Tyr-tRNATyr, 5 mM MgCl2 and either 250 μM EDTA or 40 μM zinc acetate. Filled square represents zinc acetate incubation followed by activity measurement in the presence of zinc acetate, filled triangle represents Zinc acetate incubation followed by activity measurement in the presence of EDTA, filled circle represents EDTA incubation followed by activity measurement in the presence of zinc acetate, filled diamond represents EDTA incubation followed by activity measurement in the presence of EDTA.
Strong association of zinc to GEK1
Archaeal DTD2 enzymes appear to contain two zinc ions. Thus, we examined whether GEK1 also carried zinc. For this purpose, a sample of GEK1 was dialyzed overnight against 10 mM ammonium acetate buffer (pH 7.5) containing 10 μM zinc acetate and submitted to mass spectrometry under denaturing or non-denaturing conditions. The difference between the denatured mass (34 597 Da) and the native mass (34 726 Da) exactly corresponded to the mass of two zinc atoms (65 Da).
The strong association of zinc to GEK1 was confirmed by flame atomic absorption spectroscopy. A sample of GEK1 was subjected to gel filtration in 20 mM Tris-HCl (pH 7.8) buffer containing 300 mM KCl. We verified that the specific activity of GEK1 was not modified during this chromatographic step. Then, active collected fractions were analyzed for zinc content. Based on the theoretical A280 extinction coefficient of GEK1, a stoechiometry of 2.0 ± 0.2 mol of zinc per mol of GEK1 was found.
Altogether, the above results establish that GEK1 is tightly associated with two zinc ions.
Specificity and catalytic constants of GEK1
Rates of d- and l-Tyr-tRNATyr hydrolysis were compared at the same substrate concentration (100 nM) in assay conditions including 20 mM Tris-HCl (pH 7.8), 5 mM MgCl2, 40 μM zinc acetate, 50 μg/ml BSA, 2.5 mM 2-mercaptoethanol, and 3 pM–30 nM of GEK1 (Table 4). The rate of l-Tyr-tRNATyr deacylation was at least 25 000-fold lower than that of d-Tyr-tRNATyr hydrolysis. Thus, GEK1 displays strict specificity towards the d-isomer of the amino acid. Diacetyl-l-Lys-tRNALys also fully resisted the action of the deacylase. When assayed in the same conditions as above with 3–30 nM of enzyme, the specific activity of GEK1 towards this model substrate of peptidyl-tRNA hydrolases was at least 150 000-fold lower than that towards d-Tyr-tRNATyr.
Table 4.
Specificity of GEK1 towards the amino acid esterifed to tRNA
| Substrate | Initial rate (s−1) |
|---|---|
| d-Tyr-tRNATyr | 25 |
| l-Tyr-tRNATyr | <10−3 |
| Diacetyl-L-Lys-tRNALys | <1.6 × 10−4 |
Initial rates of hydrolysis of d-[3H]Tyr-tRNATyr, l-[14C]Tyr-tRNATyr or diacetyl-l-[14C]Lys-tRNALys catalyzed by purified GEK1 (3–6 pM) were measured at 28°C for 5 min in the presence of 20 mM Tris-HCl (pH 7.8), 100 nM substrate, 5 mM MgCl2, 40 μM zinc acetate, 50 μg/ml BSA and 2.5 mM 2-mercaptoethanol. Prior to the assay, GEK1 was diluted in 20 mM Tris-HCl (pH 7.8) containing 160 μM zinc acetate and 200 μg/ml BSA. Shown values are within ± 15%.
Measurement of the rate of hydrolysis as a function of d-Tyr-tRNATyr concentration (100–4000 nM) enabled us to deduce Km and kcat values of 1.1 ± 0.1 μM and 300 ± 12 s−1, respectively (data not shown). In the same conditions, the spontaneous chemical hydrolysis of d-Tyr-tRNATyr occurred at a rate of 0.012 s−1.
Therefore, in vitro, GEK1 displays a markedly high activity (28°C). Its kcat value for hydrolysis is 50-fold higher than that of E. coli deacylase assayed under similar conditions (5). It is nearly 500-fold higher than that of P. abyssi deacylase as measured at 37°C (7). Possibly, such a high activity compensates for reduced GEK1 expression in a plant cytoplasm. Expression of GEK1 in various tissues of wild-type Arabidopsis was followed using northern blot analysis (9). The mRNA of GEK1 was ubiquitously detected. However, its exact abundance was not evaluated.
To further examine the specificity of GEK1, we tested whether this protein was able to also use d-Asp-tRNAAsp as substrate. The initial rate of hydrolysis of 50 nM d-Asp-tRNAAsp was equal to 8.6 s−1 (Table 5), a value very close to that obtained with 50 nM d-Tyr-tRNATyr (12.7 s−1). The specificity of GEK1 towards its substrate was compared to those of E. coli DTD1 and P. abyssi DTD2 under the same conditions of d-aminoacyl-tRNA concentration (50 nM). As shown in Table 5, these two proteins also hydrolyzed d-Asp-tRNAAsp almost as efficiently as they hydrolyzed d-Tyr-tRNATyr. We could therefore conclude that, similarly to a DTD1 enzyme (2), the DTD2 protein from archaea and the GEK1 protein from plants display broad specificity towards substrates carrying very different d-amino acid moieties.
Table 5.
Comparison of the substrate specificity of A. thaliana, P. abyssi and E. coli d-aminoacyl-tRNA deacylases
| Deacylase activity (s−1) | ||
|---|---|---|
| d-Tyr-tRNATyr | d-Asp-tRNAAsp | |
| A. thaliana GEK1a | 12.7 ± 1.2 | 8.6 ± 2.4 |
| P. abyssi DTD2b | 0.33 ± 0.05 | 0.12 ± 0.03 |
| E. coli DTD1c | 0.25 ± 0.02 | 0.6 ± 0.05 |
Initial rates of hydrolysis were measured in the presence of 50 nM d-aminoacyl-tRNA.
a28°C, 3–6 pM GEK1, 20 mM Tris-HCl, pH 7.8, 5 mM MgCl2, 40 μM zinc acetate, 50 μg/ml BSA, 2.5 mM 2-mercaptoethanol. b37°C, 200–400 pM DTD2, 20 mM Tris-HCl, pH 7.8, 4 mM MgCl2, 40 μM zinc acetate, 50 μg/ml BSA, 2.5 mM 2-mercaptoethanol. c28°C, 70–200 pM DTD1, 20 mM Tris-HCl, pH 7.8, 5 mM MgCl2, 0.025 mM EDTA, 50 μg/ml BSA, 2.5 mM 2-mercaptoethanol.
Complementation of an E. coli Δdtd null mutant by GEK1 expression
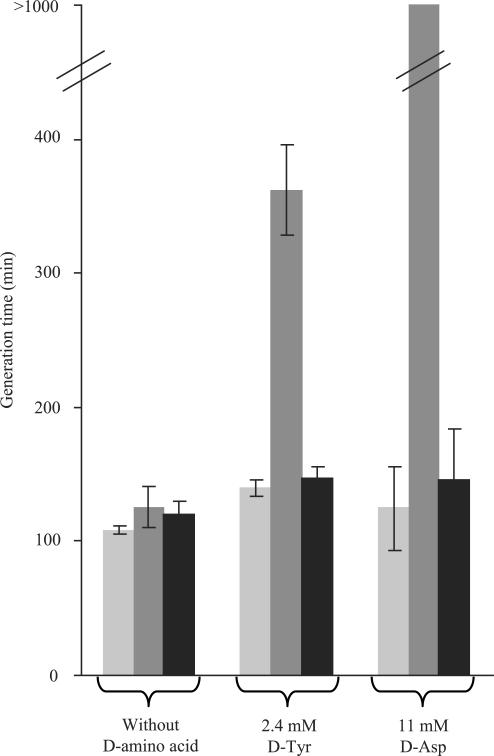
To confirm the activity of the GEK1 product in vivo, we studied the effect of GEK1 expression on the sensitivity of an E. coli Δdtd strain to d-tyrosine or d-aspartic acid. In minimal growth medium, the generation time of the strain was significantly increased in the presence of 2.4 mM d-tyrosine (2,3,5). However, our attempts to cultivate strain K37ΔrecAΔtyrHλDE3 containing pET3alpa::GEK1 failed. It is likely that excess production of d-aminoacyl-tRNA deacylase activity from this plasmid impairs E. coli growth in minimal medium, as already reported (5). To alleviate this difficulty and obtain viable cells containing the GEK1 product, we used strain K37ΔrecAΔtyrHλDE3 carrying pET15blpa::GEK1. The pET15blpa plasmid differs from the pET3alpa one by the presence of lacI. The K37ΔrecAΔtyrHλDE3 derivative harboring the pET15blpa::GEK1 plasmid was found to grow normally in minimal medium. Attenuation of GEK1 transcription by the lactose repressor may explain this behavior. Generation times of this strain in the absence or presence of d-amino acid (2.4 mM d-tyrosine or 11 mM d-aspartic acid) were compared with those of strains K37ΔrecAλDE3 and K37ΔrecAΔtyrHλDE3 containing the control plasmid pET15blpa (Figure 2). In the absence of d-amino acid, the generation times of all three strains were identical (118 ± 7 min). As expected, the generation time of the Δdtd mutant was longer in the presence of d-amino acid (2.3-fold in the presence of d-tyrosine and more than 9-fold in the presence of d-aspartic acid). Upon introduction of the plasmid producing GEK1, the Δdtd mutant exposed to d-tyrosine or d-aspartic acid recovered the generation time displayed in the absence of d-amino acid.
Figure 2.
Generation time of various E. coli strains in the presence or absence of d-amino acids in the growth medium. Strains K37ΔrecAλDE3 (dtd+) or K37ΔrecAΔtyrHλDE3 (Δdtd) cells containing pET15blpa or pET15blpa::GEK1 were grown at 37°C in M9-glucose minimal medium containing 100 μg/ml ampicillin and either 2.4 mM d-tyrosine or 11 mM d-aspartic acid. A control without d-amino acid was also performed. Cells were pre-grown overnight in the growth medium under study. For generation time measurements, inoculations were adjusted to an OD650 of 0.05. Samples were withdrawn from the cultures every 90 min during 540 min, and generation times were deduced from OD650 measurements. Shown experiments were performed using wild-type strain (K37ΔrecAλDE3) carrying pET15blpa (light bars), Δdtd mutant strain (K37▵recA▵tyrHλDE3) carrying pET15blpa (middle dark bars) and Δdtd mutant strain (K37▵recA▵tyrHλDE3) carrying pET15blpa::GEK1 (dark bars). Errors bars represent standard deviations calculated from two independent experiments. In the case of the strain K37ΔrecAΔtyrHλDE3 carrying pET15blpa grown in the presence of 11 mM d-aspartic acid, the increase in the OD650 value was less than 35% after 540 min. Therefore, only an approximate value of the generation time could be estimated.
Thus, expression of GEK1 in trans does rescue the sensitivity of a Δdtd mutant to d-tyrosine as well as to d-aspartic acid.
E. coli and S. cerevisiae strains lacking endogeneous d-Tyr-tRNATyr deacylase do not display any particular sensitivity to ethanol or to acetaldehyde
Previous results have established a link between the in vivo function of GEK1 and sensitivity of a plant to ethanol or acetaldehyde (8,9). This link appears to be plant specific. Indeed, overexpression of GEK1 in E. coli or yeast strains does not confer any particular phenotype in response to the addition of ethanol (9). However, since the latter experiments were performed in wild-type contexts, a dominant effect of the E. coli or yeast endogeneous deacylases on ethanol tolerance cannot be excluded.
In the present study, to determine whether the endogeneous deacylase function in E. coli or S. cerevisiae has any link with the ethanol response, we compared cells expressing or not their native dtd/DTD1 gene. Wild-type E. coli strain and a derived Δdtd mutant (K37ΔrecAλDE3 and K37ΔrecAΔtyrHλDE3, respectively) were grown in the presence of 0–16% ethanol or of 0–12% acetaldehyde in minimal medium at 37°C. Plates were surveyed for a week. We found that the growth rates of the two strains were identical whatever the added concentration of ethanol or acetaldehyde. Minimal inhibitory concentrations (MIC) were 12 and 2%, respectively.
Similar observations were made with S. cerevisiae. The growth rates of a dtd1 mutant (DBY2057ΔDTD1) and of its parental strain (DBY2057), at 30°C, in minimal medium containing various concentrations of ethanol or acetaldehyde, were indistinguishable. With both strains, MICs were 16% ethanol and 4% acetaldehyde.
Thus, we conclude that, in E. coli and yeast, d-Tyr-tRNATyr deacylase activity does not sustain the metabolic link with ethanol that the GEK1 product does in A. thaliana.
DISCUSSION
The results of this study establish that the A. thaliana GEK1 protein behaves as a broad-specific, markedly active d-aminoacyl-tRNA deacylase. Therefore, the sequence homology between GEK1 and archaeal d-Tyr-tRNATyr deacylases is accompanied by a functional homology. The resemblance between GEK1 and archaeal deacylases is reinforced by the occurrence in all proteins of zinc ions involved in enzymatic activity (7).
The genome of A. thaliana also contains a homolog of dtd, the E. coli deacylase gene. Therefore, A. thaliana is likely to express at least two enzyme species carrying d-Tyr-tRNATyr deacylase activity. However, because cells contain distinct compartments, the functions of these two enzymes may not be redundant. Indeed, a GEK1-green-fluorescent-protein fused protein was detected in the whole cell, including the nucleus (8), while, according to the computer program TargetP (17), the homolog of E. coli DTD1 should be localized to the chloroplast, with the help of a signal peptide of 42 amino acids. The endosymbiotic origin of chloroplasts and the resemblance between archaea and the cytosol of eukaryotes may account for segregation in two different compartments of two distinct enzymes sharing a same function.
GEK1 and the homolog of bacterial dtd co-exist in the genomes of all plants documented so far. This suggests that expression, in plant cells, of d-aminoacyl-tRNA deacylase activity is important to confer protection against d-amino acid toxicity. Sensitivity of plants to d-amino acids has already been documented. Growth and morphology are affected by exposure to d-amino acids (18). d-leucine causes ‘leucinosis’ in sunflowers, a disease characterized by chlorosis, smaller leaves and the appearance of necrotic regions on them (19). d-alanine, d-aspartic acid, d-glutamic acid and d-serine induce leaf shedding (20). Salt uptake by different plant tissues is inhibited by d-amino acids (18). Toxicity of d-amino acids depends on their side chain. For example, in A. thaliana, d-alanine and d-serine are very toxic whereas d-valine and d-isoleucine are not (21). In spite of these data, the d-amino acid metabolism of plants is still poorly understood. Most eukaryotic and prokaryotic organisms possess d-amino acid oxidase, a catabolic enzyme of d-amino acids (22). Such an enzyme has not been found in plants. Instead, plants possess a specific enzyme called 1-aminocyclopropane-1-carboxylate N-malonyltransferase, which insures malonylation of d-amino acids, but not of l-amino acids (23–25). This modification is thought to be involved in the detoxification of d-amino acids.
Originally, GEK1 was identified because of its role in the ethanol stress response of A. thaliana (8,9). The deacylase function of GEK1 suggests a link between this ethanol stress response, d-amino acid toxicity and protein synthesis. Notably, several studies indicate that ethanol could affect protein synthesis in eukaryotes (26–29). On the basis of the present study, we have now to discover functional links between ethanol or acetaldehyde addition to a plant cell and the extent of d-amino acid transfer onto tRNA.
Several speculations can be made based on the property of acetaldehyde to react with NH2 or SH groups (30–32). (i) A first possibility is an inhibition by acetaldehyde of the malonylating reaction mentioned above, with a resulting increase in the concentration of the d-amino acid pool in the cell. The malonyltransferase does not exist in E. coli or yeast. Thus, a participation of this enzyme in the behavior of the gek1 mutant would account for a restriction to plants of the link between ethanol and d-amino acids. Possibly, inhibition of the N-malonyltransferase involves inactivation of its cofactor coenzyme A. In mouse brain extracts, coenzyme A has been found to be sensitive to relatively low concentrations of acetaldehyde (0.2 mM) (33). (ii) A second possibility is an upregulation of the production of d-amino acids as side-products of l-amino acid synthetic pathways. For instance, chemical modification by acetaldehyde of an enzyme sustaining l-amino acid synthesis might change the ratio between the concentrations of d- and l-amino acids produced. (iii) A third hypothesis involves a change of the stereospecificity in the charging reaction of a tRNA, through chemical modification of the cognate aminoacyl-tRNA synthetase or of the tRNA itself. Improved transfer to the tRNA of the d-isomer of the amino acid would account for the toxicity of acetaldehyde towards a plant cell deprived of cytosolic d-aminoacyl-tRNA deacylase. (iv) A fourth possible hypothesis is a blocking reaction by acetaldehyde of the α-NH2-groups of the amino acids esterified to tRNA. Such a modification is expected to confer improved resistance against spontaneous deacylation (5). Possibly, in vivo, such N-blocked l-aminoacyl-tRNA species are enzymatically recycled, but not the d-aminoacyl-tRNA ones (34). Thus, under this hypothesis, the cell would take advantage of the presence of a d-Tyr-tRNATyr deacylase to ensure hydrolysis of d-aminoacyl-tRNAs before they are converted into long-lived, inactive species, by acetaldehyde attack.
Further studies must now be undertaken to identify which reaction(s) link d-aminoacyl-tRNAs to ethanol stress in A. thaliana. It is likely that such studies will shed new light on d-amino acid metabolism in plants.
Acknowledgements
We thank François Parcy for helpful discussions, Thomas Simonson for critical reading of the manuscript, Anne-Pascale Bouin for mass spectrometry analysis and Michel Fromant for technical support. The Arabidopsis Biological Resource Center (USA) is gratefully acknowledged for providing us with clone S81170. Funding to pay the Open Access publication charge was provided by CNRS.
Conflict of interest statement. None declared.
References
- 1.Calendar R, Berg P. The catalytic properties of tyrosyl ribonucleic acid synthetases from Escherichia coli and Bacillus subtilis. Biochemistry. 1966;5:1690–1695. doi: 10.1021/bi00869a034. [DOI] [PubMed] [Google Scholar]
- 2.Soutourina J, Plateau P, Blanquet S. Metabolism of D-aminoacyl-tRNAs in Escherichia coli and Saccharomyces cerevisiae cells. J. Biol. Chem. 2000;275:32535–32542. doi: 10.1074/jbc.M005166200. [DOI] [PubMed] [Google Scholar]
- 3.Soutourina O, Soutourina J, Blanquet S, Plateau P. Formation of D-tyrosyl-tRNATyr accounts for the toxicity of D-tyrosine toward Escherichia coli. J. Biol. Chem. 2004;279:42560–42565. doi: 10.1074/jbc.M402931200. [DOI] [PubMed] [Google Scholar]
- 4.Calendar R, Berg P. D-Tyrosyl RNA: formation, hydrolysis and utilization for protein synthesis. J. Mol. Biol. 1967;26:39–54. doi: 10.1016/0022-2836(67)90259-8. [DOI] [PubMed] [Google Scholar]
- 5.Soutourina J, Plateau P, Delort F, Peirotes A, Blanquet S. Functional characterization of the D-Tyr-tRNATyr deacylase from Escherichia coli. J. Biol. Chem. 1999;274:19109–19114. doi: 10.1074/jbc.274.27.19109. [DOI] [PubMed] [Google Scholar]
- 6.Ferri-Fioni ML, Schmitt E, Soutourina J, Plateau P, Mechulam Y, Blanquet S. Structure of crystalline D-Tyr-tRNATyr deacylase. A representative of a new class of tRNA-dependent hydrolases. J. Biol. Chem. 2001;276:47285–47290. doi: 10.1074/jbc.M106550200. [DOI] [PubMed] [Google Scholar]
- 7.Ferri-Fioni ML, Fromant M, Bouin AP, Aubard C, Lazennec C, Plateau P, Blanquet S. Identification in archaea of a novel D-Tyr-tRNATyr deacylase. J. Biol. Chem. 2006;281:27575–27585. doi: 10.1074/jbc.M605860200. [DOI] [PubMed] [Google Scholar]
- 8.Fujishige N, Nishimura N, Iuchi S, Kunii T, Shinozaki K, Hirayama T. A novel Arabidopsis gene required for ethanol tolerance is conserved among plants and archaea. Plant Cell. Physiol. 2004;45:659–666. doi: 10.1093/pcp/pch086. [DOI] [PubMed] [Google Scholar]
- 9.Hirayama T, Fujishige N, Kunii T, Nishimura N, Iuchi S, Shinozaki K. A novel ethanol-hypersensitive mutant of Arabidopsis. Plant Cell. Physiol. 2004;45:703–711. doi: 10.1093/pcp/pch078. [DOI] [PubMed] [Google Scholar]
- 10.Miller HI, Friedman DI. An E. coli gene product required for lambda site-specific recombination. Cell. 1980;20:711–719. doi: 10.1016/0092-8674(80)90317-7. [DOI] [PubMed] [Google Scholar]
- 11.Adams AE, Botstein D. Dominant suppressors of yeast actin mutations that are reciprocally suppressed. Genetics. 1989;121:675–683. doi: 10.1093/genetics/121.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soutourina J, Blanquet S, Plateau P. D-tyrosyl-tRNATyr metabolism in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:11626–11630. doi: 10.1074/jbc.275.16.11626. [DOI] [PubMed] [Google Scholar]
- 13.Guillon L, Schmitt E, Blanquet S, Mechulam Y. Initiator tRNA binding by e/aIF5B, the eukaryotic/archaeal homologue of bacterial initiation factor IF2. Biochemistry. 2005;44:15594–15601. doi: 10.1021/bi051514j. [DOI] [PubMed] [Google Scholar]
- 14.Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 15.Schmitt E, Mechulam Y, Fromant M, Plateau P, Blanquet S. Crystal structure at 1.2 Å resolution and active site mapping of Escherichia coli peptidyl-tRNA hydrolase. EMBO J. 1997;16:4760–4769. doi: 10.1093/emboj/16.15.4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dardel F. MC-Fit: using Monte-Carlo methods to get accurate confidence limits on enzyme parameters. Comput. Appl. Biosci. 1994;10:273–275. doi: 10.1093/bioinformatics/10.3.273. [DOI] [PubMed] [Google Scholar]
- 17.Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- 18.Robinson T. D-amino acids in higher plants. Life Sci. 1976;19:1097–1102. doi: 10.1016/0024-3205(76)90244-7. [DOI] [PubMed] [Google Scholar]
- 19.Gamburg KZ, Rekoslavskaya NI. Formation and functions of D-amino acids in plants. Fiziologiya Rastenii. 1991;38:1236–1246. [Google Scholar]
- 20.Valdovinos JG, Muir RM. Effects of D and L amino acids on foliar abscission. Plant Physiol. 1965;40:335–340. doi: 10.1104/pp.40.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erikson O, Hertzberg M, Nasholm T. A conditional marker gene allowing both positive and negative selection in plants. Nat. Biotechnol. 2004;22:455–458. doi: 10.1038/nbt946. [DOI] [PubMed] [Google Scholar]
- 22.Pilone MS. D-amino acid oxidase: new findings. Cell. Mol. Life Sci. 2000;57:1732–1747. doi: 10.1007/PL00000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosa N, Neish AC. Formation and occurrence of N-malonylphenylalanine and related compounds in plants. Can. J. Biochem. 1968;46:799–806. [PubMed] [Google Scholar]
- 24.Liu Y, Su LY, Yang SF. Ethylene promotes the capability to malonylate 1-aminocyclopropane-1-carboxylic acid and D-amino acids in preclimacteric tomato fruits. Plant Physiol. 1985;77:891–895. doi: 10.1104/pp.77.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo L, Phillips AT, Arteca RN. Amino acid N-malonyltransferases from mung beans. Action on 1-aminocyclopropane-1-carboxylic acid and D-phenylalanine. J. Biol. Chem. 1993;268:25389–25394. [PubMed] [Google Scholar]
- 26.Majumdar AP, Haiman MJ, Zylbert BA, Billy HT, Vesenka GD, Geokas MC. Acetaldehyde inhibition of protein synthesis in isolated rat pancreatic acini. Toxicol. Appl. Pharmacol. 1986;83:86–94. doi: 10.1016/0041-008x(86)90325-x. [DOI] [PubMed] [Google Scholar]
- 27.Preedy VR, Peters TJ. Acute effects of ethanol on protein synthesis in different muscles and muscle protein fractions of the rat. Clin. Sci. (Lond.) 1988;74:461–466. doi: 10.1042/cs0740461. [DOI] [PubMed] [Google Scholar]
- 28.Tiernan JM, Ward LC. Acute effects of ethanol on protein synthesis in the rat. Alcohol Alcohol. 1986;21:171–179. [PubMed] [Google Scholar]
- 29.David ET, Fischer I, Moldave K. Studies on the effect of ethanol on eukaryotic protein synthesis in vitro. J. Biol. Chem. 1983;258:7702–7706. [PubMed] [Google Scholar]
- 30.Hernandez-Munoz R, Baraona E, Blacksberg I, Lieber CS. Characterization of the increased binding of acetaldehyde to red blood cells in alcoholics. Alcohol Clin. Exp. Res. 1989;13:654–659. doi: 10.1111/j.1530-0277.1989.tb00399.x. [DOI] [PubMed] [Google Scholar]
- 31.Niemela O. Aldehyde-protein adducts in the liver as a result of ethanol-induced oxidative stress. Front. Biosci. 1999;4:D506–513. doi: 10.2741/niemela. [DOI] [PubMed] [Google Scholar]
- 32.Mauch TJ, Donohue TM, Jr, Zetterman RK, Sorrell MF, Tuma DJ. Covalent binding of acetaldehyde selectively inhibits the catalytic activity of lysine-dependent enzymes. Hepatology. 1986;6:263–269. doi: 10.1002/hep.1840060218. [DOI] [PubMed] [Google Scholar]
- 33.Ammon HP, Estler CJ, Heim F. Inactivation of coenzyme A by ethanol. I. Acetaldehyde as mediator of the inactivation of coenzyme A following the administration of ethanol in vivo. Biochem. Pharmacol. 1969;18:29–33. doi: 10.1016/0006-2952(69)90005-7. [DOI] [PubMed] [Google Scholar]
- 34.Kössel H, RajBhandary UL. Studies on polynucleotides. LXXXVI. Enzymic hydrolysis of N-acylaminoacyl-transfer RNA. J. Mol. Biol. 1968;35:539–560. doi: 10.1016/s0022-2836(68)80013-0. [DOI] [PubMed] [Google Scholar]