Abstract
Small non-coding RNAs (sRNAs) are an emerging class of regulators of bacterial gene expression. Most of the regulatory Escherichia coli sRNAs known to date modulate translation of trans-encoded target mRNAs. We studied the specificity of sRNA target interactions using gene fusions to green fluorescent protein (GFP) as a novel reporter of translational control by bacterial sRNAs in vivo. Target sequences were selected from both monocistronic and polycistronic mRNAs. Upon expression of the cognate sRNA (DsrA, GcvB, MicA, MicC, MicF, RprA, RyhB, SgrS and Spot42), we observed highly specific translation repression/activation of target fusions under various growth conditions. Target regulation was also tested in mutants that lacked Hfq or RNase III, or which expressed a truncated RNase E (rne701). We found that translational regulation by these sRNAs was largely independent of full-length RNase E, e.g. despite the fact that ompA fusion mRNA decay could no longer be promoted by MicA. This is the first study in which multiple well-defined E.coli sRNA target pairs have been studied in a uniform manner in vivo. We expect our GFP fusion approach to be applicable to sRNA targets of other bacteria, and also demonstrate that Vibrio RyhB sRNA represses a Vibrio sodB fusion when co-expressed in E.coli.
INTRODUCTION
Small non-coding RNAs (sRNAs) that act as regulators of gene expression are wide-spread in bacteria. Typically, these molecules are 50–200 nt in size, and do not contain expressed open reading frames (ORFs). Using a diverse array of approaches [reviewed in (1)], >70 Escherichia coli sRNAs have been identified in numerous screens [e.g. (2–7)] over the past five years, while hundreds of additional sRNA candidate genes still await experimental validation (8).
Two main modes of action have been established for the E.coli sRNAs. Some sRNAs modify the activity of proteins (9–11), while the majority act on trans-encoded target mRNAs to modulate their translation and/or stability. Several key features of antisense regulation by chromosomal sRNAs have emerged: (i) Unlike the cis-encoded antisense RNAs of plasmids and phages [reviewed in (12)], these trans-encoded antisense RNAs typically have only short and imperfect complementarity to their target(s). (ii) Base pairing most often occurs in the 5′-untranslated region (5′-UTR) of the target mRNA, and is aided by the bacterial Sm-like protein, Hfq. (iii) Binding may result in either the blockage of ribosome entry (translational repression), or the melting of inhibitory secondary structures, which sequester the ribosome binding site (RBS) of the mRNA (translational activation). (iv) Regulation is frequently coupled to nuclease-mediated cleavage of the mRNA, e.g. RNase E cleavage of sodB mRNA upon RyhB binding (13), and RNase III cleavage of tisAB mRNA upon IstR-1 binding (14).
Several E.coli sRNA target interactions have been well-defined. For example, the porin-regulating sRNAs, MicC and MicF, form an extended though imperfect RNA duplex with the 5′-UTRs of the ompC and ompF mRNAs, respectively (15,16), whereas MicA forms an almost perfect 16 bp duplex encompassing the RBS region of ompA mRNA (17,18). Shorter interactions underlie the repression of the ptsG message by SgrS (19), and of the sodB message by RyhB (20); in the latter case, 9 nt of either RNA are involved in duplex formation (21). Repression of the fhlA mRNA by OxyS is mediated by two short kissing complexes of 9 and 7 bp, respectively; the two target regions in OxyS and in fhlA mRNA are each separated by long spacers (22). DsrA was proposed to repress hns mRNA by binding it at both the start and the stop codon region; in other words, a bipartite interaction that would involve regions within the hns mRNA that are ∼400 nt apart (23).
Some sRNAs are known to activate translation of mRNAs. The DsrA and RprA sRNAs pair with the rpoS mRNA leader, thereby preventing the formation of an inhibitory structure around the rpoS RBS that would otherwise repress rpoS translation (24–26). There are two other examples of E.coli sRNAs that function as mRNA activators, i.e. GadY and RydC (27,28).
In the early days of E.coli sRNA identification, these molecules were frequently recognized through their effect on a certain mRNA. For example, the micF gene was found within a multi-copy library insert that caused OmpF depletion, while the MicF/ompF mRNA interaction was shown in subsequent analysis (15,29). That is a (main) target was known before the regulator itself was identified. In contrast, the sheer numbers of new sRNAs recently identified in systematic genome-wide searches (1), which are a priori of unknown function, require tools to efficiently predict and study interactions with target mRNAs. Besides, since sRNAs may also regulate multiple targets [e.g. (26,30,31)], knowing a single target may not fully reflect the regulatory potential of a given sRNA.
Traditionally, genome-wide screens of randomly inserted reporter gene fusions as well as global protein pattern changes upon deletion or overexpression of an sRNA have played a major role in target identification [e.g. (17,18,32)]. However, these approaches strictly require the target gene to be expressed at a measurable level under the assay condition, with the additional caveat that they provide little means to distinguish primary target effects from secondary pleiotropic changes of gene expression. Recently, several approaches were taken to narrow target searches to those mRNAs that directly interact with a given sRNA, e.g. monitoring mRNA decay on microarrays following sRNA overexpression (30,33,34), selective capture of cellular mRNAs with in vitro-synthesized sRNAs (28,35), and biocomputational target predictions (30).
Regardless of the route taken for identification, the in vivo assessment of putative target regulation remains a critical issue. Of the various reporters of bacterial gene expression (16,36), chromosomal or plasmid-borne translational fusions of the target 5′-UTR to E.coli lacZ, encoding β-galactosidase, have been the most common tool to study target gene regulation by sRNAs. However, since the fusion is typically driven by the target gene promoter, a specific effect on translation rather than on transcription has to be confirmed in independent experiments. Generally, lacZ fusions represent a robust and well-established reporter system, however, come with the disadvantages of an enzymatic assay involving cell lysis to measure β-galactosidase activity.
Using the green fluorescent protein (GFP) from the jellyfish Aequorea victoria (37), which permits a non-invasive reporter assay, we have here studied a great number of E.coli sRNAs and their targets in a uniform reporter system. This study has revealed novel aspects of regulation for several of these pairs. Furthermore, our GFP-based reporters will be helpful to rapidly validate bacterial sRNA targets of other prokaryotes.
MATERIALS AND METHODS
DNA oligonucleotides
The complete list of oligonucleotides used for cloning and as probes in hybridization is provided as Supplementary Table S2.
Bacterial strains, media and growth conditions
E.coli strain Top10 (Invitrogen) was used to clone GFP fusions, and in all experiments that involved co-expression of GFP fusions and sRNAs. E.coli strain Top10 F′ (Invitrogen) was used to clone sRNA expression plasmids. All established mutant strains are derived from E.coli Top10. Strains JVS-2001 (Δhfq::KmR) and JVS-2002 (rne701-KmR) were constructed by the one-step inactivation protocol (38) with PCR products obtained with primer pairs JVO-0515/-0516 or JVO-0856/-0857, respectively, using a modified KmR cassette of plasmid pKD4 as template (J. Vogel, unpublished data). Strain JVS-2003 (Δrnc14::TetR) was constructed similarly, using primer pair JVO-0884/-0885 and chromosomal DNA of strain W3310 rnc14::Tn10 (39). Verification of the mutant strains was carried out by colony PCR using primer pairs JVO-0517/-0518 (for JVS-2001), JVO-0858/-0859 (for JVS-2002) and JVO-0886/-0887 (for JVS-2003). C-terminal truncation of RNase E in JVS-2002 was also verified by western blot using an RNase E antiserum (kindly provided by A. G. Carpousis). Details of the aforementioned bacterial strains are given in Supplementary Table S1.
Growth in Luria–Bertani (LB) broth or on LB plates at 37°C was used throughout this study. Antibiotics were applied at the following concentrations: 100 μg/ml ampicillin, 50 μg/ml kanamycin and 20 μg/ml chloramphenicol.
Plasmids
Fusion plasmids: To construct plasmid pXG-0 (control plasmid to determine cellular autofluorescence), the p15A replicon of pZA31-luc was removed by SacI/AvrII restriction digest and replaced with a SacI/AvrII fragment containing the low-copy pSC101* replicon of pZS*24-MCS1.
Plasmid pXG-10, the standard plasmid for gfp fusion cloning was constructed as follows. A DNA fragment containing the pSC101* origin of replication, chloramphenicol resistance cassette and the PLtetO promoter was amplified from pXG-0 by PCR using primer pair JVO-0154/-0156, which adds BfrBI and NheI restriction sites right downstream of the promoter. The PCR product was digested with XbaI/NheI, and ligated to a gfp+ encoding fragment amplified from plasmid pWH601 (40) with primer pair JVO-0152/-0153. Insertion of a BfrBI/NheI-digested PCR fragment (containing the lacZ 5′-UTR and the first 186 coding residues), amplified from chromosomal E.coli MC4100 DNA using primer pair JVO-0274/-0328, into the corresponding sites gave plasmid pXG-10.
To construct plasmid pXG-1, the PLtetO promoter and RBS region of plasmid pZA31-luc was amplified with primers pZE-CAT/JVO-0330. Upon AatII/NheI digest, the fragment was inserted into plasmid pXG-10 digested with the same enzymes. Consequently, in plasmid pXG-1 an ATG start codon precedes the NheI site, and thus results in expression of full-length GFP.
To construct plasmid pXG-20, which is used for 5′-RACE product cloning, the PLtetO promoter from pZA31-luc was amplified with primers pZE-Cat/JVO-0339. The latter oligo introduces a point mutation at the promoter (changing the C at −1 to A) and adds a BfrBI restriction site to position +2. The fragment was digested with AatII/BfrBI and inserted into plasmid pXG-10 digested with the same enzymes. BfrBI/NheI cloning of a PCR product obtained on chromosomal E.coli MC4100 DNA with primers JVO-0368/-0369 resulted in an insert that contains an internal fragment of the lacZ coding region (651–976 amino acid) and a BsgI site; digest with BsgI will result in cleavage at the +1 site of PLtetO.
Plasmid pXG-30, the plasmid for operonic gfp fusion cloning, was constructed as follows. First, an E.coli DNA fragment spanning codons 2–59 of the lacZ gene was amplified with primers JVO-0642/-0685; oligo JVO-0685 added a FLAG epitope preceded by ATG to the N-terminus of the LacZ fragment. Upon KpnI/NheI digest, the fragment was inserted into plasmid pXG-1 digested with the same restriction enzymes, resulting in plasmid pJU-083. An internal fragment of the E.coli galETKM locus, from the last 58 codons of the galT C-terminal region to the 47th codon of galK, was PCR-amplified from E.coli MC4100 with primer pair JVO-0490/-0491, and inserted into pJU-083 by BfrBI/NheI cloning, which gave plasmid pJU-088. E.coli Top10 transformed with pJU-088 showed high fluorescence levels of GalK::GFP, but only low signals for the LacZ::GalT fusion were detectable in western blots with antibodies against the FLAG epitope contained in the fusion. Therefore, a DNA fragment containing the RBS of plasmid pZA31-luc was PCR-amplified with primers JVO-1102/-1103, and fused in a subsequent PCR step to a DNA fragment containing a 3× FLAG epitope proceeded by an ATG start codon obtained by PCR on plasmid pSUB11 (41) with primers JVO-1100/-1101. The resulting DNA fragment was directly ligated to a PCR product obtained on pJU-088 with primers pZE-tetO/JVO-1104 to yield plasmid pXG-30.
For cloning of gfp fusions in pXG-10, chromosomal DNA fragments were amplified by PCR with a sense oligonucleotide which anneals to the transcriptional start-site (for many E.coli K12 genes annotated at http://ecocyc.org/) of the gene of interest and adds a BfrBI restriction site and an antisense oligonucleotide which anneals in the N-terminal coding region of the gene and adds an in frame NheI restriction site. The corresponding primers for each gene are listed in Supplementary Table S2. Typically, the full-length 5′-UTR (from +1 of the most proximal promoter of a gene) and 30–150 bp (10–50 amino acid residues) of the N-terminal coding region were cloned. Inclusion of extracytoplasmic signal sequences (where known) were generally avoided to prevent traffic of the fusion proteins to the periplasm/membrane.
Intra-operonic fusions established in pXG-30 were cloned as above but the sense oligonucleotide annealed to the C-terminal coding region of the upstream gene and adds the BfrBI restriction site in frame.
sRNA plasmids: All sRNA plasmids constructed here are based on plasmid pZE12-luc. First, a DNA fragment of pZE12-luc was amplified by PCR using pfu-polymerase (Fermentas) and primers PLlacoB and PLlacoD, and subsequently digested with XbaI. This digest results in two DNA fragments of ∼2.2 kb and ∼1.7 kb, respectively. The ∼2.2 kb fragment carries the PLlacO promoter (from the position −1), an ampicillin resistance cassette, a ColE1 replicon and a strong rrnB terminator followed by the sticky end created by XbaI digestion. After gel-purification, it served as the vector backbone for sRNA cloning. The E.coli micC sRNA gene was PCR-amplified using primers JVO-0486/-0489. The sense primer (JVO-0486) anneals to the +1 site of micC and carries a 5′ monophosphate for cloning. The antisense primer (JVO-0489) binds to the region downstream of the micC terminator and will add an XbaI site to the PCR product. Following XbaI digest, the product was ligated to the 2.2 kb XbaI fragment of pZE12-luc, to yield plasmid pSK-017 upon transformation. Plasmid pSK-019 expressing DicF sRNA was constructed similarly using primers JVO-0487/-0488. To construct sRNA plasmids pJVgcvB-6 (gcvB), pJV100IA-T4 (rprA) and pJV107-8 (ryhB), the sRNA genes were amplified with primer pairs gcvB6/gcvB7, jb-100-L/jb-100-IA, and jb-107-G/jb-107-H, respectively. Different to the micC cloning described above, however, these fragments were cloned by inserting them at the KnpI site (pJV107-8, pJVgcvB-6), or the EcoRI site (pJV 100IA-T4) of pZE12-luc.
To lower the copy number, the ColE1 origin of pJV107-8 was swapped for the p15A origin of pZA31-luc by SpeI/AvrII cloning, resulting in plasmid pJU-002. Similarly, the ColE1 origin of pJVgcvB-6 was swapped for the p15A origin of pJU-002 by PacI/SpeI cloning, yielding plasmid pJU-014.
To construct control plasmid pTP-011, the ColE1 origin of pJV300 was replaced by the p15A origin of pZA31-luc by SpeI/AvrII cloning.
5′ RACE and direct cloning of full-length gfp fusions
5′ RACE was carried out as described previously (4) but with modifications, the major being a new 5′ RNA adapter (A4: 5′-GACGAGCACGAGGACACUGACAUGGAGGAGGGAGUAGAAA-3′OH), which contains a BseRI recognition site (underlined) to facilitate cleavage of the obtained cDNA at the 5′ end of the ligated RNA. 5′ triphosphates were converted to 5′ monophosphates by treatment of 6 μg total RNA (obtained on strain E.coli MC4100 grown to an OD600 of 2) with 10 U of tobacco acid pyrophosphatase (TAP, Epicentre Technologies) at 37°C for 30 min. Control RNA was incubated in the absence of the TAP. Reactions were stopped by phenol chloroform extraction, followed by ethanol/sodium acetate precipitation. Pellets were dissolved in water, mixed with 300 pmol of 5′ RNA adapter A4, heat-denatured at 95°C for 5 min, followed by a 5 min quick-chill step on ice. The adapter was ligated at 17°C for 12 h with 40 U T4-RNA ligase (New England Biolabs) in the recommended buffer and 10% dimethyl sulfoxide (DMSO). Phenol chloroform-extracted, ethanol-precipitated RNA (2 μg) was then reverse-transcribed using 100 pmol random DNA hexamers and the SuperScriptIII RT system (Invitrogen) in a total volume of 20 μl. Reverse transcription was performed in four subsequent 15 min steps at 42°C, 50°C, 55°C and 60°C. The RT enzyme was inactivated at 85°C for 5 min, followed by RNase H (New England Biolabs, 1 U)-treatment for 20 min at 37°C.
For direct cloning of full-length gfp fusions, 1 μl cDNA served as template in a standard PCR using taq polymerase (New England Biolabs), and 25 pmol each of a gene-specific primer (antisense to the N-terminal coding region of the gene of interest and with a NheI site extension) and the adapter-specific primer JVO-0367. Products were separated on 3% agarose gels, bands of interest excised (stronger bands in TAP-treated samples compared to mock-treated samples indicated full-length transcripts), gel-eluted (Jetsorb, Genomed) and digested with BseRI and NheI. The digested DNA fragment was cloned into the BsgI/NheI digested plasmid pXG-20. In some cases, weak TAP-specific PCR products required a second PCR amplification step using the same primer combinations to increase DNA yields for cloning.
In vivo whole-cell colony plate fluorescence imaging
E.coli Top10 cells expressing plasmid-borne gfp fusions were streaked on standard LB plates supplemented with the appropriate antibiotics. After over night growth colonies were photographed in a FUJI LAS-3000 image analyzer using a CCD camera with a 510 nm emission filter and excitation at 460 nm.
Liquid culture whole-cell fluorescence measurements and data processing
To measure whole-cell fluorescence in liquid culture, E.coli strains harboring gfp fusion plasmids were inoculated 1/100 from overnight cultures into 20 ml fresh LB medium in erlenmeyer flasks. Three independent overnight cultures were used throughout the study for each strain. Cultures were incubated with aeration at 37°C/220 r.p.m. and cell density was followed by measuring OD600. At the indicated cell density, three aliquots (150 μl) of each culture were transferred to a 96-well microtiter plate (Nunc, cat# 167008), and fluorescence measured at 37°C (optical excitation filter 480/31 nm, emission filter 520/10 nm, 0.2 s, CW lamp energy 21673, measurement height 8.0 mm) in a Victor3 machine (1420 Multilable Counter, Perkin Elmer).
To calculate absolute fluorescence of a given strain, the mean fluorescence of the three aliquots from each of the three independently grown cultures was determined. Unless stated otherwise, cellular autofluorescence was subtracted to obtain the specific fluorescence of the gfp fusion. Herein, the fluorescence of strains harboring the same sRNA expression or control plasmid in combination with the negative control plasmid pXG-0 (expressing luciferase, i.e. no gfp) was measured as described above and subtracted from absolute fluorescence values obtained in presence of the gfp fusion plasmid of interest.
The regulatory effect of a sRNA on a given gfp fusion was calculated as follows. Strains harboring the fusion of interest in combination with a specific negative control plasmid (i.e. without sRNA expression), were measured to obtain absolute fluorescence values and autofluorescence of strains harboring the same negative control plasmid in combination with pXG-0 was subtracted resulting in the ‘unregulated gfp fusion specific fluorescence’. Strains harboring the same gfp fusion of interest in combination with a specific sRNA expression plasmid were measured and the autofluorescence of strains harboring the same sRNA expression plasmid in combination with pXG-0 was subtracted to give the ‘regulated gfp fusion specific fluorescence’. Fold regulation mediated by expression of a sRNA was calculated by dividing the ‘unregulated gfp fusion specific fluorescence’ by the ‘regulated gfp fusion specific fluorescence’.
Fluorescence measurements in microtiter plates and data processing
Fluorescence measurements in 96-well microtiter plates was carried out as described in (42) with modifications. Single colonies (triplicate) of E.coli strains harboring gfp fusion and sRNA expression plasmids were inoculated in 150 μl LB in a 96-well microtiter plate and these cultures were overlayed with 50 μl mineral oil (Sigma) to prevent evaporation. Cultures were grown in a Victor3 fluorimeter set at 37°C and assayed with an automatically repeating protocol of shaking (2 mm orbital, normal speed, 900 s), absorbance (OD) measurements (600 nm, P600 filter, 0.1 s) and fluorescence readings (optical excitation filter 480/31 nm, emission filter 520/10 nm, 0.2 s, CW lamp energy 21 673). OD600 and flurorescence were measured at 17 min intervals (60 in total).
To plot fluorescence over OD600, curves of all three cultures within a triplicate were independently established first. The linear range of increasing fluorescence during growth covered by all members within a triplicate was selected individually and a cut-off set at the OD600 were at least one member showed non-linear increase of fluorescence. An average curve was calculated for each triplicate and the cellular autofluorescence curve of a strain harboring pXG-0 and pJV300 negative control plasmids subtracted. The OD600 range in which all measured cultures showed near-linear fluorescence increase is shown in Figure 9A.
To calculate the regulatory effect of sRNA expression on the ompC fusion (Figure 9B), fluorescence of E.coli strains harboring the ompC fusion in combination with a sRNA expression plasmid was divided by the fluorescence of a strain harboring the ompC fusion in combination with the sRNA control plasmid, pJV300.
Whole-cell protein fractions and western blot
Culture samples were taken according to 1 OD600 if not stated otherwise. Samples were spun 2 min at 16 100 g at 4°C. The cell pellet was resuspended in 1× sample loading buffer (Fermentas, #R0891) to a final concentration of 0.01 OD/μl. Samples were heated 5 min at 95°C.
A total of 0.01 or 0.05 OD of whole-cell protein fractions of strains expressing highly or weakly fluorescent GFP fusions, respectively, were separated by 15% SDS–PAGE. Gels were blotted for 60 min at 100 V at 4°C in a cable tank blotter (Peqlab) onto PVDF (PerkinElmer) membrane in transfer buffer (25 mM Tris base, 190 mM Glycin and 20% Methanol). After rinsing in TBST20 buffer (20 mM Tris base, 150 mM NaCl and 0.1% Tween 20), membranes were blocked for 1 h in 10% dry milk in TBTS20, followed by incubation with α-GFP monoclonal (Roche #11814460001) or α-FLAG monoclonal antibodies (Sigma #F1804; 1:1000 in 3% BSA, TBST20) for 1 h at RT, 5 × 6 min wash in TBST20, α-mouse-horseradish peroxidase (HRP) (Amersham Biosciences #NXA931; 1:5000 in 3% BSA in TBST20) for 1 h at RT, 6 × 10 min wash in TBST20. For simultaneous detection of GroEL (loading control), membranes were cut after the blocking step at the 47.5 kDa band indicated by the prestained protein marker (Fermentas), GroEL was detected using α-GroEL antisera conjugated with HRP (Sigma #A8705, 1:1000 in 3% BSA, TBST20; 2 h incubation at RT, followed by 6 × 10 min washes in TBST20). Blots were developed using Western Lightning reagent (PerkinElmer), and signals detected with a Fuji LAS-3000 CCD camera.
RNA isolation and northern detection
TRIZOL reagent (Invitrogen) or the Promega SV total RNA purification kit were used according to the manufacturer's protocol or as described at www.ifr.ac.uk/safety/microarrays/protocols.html, respectively, to isolate total RNA. Unless stated otherwise, RNA was isolated from cells grown to an OD600 of 1.
To detect gfp fusion mRNAs or sRNAs, RNA samples (corresponding to 0.7 OD culture volume) were denatured for 5 min at 95°C in loading buffer (containing 95% formamide), separated on 8.3 M urea −5 or 6% polyacrylamide gels, and transferred to Hybond-XL membranes (GE Healthcare) by electro-blotting (1 h, 50 V, 4°C) in a tank blotter (Peqlab).
For detection of the chromosomal sdhCDAB polycistronic mRNA 20 μg total RNA was separated on a 1.5% Agarose gel containing 2.2 M formaldehyde and transferred to a Hybond-XL membrane by upward capillary transfer in 10× SSC overnight as described (43).
DsrA, GcvB, MicA, MicC, MicF, RyhB, SgrS and Spot42 were detected using 5′ end-labeled oligodeoxyribonucleotides JVO-1367, JVO-0321, JVO-1371, JVO-1369, JVO-0909, JVO-0223, JVO-1366 and JVO-1368, respectively. 5S rRNA and gfp fusion mRNAs were detected with end-labeled oligodeoxyribonucleotide JVO-0322 and JVO-155, respectively. The sdhCD fusion mRNA was detected with a random-labeled ([32P] dCTP; Readiprime II labeling kit, GE Healthcare) PCR fragment generated with primer pair JVO-0642/-1101. To detect the chromosomally expressed sdhCDAB polycistronic mRNA, a PCR fragment generated with primer pair JVO-1360/1361 was in vitro-transcribed from the T7 promoter (added by primer JVO-1361) in the presence of [α-32P]UTP using Ambion's T7 polymerase Maxiscript kit. Riboprobes were purified over a G50 column.
Prehybridization and hybridization of membranes with riboprobes, DNA probes, or oligonucleotides was carried out in Roti-Hybri-Quick buffer (Roth, #A981.1) at 70°C, 65°C, or 42°C, respectively, for 2 h. Membranes hybridized with riboprobes were washed at 65°C in three subsequent 15 min steps in SSC (2×, 1× and 0.5×)/0.1% SDS solutions, after rinsing the membrane first in 2× SSC/0.1% SDS. Membranes hybridized with PCR fragments were rinsed in 2× SSC/0.1% SDS, followed by 15 min washes in 2× (65°C), 1× and 0.5× (42°C) SSC/0.1% SDS. For end-labeled oligodeoxyribonucleotides hybridization membranes were rinsed in 5× SSC followed by three wash steps at 42°C in SSC (5×, 1× and 0.5×, respectively). Signals were visualized on a phosphorimager (FLA-3000 Series, Fuji), and band intensities quantified with AIDA software (Raytest, Germany).
RESULTS
General approach
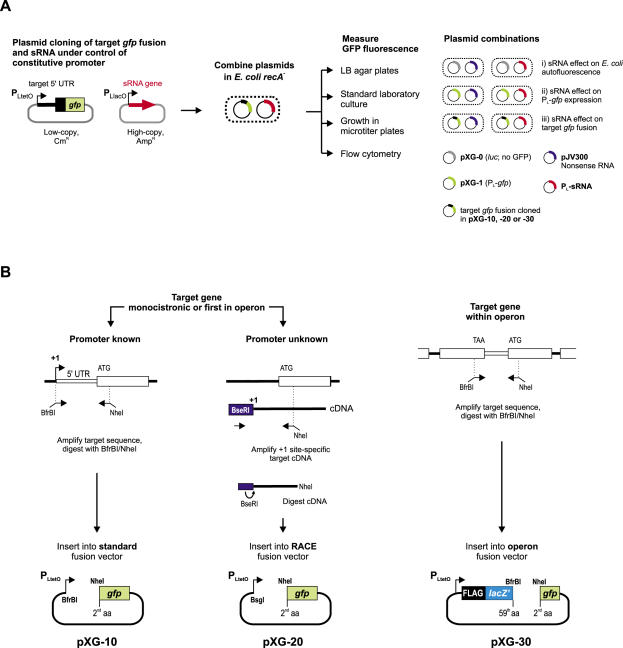
To study sRNA-mediated translational control at the 5′ region of a given target mRNA (from here on: target), we use two compatible plasmids derived from the pZE series of expression vectors (44) that can be stably maintained in an E.coli recA- strain (Figure 1A). The target plasmid is a low-copy vector that carries a pSC101* origin of replication (3–4 plasmid copies/cell), a cat chloramphenicol resistance marker, and the 5′ sequence of the target as a translational fusion to the N-terminus of GFP. Transcription of the gfp fusion gene is driven by PLtetO-1, a constitutive promoter that is derived from the native phage λ PL promoter (44). The sRNA plasmid is a high-copy vector carrying a ColE1 origin of replication (∼70 copies/cell) and a bla ampicillin resistance gene. The sRNA gene of interest is cloned under control of the constitutive PLlacO-1 promoter [another modified version of λ PL; (44)] such that transcription will precisely start at the native +1 site of the sRNA.
Figure 1.
Principle approach and gfp fusion cloning strategies. (A) Putative sRNA target sequences are cloned as translational fusions to gfp on a low-copy vector that carries a pSC101* origin of replication (3–4 plasmid copies/cell) and confers chloramphenicol resistance. The fusion is transcribed from a constitutive λ PLtetO-1 promoter (PL derivative). Regulatory sRNAs are cloned on a high-copy vector that carries a ColE1 origin of replication (∼70 copies/cell) and confers ampicillin resistance. The sRNA gene is cloned under control of the constitutive PLlacO-1 promoter (another derivative of λ PL) such that transcription will precisely start at the native +1 site of the sRNA. For E.coli cells that carry both plasmids, the effect of a given sRNA on a target fusion can be determined by monitoring GFP fluorescence of colonies grown on agar plates, of liquid cultures grown in standard laboratory flasks or in microtiter plates, or by flow cytometry. Combinations of fusion and sRNA expression plasmids with control plasmids are used to determine (i) the basal fluorescence of E.coli cells and how it is affected by sRNA overexpression, (ii) the general effect of the plasmid-borne sRNA gene on GFP expression and (iii) the specific effect of an sRNA on a target fusion of interest. (B) Putative target sequences are PCR-amplified and cloned into specialized gfp fusion vectors. If the target sequence is derived from a monocistronic gene or the first gene of an operon, and its promoter is known (left panel), it is amplified with an upstream primer that binds at the +1 site of the target gene and adds a BfrBI site, and a downstream primer that binds in the N-terminal region of the target gene and adds an NheI site in frame with the target gene coding region. The resulting PCR product is inserted into the standard fusion vector, pXG-10, digested with BfrBI/NheI. If the promoter +1 site is unknown (middle panel), the target sequence is amplified from cDNA of total E.coli RNA that was ligated to a 5′ RNA linker oligo upon treatment with TAP (this enzyme converts the 5′ PPP group of primary transcripts to 5′ P and thus allows the differential amplification of cDNAs that correspond to the native +1 site of an mRNA). The amplified cDNA will carry a 5′ BseRI site (contained in the RNA linker sequence). Insertion of the NheI/BseRI-digested cDNA into NheI/BsgI-digested RACE fusion plasmid, pXG-20, ensures that transcription of the fusion mRNA starts at the native +1 site of the target gene. Target sequences that are derived from within polycistronic mRNAs are amplified and cloned into the operon fusion vector pXG-30 (right panel). The upstream primer adds a BfrBI site in frame with the C-terminus of the upstream ORF; cloning into pXG-30 will create a C-terminal fusion to a short artificial reading frame composed of a FLAG epitope and a truncated lacZ gene, thus mimicking operon mRNA expression. See text and Figure 6A for more details.
The constant transcription rate of both the regulatory sRNA and the target fusion is a key feature of this system. It uncouples both players from the chromosomal transcriptional network, and diminishes the possible pleiotropic effects of sRNA expression on target fusion transcription. It also ensures high yields of the expressed RNAs, thus minimizing the contribution of any transcripts from the chromosomal copies of the respective sRNA or target genes. In case the high transcription rate of either promoter yields toxic RNA levels, it may be controlled in E.coli strains that encode the LacI or TetR repressor proteins [repressing PLlacO-1 or PLtetO-1, respectively; (44)] by addition of an appropriate inducer. Alternatively, sRNA genes are cloned on a plasmid carrying a p15A origin of replication, thus lowering the copy number to ∼20 per cell (44).
E.coli cells carrying a target fusion plasmid of interest are transformed with plasmids expressing either the cognate regulatory sRNA or a nonsense (control) RNA. The GFP fluorescence of the resulting transformants is subsequently read out from colonies on LB agar plates or from cells grown in liquid culture, and corrected for the autofluorescence of E.coli. Fusions that exhibit higher GFP activity in the presence of a sRNA plasmid are considered activated, whereas lower GFP fluorescence indicates target repression. In cases where GFP activity is low, i.e. close to autofluorescence, western blotting with an anti-GFP antibody provides a more sensitive measure for quantification of fusion protein levels.
Cloning and activity of translational gfp fusions
All gfp fusions described carry the gfp+ allele, which encodes a GFP variant that combines mutations for higher fluorescence yield and increased folding efficiency (45). Plasmid pXG-10 is the standard plasmid for directional cloning of a potential target mRNA sequence as N-terminal translational fusion to GFP (Figure 1B). Selected target regions are PCR-amplified using a sense primer that binds to the +1 site (if known) of the target gene and adds a BfrBI restriction site to this sequence, and an antisense primer that binds in the 5′ coding region and adds a NheI restriction site in frame with the target gene. Plasmid pXG-10 has a single BfrBI site at the +1 position of the PLtetO-1 promoter hence all fusion transcripts will have a uniform AUGCAU 5′ end. The single NheI site (GCTAGC) of pXG-10 represents the 2nd and 3rd codon of the gfp reading frame, hence the translational target gene fusion will be to full-length GFP protein. Control plasmid pXG-0 does not contain a gfp gene but instead constitutively expresses luciferase, and is used to determine the autofluorescence background of E.coli cells. Control plasmid pXG-1 expresses full-length GFP and carries an artificial 5′-UTR containing a strong RBS that is derived from the pZE family of expression vectors (44). Two additional fusion vectors, pXG-20 and pXG-30, were constructed for cloning of 5′ RACE products and of intra-operonic target sequences, respectively (Figure 1B). The cloning strategies for these plasmids are described along with their applications further below.
Using these vectors, we have thus far constructed >80 translational gfp fusions to diverse genes of E.coli, Salmonella typhimurium and Vibrio cholerae, which are listed in Table 1 and in Supplementary Table S3. These fusions include several known targets of E.coli sRNAs, which were the focus of this study, as well as various mRNAs that were predicted as sRNA targets in our laboratory (C. M. Sharma and J. Vogel, unpublished data). For simplicity, the fusions listed throughout this paper refer to E.coli genes unless stated otherwise.
Table 1.
Overview of relevant gfp fusion plasmids
| Target genea | Plasmid trivial nameb | Plasmid original namec | Insert 5′ endd | Fused codone | Fusion vectorf | Fluorescence on plateg | Western blot detectionh | Comment |
|---|---|---|---|---|---|---|---|---|
| E.coli | ||||||||
| dppA | pDppA::gfp | pSK-015 | −165 | 14 | pXG-10 | + | + | Predicted GcvB target |
| galK | pGalTK::gfp | pJU-147 | −180 | 47 | pXG-30 | ++++ | + | Repressed by Spot42 |
| pGalK::gfp | pSK-028 | −180 | 47 | pXG-10 | +++ | + | Repressed by Spot42 | |
| hns | pHns::gfp | pSK-009 | −36 | 28 | pXG-10 | ++++ | + | Repressed by DsrA |
| lacZ | pLacZ29::gfp | pJV-861-9 | −37 | 29 | pXG-10 | ++ | + | |
| pLacZ186::gfp | pJV-862-13 | −37 | 186 | pXG-10 | − | + | ||
| ompC | pOmpC::gfp | pSK-003 | −81 | 12 | pXG-10 | +++ | + | Repressed by MicC |
| ompF | pOmpF::gfp | pSK-005 | −50 | 13 | pXG-10 | ++++ | + | Repressed by MicF |
| ompA | pOmpA::gfp | pSK-008 | −133 | 16 | pXG-10 | + | + | Repressed by MicA |
| pOmpA*::gfp | pJU-023 | −133 | 16 | pXG-20 | + | + | Repressed by MicA | |
| pOmpA-M6::gfp | pJU-094 | −133 | 16 | pXG-10 | + | + | ||
| pOmpA-95::gfp | pJU-096 | −95 | 16 | pXG-20 | + | + | Repressed by MicA | |
| pOmpA-30::gfp | pJU-099 | −30 | 16 | pXG-20 | − | − | ||
| ptsG | pPtsG::gfp | pSK-024 | −103 | 26 | pXG-10 | + | + | Repressed by SgrS |
| rpoS | pRpoS::gfp | pSK-031 | −564 | 41 | pXG-10 | + | + | Activated by DsrA, RprA |
| sdhD | pSdhD::gfp | pSK-042 | −60 | 21 | pXG-10 | − | − | Repressed by RyhB |
| pSdhCD::gfp | pJU-162 | −59 | 21 | pXG-30 | ++ | + | Repressed by RyhB | |
| sodB | pSodB::gfp | pJV-863-18 | −56 | 47 | pXG-10 | ++++ | + | Repressed by RyhB |
| Vibrio | ||||||||
| sodB | pV.c.SodB::gfp | pJU-066 | −82 | 69 | pXG-10 | ++++ | + | Repressed by RyhB |
aGene whose N-terminal coding sequence was fused to GFP. Gene names refer to the following genome annotations. E.coli K12 (NC_000913), V.cholerae O1 biovar eltor (NC_002505). Known sRNA targets are set in boldface.
bFusion plasmid name used throughout the manuscript.
cOriginal plasmid name used for construction and storage (to be cited when requesting plasmids).
d5′ End of the target gene insert relative to annotated ATG.
eTarget gene codon that is fused to the NheI site preceding the gfp reading frame in the cloning vectors.
fVector type used for cloning.
gFluorescence on LB agar plates of E.coli strains carrying a fusion plasmid as shown in Figure 2A. (−) denotes background fluorescence, (+) weak but detectable fluorescence, (++) intermediate fluorescence, (+++) fluorescence similar to full-length GFP (control plasmid pXG-1), and (++++) stronger fluorescence than that of a pXG-1 strain.
hWestern blot detection of the fusion protein in cells grown to OD600 of 1. (−) = no detection, (+) = protein detected.
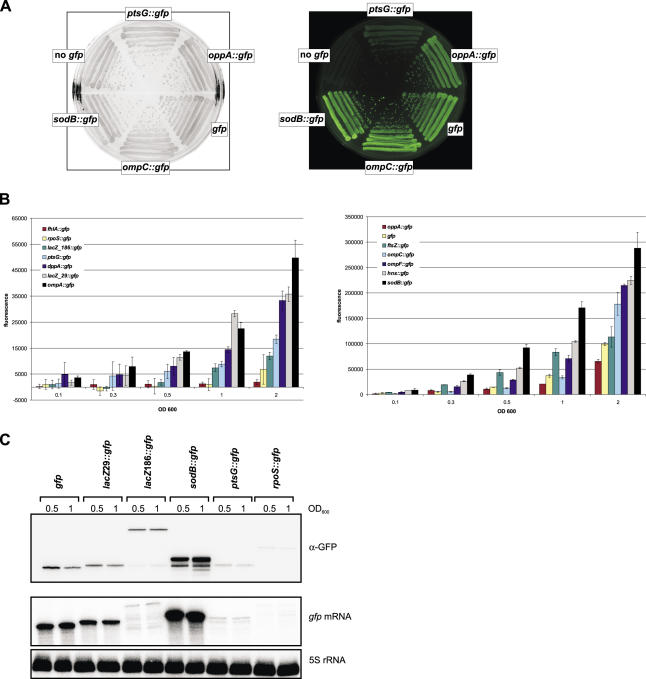
A preliminary determination of fluorescence on standard LB agar plates by visual inspection revealed large variations of GFP activity among these reporter strains. Figure 2A shows images of five representative reporter strains that were used for a rough classification of GFP activity (Table 1). Fusions that show fluorescence similar to full-length GFP (control plasmid pXG-1), e.g. ompC, were classified +++. Interestingly, a sodB fusion exhibited a higher fluorescence than pXG-1, and was thus classified ++++. Fusions with intermediate yet readily detectable fluorescence, e.g. oppA were marked ++, whereas fusions, such as ptsG with levels just above the autofluorescence of pGX-0 cells were classified +. Altogether, >80% of the 68 E.coli fusions listed in Table 1 and Supplementary Table S3 had detectable fluorescence on agar plates, and all but the sodB fusion (smaller colonies) formed colonies of regular size.
Figure 2.
Fluorescence and expression characteristics of representative gfp fusions. See Table 1 and Supplementary Table S3 for details of fusion plasmids. (A) E.coli strains carrying control plasmids (no gfp = pXG-0; gfp = pXG-1) or target fusion plasmids (as indicated) were grown on LB agar. The left image was obtained in the visible light mode and shows the colony morphology of these strains. The right image shows the same plate in the fluorescence mode. GFP fluorescence was excitated at 460 nm, and light emission was recorded using a 510 nm-filter. (B) Fluorescence of E.coli cells carrying the indicated gfp fusions at different cell densities. Bacteria were grown aerobically in liquid culture in triplicates and aliquots were measured at the indicated cell density (OD600). Fluorescence values are given in arbitrary units and were corrected for the basal fluorescence of an E.coli strain harboring plasmid pXG-0 (∼40 000 U). The left panel shows a set of low fluorescence fusions; the right panel shows fusions that yielded high fluorescence and includes plasmid pXG-1 expressing full-length GFP. (C) Detection of GFP fusion proteins and gfp fusion mRNAs. Samples were taken from liquid cultures of strains carrying the gfp control plasmid pXG-1 or the indicated fusion plasmids at OD600 of 0.5 and 1, and were subjected to western blot analysis with monoclonal α-GFP antibodies (upper panel) and to northern analysis (middle panel). The same northern blot was probed for 5S rRNA as a loading control (lower panel).
We next determined the GFP activity of a broad set of fusions in liquid culture (Figure 2B). Overnight cultures were diluted into fresh LB media, and overall culture fluorescence was determined at five growth stages, i.e. at a cell density of OD600 of 0.1, 0.3, 0.5, 1 and 2. We observed an almost linear correlation of cell number and culture fluorescence, as well as small standard deviations within triplicates, with fusions that had shown high GFP activity on plates (Figure 2B, right panel). Several fusions surpassed the full-length GFP expressed from pXG-1 in terms of fluorescence, i.e. ftsZ, hns, ompC, ompF, and sodB. In contrast, many fusions with low GFP activity required growth to an OD600 ≥ 0.5 for reliable detection (Figure 2B, left panel). Interestingly, some of the target genes that yielded low GFP activity had previously been fused to lacZ, e.g. fhlA or rpoS, and similarly small (<200) Miller unit numbers had been reported (22,25,26). However, we need to caution against a general comparison to previously published lacZ fusion results since these fusions greatly vary in their way of construction (chromosomal versus plasmid-borne fusions), the growth stage at which β-galactosidase activity was determined, as well as enzyme activity units.
We next sought to determine a correlation among reporter fluorescence, steady-state fusion mRNA levels and fusion protein accumulation. Northern blots of RNA samples taken at two growth stages were probed for the gfp portion of the fusion mRNAs, and likewise fusion protein levels were determined on western blots with a mixture of two monoclonal antibodies that recognize GFP. A cross-comparison of GFP fluorescence (Figure 2B) with the corresponding mRNA and protein levels (Figure 2C) indicates a good correlation in five cases, i.e. the sodB, ptsG, rpoS and lacZ29 fusions, and wild-type GFP. For example, both the mRNA and fusion protein levels of the bright sodB fusion far exceed those of wild-type GFP. In contrast, the ptsG and rpoS fusions, both being in the lower fluorescence range, are hardly detectable at the mRNA and protein level. However, the case of the two different lacZ::gfp fusions included here merits further description. Fusion lacZ186 differs from lacZ29 by the additional inclusion of residues 30–186 of LacZ (Table 1). Even though there were drastic differences between the mRNA levels and processing patterns of the two fusions, comparable amounts of fusion protein were detected (Figure 2C). Fluorescence still differed by a factor of 3 (Figure 2B), indicating that the larger LacZ portion of lacZ186 may affect proper folding or solubility of the GFP fusion protein. It may thus be advisable to keep the fused target sequence as short as possible, thereby also avoiding the inclusion of intact signal peptides of extracytoplasmic target proteins.
Repression of target fusions by sRNAs
We cloned several regulatory sRNAs previously reported by us and others (see Table 2) into a ColE1-based vector that is compatible with the aforedescribed gfp fusion plasmids. Our strategy ensures transcription from the plasmid-borne constitutive PLlacO promoter to start precisely at the native +1 site of the sRNA (see Materials and Methods). Plasmid pJV300, which expresses a ∼50 nt nonsense RNA derived from the rrnB terminator region (46), is the standard control vector for these PL-driven sRNA expression plasmids.
Table 2.
Regulatory sRNA plasmids used in this study
| Plasmid trivial namea | Plasmid original nameb | Promoterc | Origind | Source/commente | Control plasmidf |
|---|---|---|---|---|---|
| E.coli | |||||
| pDicF | pSK-019 | PLlacO-1 | ColE1 | This study | pJV300 |
| pDsrA | pBRdsrA | PdsrA | pMB1 | (84) | pBR322 |
| pGcvB | pJU-014 | PLlacO-1 | p15A | This study | pTP-011 |
| pIstR | pJV3H-22 | PLlacO-1 | ColE1 | (14) | pJV300 |
| pMicA | pJV150IG-34 | PLlacO-1 | ColE1 | (17) | pJV300 |
| pMicA_M6 | pMicA_M6 | PLlacO-1 | ColE1 | (17) | pJV300 |
| pMicC | pSK-017 | PLlacO-1 | ColE1 | This study | pJV300 |
| pMicF | pMI | PLlacO-1 | ColE1 | J. Slaghter-Jäger and E. G. Wagner, unpublished data | placIq-micF |
| pOxyS | pOxyS | Ptac | pMB1 | (32) | pKK177-3 |
| pRyhB | pJU-002 | PLlacO-1 | p15A | This study | pTP-011 |
| pRprA | pJV-100IA-T4 | PLlacO-1 | ColE1 | This study | pJV300 |
| pSgrS | pLCV1 | PLlacO-1 | pMB1 | (48) | pHDB3 |
| pSpot42 | pISpf | PAl/O4 | pMB1 | P. Valentin-Hansen, unpublished data | pBR322 |
| Vibrio | |||||
| pVC-ryhB | pJU-073 | PLlacO-1 | ColE1 | This study | pJV300 |
asRNA expression plasmid names as used throughout the manuscript.
bName of original plasmid as provided by others or as described in Materials and Methods.
cPromoter that drives sRNA gene expression in a given plasmid.
dOrigin of replication.
eOriginal publication and/or source of a plasmid unless contructed in this study.
fCorresponding control plasmid for a given sRNA plasmid.
The RyhB and the GcvB plasmids gave aberrantly small colonies after transformation of E.coli; we thus lowered their copy number by replacing the ColE1 origin with p15A. We also note that on three sRNA expression plasmids that we obtained from other labs (Table 2), OxyS and Spot42 are expressed from a different constitutive promoter and DsrA from its native promoter.
We first checked possible effects of these plasmid-expressed sRNAs on the activity of full-length GFP (plasmid pXG-1). Figure 3A, upper left panel, shows the changes of fluorescence in the presence of diverse sRNA plasmids normalized to fluorescence obtained with the corresponding control plasmid (see Table 2). Most of these sRNA plasmids had a negligible effect on GFP fluorescence, whereas DsrA, RyhB and Spot42 positively changed fluorescence up to 1.5-fold. This unspecific effect will have to be taken into account when calculating the regulation of target mRNA fusions. Although some of these plasmids (DsrA, RyhB and Spot42) were observed to affect bacterial growth, either causing a longer lag phase or earlier entry into stationary phase (data not shown), this does not seem to influence GFP expression.
Figure 3.
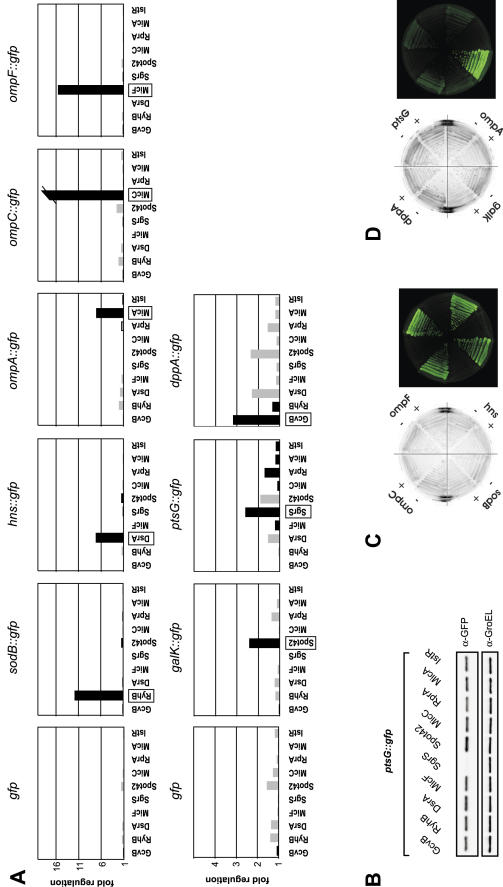
Specific regulation of target fusions by co-expression of cognate sRNAs. (A) E.coli strains carrying the full-length GFP control plasmid pXG-1 (upper and lower left panel) or target gene fusions (as indicated above the individual panels) were co-transformed with the sRNA expression plasmids given below each panel. Fluorescence of liquid cultures was determined at an OD600 of 1. Fold regulation was calculated from the relative fluorescence by dividing the fluorescence obtained in the presence of the sRNA plasmid by that obtained with a control plasmid that does not express a regulatory RNA. Gray and black bars indicate positive regulation (target activation) and negative regulation (target repression), respectively. The cognate regulatory sRNA for each fusion is boxed. The ompC fusion was repressed >20-fold in the presence of the MicC expression plasmid. (B) Specific regulation of the low-activity ptsG fusion by SgrS as determined by western blot analysis. Total cell protein samples of the same strains as in (A), samples were taken at an OD600 of 1. The PtsG::GFP fusion protein was detected with α-GFP antibodies (upper panel). Detection of GroEL protein with a specific antibody was used to confirm equal loading (lower panel). (C) Fluorescence of strains that carry the ompC, ompF, hns or sodB fusion in combination with either a control plasmid (−), or MicC, MicF, DsrA or RyhB expression plasmids, respectively (denoted by +). Strains were grown on agar plates and images were taken in the visible light or fluorescence mode as in Figure 2A. (D) As in (C) but showing dppA, ptsG, ompA or galK fusion regulation by GcvB, SgrS, MicA or Spot42 co-expression, respectively.
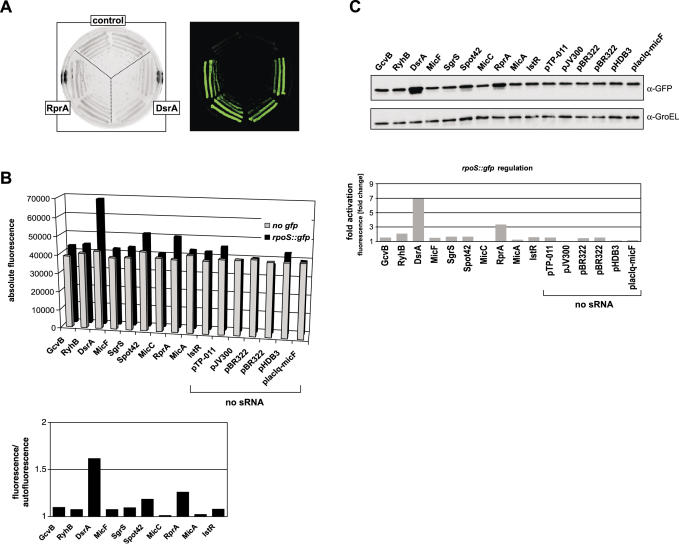
Subsequently, we combined 10 sRNA plasmids with eight target fusions. We expected to see repression with the sRNA/target pairs, DsrA/hns (47), MicA/ompA (17,18), MicC/ompC (16), MicF/ompF (29), RyhB/sodB (20), SgrS/ptsG (48,49) and Spot42/galK (50). In addition, there was some evidence of dppA mRNA being a target of GcvB (51). RprA and IstR-1, which regulate rpoS (52) and tisAB (14), respectively, were included as unspecific control RNAs (for simplicity, IstR-1 is referred to as IstR throughout this paper). Figure 3A shows that for each of the targets tested in this array, the previously described regulatory sRNA provided the highest degree of repression. Generally, repression was more pronounced for target fusions with high fluorescence yields, as it is most obvious for ompC (cf. Figure 3A), which was regulated >20-fold by MicC, but less than 2-fold by any other sRNA. The low fluorescence dppA fusion was repressed ∼3-fold by GcvB, but this repression appeared to be specific since GcvB had marginal effects on all the other targets; DsrA and Spot42 even had a positive effect on dppA in line with their aforementioned activating effect on GFP alone (Figure 3A). At first glance, the subtle regulation observed for the low fluorescent ptsG fusion by SgrS seems to be the least specific one in this array. At an OD600 of 1, ptsG had a regular fluorescence value of ∼47 000 (arbitrary units), which was close to the E.coli autofluorescence (∼38 000 to ∼40 000) and renders reliable calculations of regulation factors difficult. To measure ptsG regulation more precisely, we determined PtsG::GFP protein expression (by western blot) in the presence of all sRNAs. Figure 3B shows that SgrS reduced the PtsG::GFP signal to background levels, whereas all other tested sRNA plasmids had negligible effects. This confirms the high specificity and regulatory strength of the SgrS/ptsG interaction, while it also indicates the requirement for a minimal fusion activity to observe clear-cut regulation in liquid culture measurements.
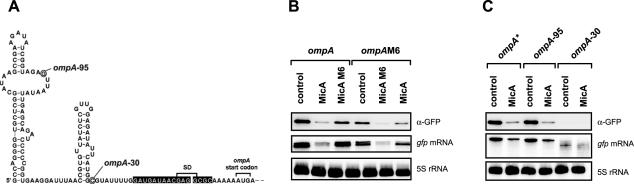
Ultimate proof for in vivo interaction is typically obtained by the introduction of compensatory base pairs in the regulatory sRNA and its mRNA target. The so-called M6 mutation in the MicA/ompA pair refers to simultaneous disruption of 6 bp in this interaction site, either obtained by mutation of MicA or ompA. Previously, introduction of six compensatory mutations in ompA to restore base pairing with the MicA_M6 mutant RNA (and vice versa) successfully restored regulation of MicA/ompA as measured by the activity of plasmid-borne ompA::lacZ fusions (17). Since this provided a means for direct comparison between a lacZ and a gfp reporter, the same mutations were introduced in ompA::gfp. Similar to the data reported by (17), wild-type ompA::gfp was hardly regulated by MicA_M6 at the fusion mRNA or protein level, whereas ompAM6::gfp was regulated by MicA_M6 but hardly responded to wild-type MicA (Figure 4B). This finding proves that gfp is as reliable a reporter of ompA regulation as lacZ.
Figure 4.
Regulation of various ompA fusions by MicA. (A) Structure of the E.coli ompA leader adapted from (17). Nucleotides that pair with MicA are indicated by black boxes. The 5′ ends of the two truncated ompA fusion, ompA-30 and ompA-95, are circled. (B) Introduction of compensatory base pair changes into MicA sRNA, yielding MicA_M6, restored regulation of the mutated ompAM6 fusion mRNA, thus confirming specific pairing of the two RNAs in vivo. The M6 mutation in ompA or MicA simultaneously disrupts base pairing between ompA and MicA at six positions, as described previously (17). E.coli strains carrying the ompA wild-type or ompAM6 mutant fusion plasmid were combined with a control plasmid (no sRNA expression), or the MicA or MicAM6 expression plasmids. Samples were taken at an OD600 of 1, and subjected to western (upper panel) and northern (lower two panels) blot analysis as in Figure 2C. (C) Effects of 5′ truncations on ompA* fusion mRNA stability and regulation by MicA. In the ompA* wild-type fusion (constructed by 5′ RACE cloning, see text), transcription from the constitutive PLtetO promoter starts at the native ompA +1 site. Destruction of the terminal stem–loop of the ompA leader neither affects stability or translation of the mutant fusion ompA-95, nor its repression by MicA. However, further shortening of the ompA leader as in mutant ompA-30, which is deprived of both stem–loops, results in partial degradation of the fusion mRNA and loss of fusion protein translation. Samples were taken and probed as in (B).
The length of the fused target mRNA sequence could be another determinant of sRNA regulation. Ideally, the cloned region would encompass the entire 5′-UTR and include a short stretch of the coding region. We reasoned that fusion cloning should be based on the native 5′-UTR to ensure a comparable stability of the fusion transcript to the parental mRNA. However, the +1 site or promoter is only known for a subset of the E.coli genes, and even less information on transcription start sites is available for other bacteria. To solve this problem, we developed vector pXG-20 as part of a cloning strategy that combines +1 site mapping and rapid fusion cloning (Figure 1B). Briefly, this includes a 5′ RACE protocol that distinguishes primary 5′ mRNA ends (carrying a tri-phosphate) group from processed mRNA species (2,4,53,54), followed by the direct insertion of a target mRNA 5′ RACE fragment into vector pXG-20 such that transcription from PLtetO will precisely start at the mapped +1 site. The full protocol and an example of 5′ RACE fusion cloning are provided in the Supplementary Data. Following this approach we were able to directly clone an ompA::gfp fusion using E.coli total cDNA as PCR template. The obtained plasmid, pOmpA*::gfp, contains the same full-length fusion as ompA::gfp, but without the 5′-ATGCAT extension added in the standard cloning procedure. To investigate how varying the length of a target 5′-UTR would affect regulation, two shorter derivatives of the ompA*::gfp fusion were constructed. The 133 nt ompA 5′-untranslated leader (Figure 4A), as contained in ompA*::gfp, is well-characterized in terms of both its 5′ end structure (55) and the MicA interaction site (17,18). Moreover, the first 115 nt containing two stem–loop structures were shown to act as a stabilizer of ompA mRNA in vivo (56,57). In ompA-95, the first stem–loop was destroyed, thus creating a 5′ end that should be single-stranded. In ompA-30, transcription was expected to start only 30 nt upstream of the ompA start codon; this mutant retained the MicA target site but not the ompA Hfq binding site (58). Figure 4C shows that destruction of the stabilization stem–loop I (ompA-95) had no effect on fusion mRNA or protein abundance, and that repression by MicA was unaffected. However, ompA-30 yielded a much less abundant and partially degraded fusion mRNA, and no detectable fusion protein, which rendered it difficult to determine regulation by MicA.
Activation of an rpoS fusion by sRNAs
While repression of target translation is the predominant mode of sRNA action, the rpoS mRNA provides an excellent example to study activation by sRNAs [reviewed in (59)]. We tested activation of an rpoS::gfp fusion with a set of sRNA plasmids, expecting higher fluorescence exclusively with DsrA and RprA, two sRNAs that act to melt the inhibitory structure that sequesters the rpoS RBS. Although rpoS::gfp fluorescence is in the lower activity range (Table 1), its activation in the presence of DsrA or RprA plasmids is already visible on agar plates (Figure 5A). Such activation was also observed in liquid culture (Figure 5B), and generally the two sRNAs elevated fluorescence stronger than any other sRNA or the control plasmid. However, in these measurements the poor fluorescence of the rpoS fusion rendered calculation of activation factors difficult. Thus, the regulation factor in this case is given as the ratio of rpoS fusion fluorescence to E.coli autofluorescence, each obtained in the presence of the same control or sRNA expression plasmid. As seen before with other low fluorescence fusions (e.g. ptsG; Figure 3A and B), direct detection of the fusion protein on western blots provided a much clearer picture of regulation (Figure 5C). The >3-fold and the 7-fold activation determined here for RprA and DsrA, respectively, are in excellent agreement with data obtained with a chromosomal rpoS::lacZ fusion (25,52).
Figure 5.
Activation of an rpoS fusion by DsrA and RprA expression as evaluated by three independent methods. (A) Colony morphology and fluoresence of E.coli strains carrying the rpoS fusion in combination with the sRNA control vector, pJV300, or DsrA or RprA expression plasmids on agar plates (left image: visible light mode; right image, fluorescence mode as in Figure 2A). (B) Total fluorescence values of liquid cultures of E.coli carrying control plasmid pXG-0 (no gfp) or the rpoS fusion plasmid in combination with 10 sRNA expression and six control plasmids (no sRNA). The rpoS fluorescence/autofluorescence ratio is shown for the ten sRNA expression plasmids in the graph below. (C) Effects of sRNA and control plasmids on RpoS::GFP fusion protein accumulation (western blot). Quantification of RpoS::GFP levels, followed by normalization to GroEL levels, was used to calculate the activation factor relative to the control plasmid pJV300, and is shown in the graph below. Fluorescence and protein levels (as in Figure 3B) were determined from cultures grown to an OD600 of 1.
Intra-operonic sRNA target sites
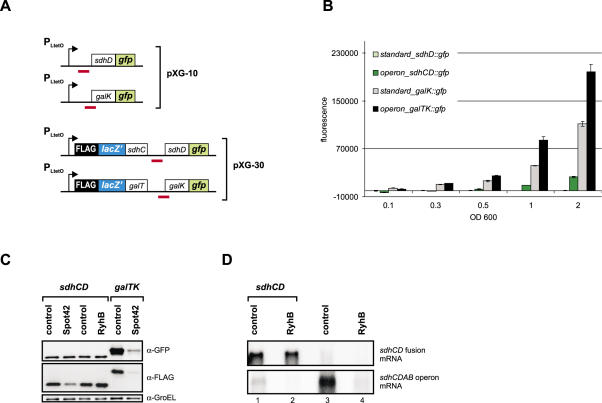
The target genes investigated so far were either monocistronic or first in an operon. Some sRNAs, however, target UTRs of downstream reading frames within a polycistronic mRNA. In the case of the polycistronic galETKM mRNA, Spot42 binds to the galK RBS region, which leads to translational repression of galK without affecting expression of the two upstream genes, galE and galT; consequently, this type of regulation was termed discoordinate operon expression (50). RyhB, which is predicted to block the sdhD RBS, may regulate the sdhCDAB operon mRNA in a similar fashion (20). Such intra-operonic targets could pose a challenge for our approach since transcription of the fusion mRNA would not start at its native +1 site, thus creating an arbitrary 5′ end that could destabilize the fusion mRNA. To solve this problem, we developed vector pXG-30, in which intra-operonic target genes are expressed as part of an artifical dicistronic mRNA (Figure 1B). Putative target genes are cloned on pXG-30 as dual fusions: the upstream coding sequence is fused to the C-terminus of a FLAG epitope-tagged, truncated lacZ gene (FlacZ′), whereas the actual target gene is fused to gfp as described above. In addition, the FlacZ′ ORF is preceded by a strong RBS derived from protein expression vector pZE12-luc (44) to ensure efficient translation initiation of the dicistronic operon mRNA. Next we cloned the sdhCD and galTK target sequences of RyhB and Spot42, respectively, in plasmids pXG10 and pXG30, and compared the GFP activity of these fusions (Figure 6A). Following the GFP activity over growth, we observed striking differences for the two vector types in the case of either fusion (Figure 6B). Specifically, when the sdhCD target site is cloned on standard vector pXG-10, the fluorescence of this fusion is close to background levels. However, if the same sequence is cloned into operon plasmid pXG-30, it yields a fusion with well-detectable GFP activity. Moreover, activity of the galTK fusion was also enhanced 2-fold by cloning into pXG30 as compared to pXG10.
Figure 6.
Regulation of targets that are derived from within polycistronic mRNAs. (A) Schematic drawing (not to scale) of sdhD and galK fusions constructed with pXG-10 (standard fusion vector), or pXG-30 (operon fusion vector). The RyhB/sdhD and Spot42/galK pairing regions are indicated by a red bar. (B) Comparison of fluorescence of E.coli strains carrying the fusion plasmids shown in (A). Fluorescence was determined at five different cell densities (OD600) of liquid cultures and was corrected for the E.coli autofluorescence as in Figure 2B. (C) Effects of RyhB and Spot42 expression on the sdhCD and galTK fusions. Shown are western blots of total protein samples taken at an OD600 of 1. Probing with α-GFP antibodies detected the SdhD::GFP and GalK::GFP proteins (upper panel), and probing with an FLAG epitope-specific antibody the FLacZ::SdhC and FLacZ::GalT proteins (middle panel). GroEL detection served as loading control (lower panel). (D) Northern blot of RNA samples taken from E.coli strains (grown to an OD600 of 1) that carry the sdhCD fusion in combination with the sRNA control plasmid pJV300 (lane 1) or the RyhB expression plasmid (lane 2), or from E.coli cells without a fusion plasmid but harboring pJV300 (lane 3) or the RyhB expression plasmid (lane 4). In the upper panel, the blot was probed for the plasmid-expressed sdhCD fusion mRNA using a labeled FlacZ′ dsDNA fragments. In the lower panel, the same blot was hybridized with an sdhC-specific probe to detect the chromosomally expressed sdhCDAB mRNA. 5 μg of total RNA were loaded in lanes 1 and 2, whereas 20 μg were loaded in lanes 3 and 4.
In the pXG30-based galTK and sdhCD constructs, the upstream and downstream fusion proteins can be specifically detected with anti-FLAG and anti-GFP antibodies, respectively (Figure 6C). According to the concept of discoordinate gal operon expression (50), we expected a reduction of GalK::GFP levels upon Spot42 co-expression but no change of FLacZ′::GalT levels. Quantification of the western blot signals shown in Figure 6C revealed an 8-fold decrease of GalK::GFP in the presence of the Spot42 plasmid, but also a 10-fold reduction of FLacZ′::GalT (Figure 6C, compare lanes 5 and 6). While the first was in keeping with the previously published model, the strong reduction of the GalT fusion protein seemed to contradict it. However, Spot42 has been consistently observed to have a ∼3-fold negative effect on the FLacZ′ moeity in other non-target constructs, e.g. FLacZ′::SdhC (Figure 6C, lanes 1 and 2, and data not shown). Since no other sRNA tested by us has shown a similar effect, we believe that this is a currently unexplained peculiarity of Spot42. If corrected for this target-independent FlacZ′-dependent effect, Spot42 regulates the galT part only 3.3-fold as opposed to 8-fold galK regulation. Notably, this is in very good agreement with (50), who reported 1.4-fold and 4.9-fold regulation of galT and galK expression, respectively.
Whereas this data confirmed discoordinate gal operon expression by Spot42, we were unable to mimic RyhB-mediated regulation of the sdhCDAB operon. As shown in Figure 6C, RyhB co-expression did not regulate the sdhCD fusion whereas it had a drastic effect on the sodB fusion (Figures 3A and 8B). However, in contrast to the sdhCD fusion, we did see a RyhB effect on the native sdhCDAB operon mRNA (Figure 6D). As outlined in the discussion section, this result does not call into question sdhCDAB as a RyhB target, or the use of pXG30 to study intra-operonic sRNA targets.
Regulation is independent of major RNA processing factors
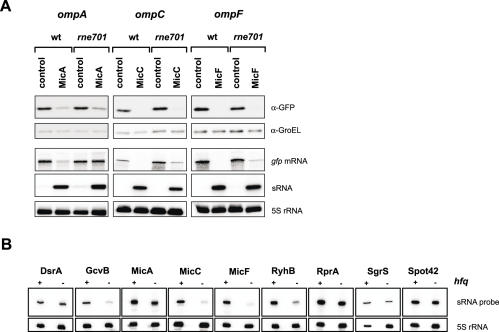
Bacterial RNA metabolism involves a large number of ribonucleases and other RNA-binding proteins, three of which—Hfq, RNase E and RNase III—are known to play prominent roles for the activity of trans-encoded antisense RNAs. In principle, our GFP system is well-suited to test the contribution of such factors by determining sRNA/target regulation in the respective hfq and RNase deletion strains. For RNase E is encoded by an essential gene (rne), we resorted to a viable rne701 mutant strain. This mutant expresses a C-terminally truncated RNase E that is defective both in interaction with Hfq and in assembly of a functional degradosome, and was recently shown to impair RyhB and SgrS action on their targets mRNAs (13,60). The rne701 and the Δhfq mutant strains were transformed with the ten sRNA/target pairs listed in Table 3. In the absence of sRNA expression plasmids, all of these fusions exhibited normal or even slightly enhanced activity in either of the two mutant strains (data not shown). Upon sRNA co-expression, none of the cognate sRNA completely failed to regulate its target in the rne701 background (Table 3), although the degree of regulation differed from the wild-type background in some cases. We also investigated if the RNase E truncation had an impact on degradation of the target fusion mRNAs. Figure 7A shows the effects of sRNA overexpression on three omp target fusions in wild-type and rne701 cells. Strikingly, although reduction of OmpA::GFP fusion protein synthesis by MicA is unaffected in rne701 cells (as compared to wild-type cells), the mutation strongly impairs degradation of the ompA fusion mRNA. The other fusion mRNAs we tested, i.e. ompC and ompF (Figure 7A) and hns, ptsG and sodB (Supplementary Figure S2) also showed some defect in fusion mRNA decay upon co-expression of the cognate sRNA in rne701 cells, although this was far less obvious than with the MicA-ompA pair.
Table 3.
Regulation in the absence of full-length RNase E or Hfq
| sRNA, fusiona | Wild-typeb | rne701c | Δhfqd | sRNA levels in Δhfq versus wild-type %e |
|---|---|---|---|---|
| DsrA, hns | −7.1 | −4.5 | −1.2 | 100 |
| DsrA, rpoSf | +7.0 | + 4.6 | +3.8 | 100 |
| GcvB, dppA | −3.2 | −4.9 | −1.6 | 19 |
| MicA, ompA | −6.0 | −2.4 | −1.5 | 63 |
| MicC, ompC | >−20 | >−20 | −1.2 | 9 |
| MicF, ompF | −14.5 | −8.1 | −1.3 | 10 |
| RprA, rpoSf | +3.4 | +3.6 | +1.3 | 69 |
| RyhB, sodB | −11.7 | −11.2 | −1.6 | 30 |
| SgrS, ptsG | −2.6 | −5.4 | 1 | 36 |
| Spot42, galK | −2.4 | −2.6 | −1.4 | 71 |
aCombination of sRNA expression and target gfp fusion plasmid.
bFold-regulation observed in wild-type background.
cFold-regulation observed in a strain that expresses a truncated RNase E.
dFold-regulation observed in an hfq deletion strain.
esRNA signals in the hfq deletion strain in % of the signal obtained in the wild-type. Signals were quantified on northern blots (Figure 7B) in strains carrying the indicated sRNA expression plasmid co-transformed with pXG-1. RNA was prepared from cells grown to an OD600 of 1. Signals were normalized to 5S rRNA detected on the same blot.
fRegulation was generally determined by measurement of fluorescence of liquid cultures grown to an OD600 of 1 but for the rpoS fusion, which was assayed on western blots.
Figure 7.
Effects of rne701 and hfq mutations. (A) Regulation of ompA, ompC and ompF fusions by MicA, MicC and MicF, respectively, in wild-type E.coli and an otherwise isogenic strain expressing a truncated RNase E (rne701). Shown are western blots of protein samples that were probed with GFP- and GroEL-specific antibodies as in Figure 2B. Below the western blot panels, northern blots show accumulation of the respective fusion mRNA (gfp probe), the overexpressed sRNA (from left to right: probing for MicA, MicC or MicF), and 5S rRNA levels as loading control. Samples were taken at an OD600 of 1. (B) Accumulation of sRNAs in the absence of Hfq. Shown are northern blots of wild-type (+) and Δhfq (−) cells that carry the sRNA expression plasmids indicated above each panel in combination with the full-length GFP control plasmid pXG-1. RNA samples were taken at an OD600 of 1; 5S rRNA probing served as loading control.
In stark contrast, the hfq deletion abrogated regulation of almost all sRNA/target pairs (Table 3). Since numerous sRNAs were previously observed to be unstable in the absence of Hfq, we compared the amounts of overexpressed sRNAs between wild-type and Δhfq cells. As shown in Table 3 and Figure 7B, the hfq deletion reduced the steady-state levels of most sRNAs, which would also contribute to the observed loss of regulation. However, the data also indicates that Hfq contributes to regulation independent of sRNA stabilization. For example, the hfq deletion reduced RyhB levels to ∼30%, yet sodB regulation to 1.6-fold as compared to ∼12-fold in wild-type cells. In addition, DsrA levels in the Δhfq strain were indistinguishable from the wild-type background; in the absence of Hfq, DsrA could still promote rpoS activation whereas it entirely failed to repress hns translation. Interestingly, this is in keeping with a previous observation that multi-copy DsrA could partially bypass the Hfq requirement for rpoS but not for hns regulation (61).
Assaying sRNA/target regulation in a strain defective of RNase III (rnc14) proved more difficult. For unknown reasons, all fusions showed dramatically lower activity, often indistinguishable from the E.coli autofluorescence. We thus selected two high fluorescence fusions, ompC and ompF, and studied their regulation in rnc14 on western blots. The cognate sRNAs, MicC and MicF, form extended duplexes with these targets, which we considered as good RNase III substrates. Furthermore, we have observed that MicC and MicF accumulate to high levels in a Salmonella rnc14 strain (Pfeiffer et al., unpublished data), which may indicate impaired interaction with target mRNAs. However, we observed that MicC/ompC and MicF/ompF regulation is as effective in E.coli rnc14 as in the isogenic wild-type strain (data not shown). Similarly, repression or activation was not impaired by rnc14 for MicA/ompA, SgrS/ptsG, and RyhB/sodB, or for RprA/rpoS, respectively (data not shown).
Assaying heterologous sRNA–target interactions
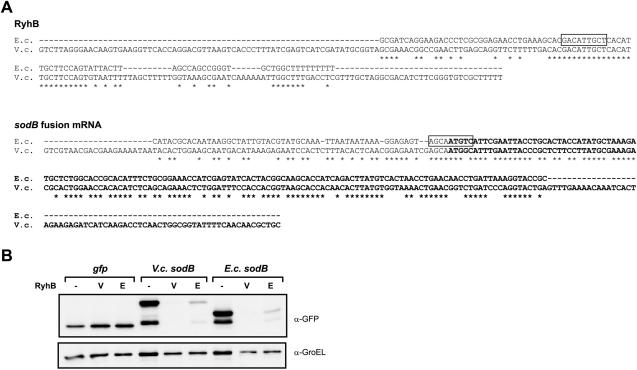
Few other bacteria in which sRNAs have been identified offer as excellent genetic tools as E.coli to study regulation of putative sRNA targets in vivo. However, in some cases sRNAs of even distantly related species were shown to regulate their target upon co-expression in E.coli, e.g. lhtA RNA of Chlamydia trachomatis (62). To test if we could use E.coli as a host to assay heterologous sRNA/target pairs, we co-expressed V.cholerae RyhB along with a sodB fusion derived from this bacterium. RyhB/sodB regulation in V.cholerae was previously suggested by (63). Interestingly, V.cholerae RyhB (∼225 nt) is more than twice as long as E.coli RyhB (∼90 nt), and the two homologues show little similarity except for the sodB interaction site (Figure 8A). Similarly, the sodB 5′-UTR differs substantially between the two bacteria. Nonetheless, we found that Vibrio RyhB effectively represses translation of both the Vibrio and the E.coli sodB fusion (Figure 8B). Reciprocally, E.coli RyhB effectively represses both the Vibrio and the E.coli sodB fusion.
Figure 8.
E.coli as a host to assay heterologous sRNA/target interactions. (A) Alignments of E.coli and V.cholerae RyhB RNA and sodB fusion mRNA (the cloned sodB DNA fragment is shown). Sequence information is based on (20,21,63). Note that the native +1 site of sodB mRNA in V.cholerae is unknown (B. Davis and M. K. Waldor, personal communication). The nucleotides of the RyhB/sodB pairing regions, as experimentally determined for E.coli (21), are boxed. Note that a different interaction has been proposed for the Vibrio RyhB/sodB pair (63). The sodB coding region is set in boldface. (B) Western blot detection of GFP and SodB::GFP fusion proteins from E.coli strains that expressed GFP (control plasmid pXG-1), a V.cholerae sodB fusion, or the E.coli sodB fusion, each in combination with the control plasmid pJV300 (−), the V.cholerae RyhB (V) or the E.coli RyhB (E) expression plasmid. Samples were taken at an OD600 of 1.
Growth in microtiter plates
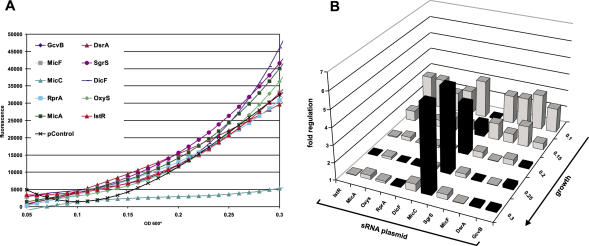
The experiments thus far described were carried out under standard laboratory growth conditions, i.e. growth in culture flasks with aeration. To test if specific regulation could also be obtained in a set-up that is more suited for high-throughput screening, we grew the ompC fusion strain co-transformed with various sRNA plasmids in small culture volumes (150 μl) overlayed with mineral oil in microtiter plates. Following inoculation from single colonies, cell density and fluorescence were monitored in 15 min intervals over a course of 16 h (Figure 9A). Fluorescence increased almost linear after an initial phase for all strains but MicC/ompC; fluorescence of the latter remained almost constant throughout and only increased slightly towards the end of the assay. Specific ompC repression by MicC is observed early in growth, and increases to 7-fold at the end of the assay (Figure 9B). Taken together, although the degree of ompC regulation is about one third as compared to culture in laboratory flasks (Figure 3A), this microtiter plate-based assay provides the same specificity.
Figure 9.
Microtiter plate-based assay of MicC-mediated ompC fusion repression. (A) E.coli strains carrying the ompC fusion in combination with one of the 10 sRNA expression plasmids or the pJV300 control plasmid (specified by the color code) were inoculated in 150 μl LB medium in a 96-well microtiter plate, overlaid with mineral oil and grown with agitation at 37°C in a Victor3 plate reader for 16 h. Cell density and fluorescence were determined in 15 min intervals. Plotting of fluorescence values over growth (OD600*) shows specific repression of the ompC fusion by the MicC expression plasmid. Cell density is given as OD600* values of 600 nm light absorption in the microtiter plate well (0.2 OD600* units corresponds to ∼1 OD600 standard units). Fluorescence values were corrected for the autofluorescence of an E.coli strain carrying control plasmids pXG-0 and pJV300. (B) Factor of ompC fusion regulation by sRNAs (relative to the control plasmid pJV300, calculated as in Figure 3A) at five selected growth stages (OD600* of 0.1, 0.15, 0.2, 0.25 and 0.3). Black bars indicate ompC repression, whereas gray bars ompC activation.
DISCUSSION
We have studied sRNA-mediated control of mRNA targets by using translational fusions to gfp, encoding a non-invasive reporter of bacterial gene expression (64,65). We have observed faithful regulation of target fusions with all sRNA/target pairs whose interactions had previously been dissected at the molecular level. Of these, several had been tested by fusions to other reporter genes. Our results show that the GFP fusions constructed here perform at least equally well in terms of sRNA regulation. For example, ∼8-fold activation of an rpoS::lacZ fusion was observed upon DsrA overexpression (25,26), whereas overexpressed MicA caused a ∼6-fold decrease in the activity of an ompA::lacZ reporter gene (17). The corresponding regulation of rpoS::gfp and ompA::gfp (Figures 3A and 5B) perfectly match these previous results. Interestingly, MicC repressed a translational ompC fusion to luciferase ∼2.5-fold (16), whereas >20-fold repression of ompC::gfp was observed here (Figure 3A). While other inhibitory sRNA/target interactions, i.e. RyhB/sodB, DsrA/hns, MicF/ompF, Spot42/galK and SgrS/ptsG, were previously validated by in vitro complex formation, the fusions used here independently confirm that sRNA-mediated mRNA repression occurs in the 5′-UTR.
Several cases merit further discussion. First, repression of hns mRNA by DsrA was proposed to involve two RNA duplexes with 13 nt at the hns RBS and with 11 nt upstream of the hns stop codon, in effect leading to a circularization of hns mRNA (23). Although our hns::gfp fusion included the 13 nt RBS target region only, it was still subject to ∼7-fold regulation by DsrA (Figure 3A). Thus, our system may report regulation even if only partial target sequences are included in a fusion. Second, a chromosomal gcvB deletion was previously shown to elevate expression of a dppA::lacZ fusion but it remained unclear if GcvB acted on the cloned dppA mRNA fragment or regulated dppA transcription (51). Our results obtained with a dppA::gfp fusion strongly suggest that GcvB post-transcriptionally regulates dppA by targeting its mRNA in the 5′-UTR (Figure 3A). This has also been confirmed by biochemical analyses of the GcvB interaction site on the dppA mRNA (C. M. Sharma, F. Darfeuille and J. Vogel, manuscript in preparation). Similarly, a GcvB target site was predicted in the dppA RBS region in biocomputational analyses (30). Third, Wagner and Darfeuille (66) evaluated the free energy values (ΔG°) of several confirmed sRNA/target duplexes as well as near-cognate and non-cognate combinations, and found that ΔG° values are rarely good predictors of unknown target interactions. For example, some cognate combinations, such as RyhB/sodB and RprA/rpoS were predicted to have ΔG° values of −17 and −24 kcal/mol−1, respectively, but the values for near-cognate combinations such as MicC/ompF and MicF/ompC were similarly low (−19.3 and −20.9 kcal/mol−1, respectively). Although the latter may indicate cross-regulation of ompF and ompC by MicC and MicF, respectively, no such regulation is seen with our GFP fusions (Figure 3A). This is of particular interest since many sRNAs target RBS regions which by default have lower sequence complexity than other mRNA parts (because of interaction with 16S rRNA). However, our data indicate that this lower complexity does not seem to compromise specificity.
By developing a specialized vector to clone fusions to intra-operonic target sites, we were able to mimic discoordinate expression of the galETKM operon as mediated by Spot42. In addition, the inclusion of an upstream fusion to the artificial FlacZ′ gene appeared to greatly enhance fusion mRNA translation or stability, resulting in a detectable activity of an sdhD fusion. Using this vector, we have meanwhile identified more polycistronic mRNAs that are subject to discoordinate regulation by E.coli sRNAs (J. H. Urban and J. Vogel, unpublished data). Although we failed to detect RyhB regulation of an sdhD fusion (Figure 6C) this does not call the sdhCDAB operon as a RyhB target into question. Parallel probing of the chromosomal sdhCDAB mRNA confirmed downregulation of this target mRNA by RyhB (Figure 6D) as previously shown by (20). Moreover, other results from our lab obtained for Salmonella RyhB regulation strongly support sdhD as a RyhB target (unpublished data). Hence, the lack of sdhCD fusion repression hints at additional determinants of this regulation. For example, RyhB targeting may require additional residues or an Hfq binding site of the sdhCDAB mRNA that are located outside of the cloned 122 bp fragment, or an unknown protein factor that associates with the native sdhCDAB transcript but not with the sdhCD fusion mRNA.
While sRNA–target complexes have been extensively studied in vitro, less is known about the factors that contribute to regulation in vivo. Most of the sRNAs studied here require the bacterial RNA chaperone, Hfq, for target interaction in vitro. However, since many sRNAs also fail to accumulate in hfq mutant strains because of largely reduced stability, the contribution of Hfq to sRNA target annealing in vivo is hard to assess. In contrast, sRNA overexpression as shown here (Figure 7B and Table 3) may provide a better means to evaluate an involvement of Hfq in sRNA function in vivo. For example, we found DsrA/hns regulation to be abrogated in Δhfq cells although plasmid-borne dsrA and the hns fusion were expressed normally. Thus this system could be used to study possible defects of Hfq-dependent ribonucleoprotein complex (RNP) formation or ribosome association of the two RNAs.
RNase E-based RNPs, either containing other degradosome components or Hfq, were recently implicated in translational repression and decay of the ptsG and sodB mRNAs in vivo (13,60). We have studied the regulation of ptsG, sodB and other target gene fusions in an rne701 mutant strain that cannot assemble either of these RNPs because of the C-terminal RNase E truncation (Figure 7A and Supplementary Figure S2). Surprisingly, fusion regulation was found to be almost as effective as in the wild-type background. Although overexpression likely results in a stoichiometry that is different from that of chromosomally expressed sRNA and targets, we still expected to see some effect of the rne mutation. Nonetheless, our results were in better keeping with a more recent report showing that sRNA-mediated repression of the ptsG and sodB mRNAs in rne701 cells does occur at the level of translation inspite of the defect in mRNA degradation (67). However, there are important differences between the experiments described by Morita et al. (67) and our experimental set-up. In the former case, the ptsG and sodB mRNA decay defects in rne701 cells were most apparent upon short-term induction of the regulatory sRNA genes, which may be closer to studying the natural kinectics of sRNA-mediated gene silencing in the absence of native RNase E (67). In contrast, the experiments described here report on steady-state levels of target (fusion) mRNAs and proteins. Similar to (67) we find that upon extended SgrS expression, the ptsG target mRNA is effectively degraded in the rne701 cells, too, and we observe the same effect on fusion mRNA decay for four other target mRNAs, i.e. hns, sodB, ompC and ompF. However, while the ompA fusion mRNA is fully degraded upon MicA overexpression in wild-type cells, it fails to get depleted when MicA is expressed in rne701 cells. That this can be seen upon long-term sRNA expression makes the MicA-ompA pair an attractive model to study the contribution of RNase E to sRNA-mediated control of target mRNAs.
Although an RNase E-homologue is found in many bacteria (68), neither its RNA recognition sequences nor its ability to form RNPs are known to be conserved. Since overexpressed sRNAs regulate their targets largely independent of RNase E-based RNPs, we expect that E.coli will be a suitable host for the validation of putative sRNA/target pairs from remotely related bacteria. Here we demonstrated the regulatory capacity of such a heterologous sRNA/target pair, i.e. Vibrio RyhB/sodB, while others recently used E.coli as a host to show translational control of a C.trachomatis sRNA/target pair (62).
The double-strand specific RNase III was shown to act on E.coli sRNAs (2,69), and to cleave IstR RNA and its target upon interaction (14). The ideal RNase III substrate is a >20 bp full duplex (equivalent to about two turns of A-form dsRNA). However, considerably shorter and/or imperfect duplexes were also shown to be substrates [e.g. (70,71)]. Since MicC and MicF form extended duplexes with their targets ompC and ompF, respectively, we chose to study the regulation of ompC/F fusions in an RNase III-deficient strain. No difference in ompC/F repression was found in this strain as compared to wild-type E.coli, suggesting that the formed duplexes are no substrates of RNase III, or that inhibition of translation initiation is sufficient for regulation.
Taken together, our translational GFP fusion approach offers a rapid and reliable tool to study sRNA control of targets that are derived from both monocistronic and polycistronic mRNAs. GFP was previously used by others as a reporter of translational control to engineer artificial riboregulators (72); the major difference to our approach being that the RNA regulator and its target were expressed from the same plasmid. However, we believe that our two-plasmid system is better suited to meet the requirements of larger screens because of the ease with which already existing sRNA and fusion plasmids can be combined. Similar two-plasmid systems with lacZ reporter genes were successfully used to study interactions of cis-encoded antisense RNAs with their targets [e.g. (73)].
The results presented here encourage further work to improve and refine our methodology. First, although fusions with low GFP activity can be easily assayed on Western blots, the use of gfp alleles with increased fusion fluorescence will facilitate screening approaches. While this work was in progress, a new GFP variant, superfolder GFP, with brighter fluorescence and higher tolerance of fusion partners was described (74). Preliminary results from our lab suggest that this variant enhances the activity of some of the fusions described here (unpublished data). Second, the vast majority of the sRNA targets sites known to date are located in mRNA 5′ regions. In contrast, E.coli GadY sRNA overlaps in antisense orientation with the 3′ end of its target mRNA (27). Since GFP tolerates fusion to its carboxy terminus, it should be possible to adapt our approach to studying sRNA interactions with the 3′ end of target mRNAs. Third, many target mRNAs encode proteins that are exported to the periplasm or which integrate into membranes. We have thus far avoided inclusion of signal sequences to ensure cytoplasmic localization of the gfp fusions. Recent work from the Aiba lab, however, showed that membrane localization of the native ptsG mRNAs is required for its repression by SgrS (49). It will thus be interesting to determine how the inclusion of extracytoplasmic signal sequences affects regulation of a fusion by its cognate sRNA. Fourth, GFP as a reporter that does not require a chromogenic substrate allows studying gene regulation at the single-cell level. Several recent studies have shown considerable heterogeneity of transcriptional responses within bacterial populations [e.g. (75,76)]. Whether this also holds true for post-transcriptional processes could be determined using the reporter system described here. On this line, preliminary results with our rpoS::gfp fusion indicate that co-expression of DsrA or RprA results in bacterial populations that can be separated from control strains by high-speed flow cytometry (unpublished data).
Small non-coding RNAs have been discovered at a staggering rate in E.coli and many other eubacteria (1,77–83). Given the hundreds of sRNAs of unknown function, target identification has become a pressing issue but has been lagging behind, mainly due to an incomplete understanding of molecular rules for sRNA/target pairing. Although a first algorithm for target prediction has been implemented and even suggested additional targets for hitherto well-studied sRNAs (30), it has created rather than obviated the need for rapid and independent methods to validate the increasing numbers of predicted targets by independent methods. We believe that GFP-based reporters as the ones constructed here will be particularly useful when having to test larger numbers of predicted sRNA targets.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
We express our sincere gratitude to H. Aiba, P. Bolouq, F. Darfeuille, B. M. Davis, S. Gottesman, W. H. Hess, P. Valentin-Hansen, R. K. Hartmann, E. G. Wagner and members of our lab for their helpful comments on a manuscript draft and to U. Alon and A. Zaslavar for discussions of GFP reporter strains. The authors thank K. P. Pleissner for his much appreciated help with data analysis and SeongJoo Koo for technical assistance. B. Suess, S. Gottesman, C. Slagther-Jäger, P. Valentin-Hansen, E. G. Wagner and R. A. Lease kindly provided plasmids. Vibrio DNA was provided by J. Reidl and the RNase E antiserum by A. G. Carpousis. This work was supported by DFG grant VO 875/1-1 (Deutsche Forschungsgemeinschaft). Funding to pay the Open Access publication charges for this article was provided by Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vogel J., Sharma C.S. How to find small non-coding RNAs in bacteria. Biol. Chem. 2005;386:1219–1238. doi: 10.1515/BC.2005.140. [DOI] [PubMed] [Google Scholar]
- 2.Vogel J., Bartels V., Tang T.H., Churakov G., Slagter-Jager J.G., Hüttenhofer A., Wagner E.G. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wassarman K.M., Repoila F., Rosenow C., Storz G., Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Argaman L., Hershberg R., Vogel J., Bejerano G., Wagner E.G., Margalit H., Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 5.Zhang A., Wassarman K.M., Rosenow C., Tjaden B.C., Storz G., Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 6.Rivas E., Klein R.J., Jones T.A., Eddy S.R. Computational identification of noncoding RNAs in E.coli by comparative genomics. Curr. Biol. 2001;11:1369–1373. doi: 10.1016/s0960-9822(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 7.Kawano M., Reynolds A.A., Miranda-Rios J., Storz G. Detection of 5'- and 3'-UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acids Res. 2005;33:1040–1050. doi: 10.1093/nar/gki256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershberg R., Altuvia S., Margalit H. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 2003;31:1813–1820. doi: 10.1093/nar/gkg297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wassarman K.M., Storz G. 6S RNA regulates E.coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 10.Weilbacher T., Suzuki K., Dubey A.K., Wang X., Gudapaty S., Morozov I., Baker C.S., Georgellis D., Babitzke P., Romeo T. A novel sRNA component of the carbon storage regulatory system of Escherichia coli. Mol. Microbiol. 2003;48:657–670. doi: 10.1046/j.1365-2958.2003.03459.x. [DOI] [PubMed] [Google Scholar]
- 11.Liu M.Y., Gui G., Wei B., Preston J.F., 3rd, Oakford L., Yuksel U., Giedroc D.P., Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J. Biol. Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 12.Wagner E.G., Altuvia S., Romby P. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 2002;46:361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- 13.Massé E., Escorcia F.E., Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogel J., Argaman L., Wagner E.G., Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Schmidt M., Zheng P., Delihas N. Secondary structures of Escherichia coli antisense micF RNA, the 5′ end of the target ompF mRNA, and the RNA/RNA duplex. Biochemistry. 1995;34:3621–3631. doi: 10.1021/bi00011a017. [DOI] [PubMed] [Google Scholar]
- 16.Chen S., Zhang A., Blyn L.B., Storz G. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 2004;186:6689–6697. doi: 10.1128/JB.186.20.6689-6697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Udekwu K.I., Darfeuille F., Vogel J., Reimegard J., Holmqvist E., Wagner E.G. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rasmussen A.A., Eriksen M., Gilany K., Udesen C., Franch T., Petersen C., Valentin-Hansen P. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol. 2005;58:1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- 19.Kawamoto H., Koide Y., Morita T., Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 20.Massé E., Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geissmann T.A., Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Argaman L., Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- 23.Lease R.A., Belfort M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl Acad. Sci. USA. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Majdalani N., Hernandez D., Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol. Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 25.Majdalani N., Cunning C., Sledjeski D., Elliott T., Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lease R.A., Cusick M.E., Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl Acad. Sci. USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Opdyke J.A., Kang J.G., Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antal M., Bordeau V., Douchin V., Felden B. A small bacterial RNA regulates a putative ABC transporter. J. Biol. Chem. 2005;280:7901–7908. doi: 10.1074/jbc.M413071200. [DOI] [PubMed] [Google Scholar]
- 29.Mizuno T., Chou M.Y., Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA) Proc. Natl Acad. Sci. USA. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tjaden B., Goodwin S.S., Opdyke J.A., Guillier M., Fu D.X., Gottesman S., Storz G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guillier M., Gottesman S. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol. Microbiol. 2006;59:231–247. doi: 10.1111/j.1365-2958.2005.04929.x. [DOI] [PubMed] [Google Scholar]
- 32.Altuvia S., Weinstein-Fischer D., Zhang A., Postow L., Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 33.Massé E., Vanderpool C.K., Gottesman S. Effect of RyhB small RNA on global iron use in Escherichia coli. J. Bacteriol. 2005;187:6962–6971. doi: 10.1128/JB.187.20.6962-6971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papenfort K., Pfeiffer V., Mika F., Lucchini S., Hinton J.C., Vogel J. SigmaE-dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 2006;62:1674–1688. doi: 10.1111/j.1365-2958.2006.05524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Douchin V., Bohn C., Bouloc P. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J. Biol. Chem. 2006;281:12253–12259. doi: 10.1074/jbc.M600819200. [DOI] [PubMed] [Google Scholar]
- 36.Wagner E.G., Vogel J. Functional analysis of identified non-mRNAs. In: Hartmann R.K., Bindereif A., Schön A., Westhof E., editors. Handbook of RNA Biochemistry. Vol. 2. Wiley-VCH; 2005. pp. 614–642. [Google Scholar]
- 37.Kain S.R., Adams M., Kondepudi A., Yang T.T., Ward W.W., Kitts P. Green fluorescent protein as a reporter of gene expression and protein localization. Biotechniques. 1995;19:650–655. [PubMed] [Google Scholar]
- 38.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takiff H.E., Baker T., Copeland T., Chen S.M., Court D.L. Locating essential Escherichia coli genes by using mini-Tn10 transposons: the pdxJ operon. J. Bacteriol. 1992;174:1544–1553. doi: 10.1128/jb.174.5.1544-1553.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suess B., Hanson S., Berens C., Fink B., Schroeder R., Hillen W. Conditional gene expression by controlling translation with tetracycline-binding aptamers. Nucleic Acids Res. 2003;31:1853–1858. doi: 10.1093/nar/gkg285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uzzau S., Figueroa-Bossi N., Rubino S., Bossi L. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl Acad. Sci. USA. 2001;98:15264–15269. doi: 10.1073/pnas.261348198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalir S., McClure J., Pabbaraju K., Southward C., Ronen M., Leibler S., Surette M.G., Alon U. Ordering genes in a flagella pathway by analysis of expression kinetics from living bacteria. Science. 2001;292:2080–2083. doi: 10.1126/science.1058758. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J., Russel D.W. Molecular Cloning: A Laboratory Manuel. 2001. Transfer and fixation of denatured RNA to membranes; pp. 7.35–7.40. 7.35–37.40. [Google Scholar]
- 44.Lutz R., Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scholz O., Thiel A., Hillen W., Niederweis M. Quantitative analysis of gene expression with an improved green fluorescent protein p6. Eur. J. Biochem. 2000;267:1565–1570. doi: 10.1046/j.1432-1327.2000.01170.x. [DOI] [PubMed] [Google Scholar]
- 46.Sittka A., Pfeiffer V., Tedin K., Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sledjeski D., Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc. Natl Acad. Sci. USA. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vanderpool C.K., Gottesman S. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 2004;54:1076–1089. doi: 10.1111/j.1365-2958.2004.04348.x. [DOI] [PubMed] [Google Scholar]
- 49.Kawamoto H., Morita T., Shimizu A., Inada T., Aiba H. Implication of membrane localization of target mRNA in the action of a small RNA: mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 2005;19:328–338. doi: 10.1101/gad.1270605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Møller T., Franch T., Udesen C., Gerdes K., Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E.coli galactose operon. Genes Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Urbanowski M.L., Stauffer L.T., Stauffer G.V. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol. Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 52.Majdalani N., Chen S., Murrow J., St John K., Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- 53.Bensing B.A., Meyer B.J., Dunny G.M. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc. Natl Acad. Sci. USA. 1996;93:7794–7799. doi: 10.1073/pnas.93.15.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogel J., Axmann I.M., Herzel H., Hess W.R. Experimental and computational analysis of transcriptional start sites in the cyanobacterium Prochlorococcus MED4. Nucleic Acids Res. 2003;31:2890–2899. doi: 10.1093/nar/gkg398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen L.H., Emory S.A., Bricker A.L., Bouvet P., Belasco J.G. Structure and function of a bacterial mRNA stabilizer: analysis of the 5′-untranslated region of ompA mRNA. J. Bacteriol. 1991;173:4578–4586. doi: 10.1128/jb.173.15.4578-4586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emory S.A., Belasco J.G. The ompA 5′-untranslated RNA segment functions in Escherichia coli as a growth-rate-regulated mRNA stabilizer whose activity is unrelated to translational efficiency. J. Bacteriol. 1990;172:4472–4481. doi: 10.1128/jb.172.8.4472-4481.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arnold T.E., Yu J., Belasco J.G. mRNA stabilization by the ompA 5′-untranslated region: two protective elements hinder distinct pathways for mRNA degradation. RNA. 1998;4:319–330. [PMC free article] [PubMed] [Google Scholar]
- 58.Moll I., Afonyushkin T., Vytvytska O., Kaberdin V.R., Bläsi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Repoila F., Majdalani N., Gottesman S. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol. Microbiol. 2003;48:855–861. doi: 10.1046/j.1365-2958.2003.03454.x. [DOI] [PubMed] [Google Scholar]
- 60.Morita T., Maki K., Aiba H. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sledjeski D.D., Whitman C., Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Grieshaber N.A., Grieshaber S.S., Fischer E.R., Hackstadt T. A small RNA inhibits translation of the histone-like protein Hc1 in Chlamydia trachomatis. Mol. Microbiol. 2006;59:541–550. doi: 10.1111/j.1365-2958.2005.04949.x. [DOI] [PubMed] [Google Scholar]
- 63.Davis B.M., Quinones M., Pratt J., Ding Y., Waldor M.K. Characterization of the small untranslated RNA RyhB and its regulon in Vibrio cholerae. J. Bacteriol. 2005;187:4005–4014. doi: 10.1128/JB.187.12.4005-4014.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Southward C.M., Surette M.G. The dynamic microbe: green fluorescent protein brings bacteria to light. Mol. Microbiol. 2002;45:1191–1196. doi: 10.1046/j.1365-2958.2002.03089.x. [DOI] [PubMed] [Google Scholar]
- 65.Zaslaver A., Bren A., Ronen M., Itzkovitz S., Kikoin I., Shavit S., Liebermeister W., Surette M.G., Alon U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nature Meth. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- 66.Wagner E.G., Darfeuille F. Small Regulatory RNAs in Bacteria. In: Wolfgang N., editor. Small RNAs: Analysis and Regulatory Functions. 2006. pp. 1–30. [Google Scholar]
- 67.Morita T., Mochizuki Y., Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc. Natl Acad. Sci. USA. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li Z., Gong X., Joshi V.H., Li M. Co-evolution of tRNA 3′ trailer sequences with 3′ processing enzymes in bacteria. RNA. 2005;11:567–577. doi: 10.1261/rna.7287505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Afonyushkin T., Vecerek B., Moll I., Blasi U., Kaberdin V.R. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dasgupta S., Fernandez L., Kameyama L., Inada T., Nakamura Y., Pappas A., Court D.L. Genetic uncoupling of the dsRNA-binding and RNA cleavage activities of the Escherichia coli endoribonuclease RNase III--the effect of dsRNA binding on gene expression. Mol. Microbiol. 1998;28:629–640. doi: 10.1046/j.1365-2958.1998.00828.x. [DOI] [PubMed] [Google Scholar]
- 71.Franch T., Thisted T., Gerdes K. Ribonuclease III processing of coaxially stacked RNA helices. J. Biol. Chem. 1999;274:26572–26578. doi: 10.1074/jbc.274.37.26572. [DOI] [PubMed] [Google Scholar]
- 72.Isaacs F.J., Dwyer D.J., Ding C., Pervouchine D.D., Cantor C.R., Collins J.J. Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 2004;22:841–847. doi: 10.1038/nbt986. [DOI] [PubMed] [Google Scholar]
- 73.Kolb F.A., Engdahl H.M., Slagter-Jager J.G., Ehresmann B., Ehresmann C., Westhof E., Wagner E.G., Romby P. Progression of a loop-loop complex to a four-way junction is crucial for the activity of a regulatory antisense RNA. EMBO J. 2000;19:5905–5915. doi: 10.1093/emboj/19.21.5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pedelacq J.D., Cabantous S., Tran T., Terwilliger T.C., Waldo G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 75.Bagge N., Hentzer M., Andersen J.B., Ciofu O., Givskov M., Hoiby N. Dynamics and spatial distribution of beta-lactamase expression in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2004;48:1168–1174. doi: 10.1128/AAC.48.4.1168-1174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hautefort I., Proenca M.J., Hinton J.C. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 2003;69:7480–7491. doi: 10.1128/AEM.69.12.7480-7491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silvaggi J.M., Perkins J.B., Losick R. Genes for small, noncoding RNAs under sporulation control in Bacillus subtilis. J. Bacteriol. 2006;188:532–541. doi: 10.1128/JB.188.2.532-541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Axmann I.M., Kensche P., Vogel J., Kohl S., Herzel H., Hess W.R. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 2005;6:R73. doi: 10.1186/gb-2005-6-9-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Livny J., Brencic A., Lory S., Waldor M.K. Identification of 17 Pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNAPredict2. Nucleic Acids Res. 2006;34:3484–3493. doi: 10.1093/nar/gkl453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Christiansen J.K., Nielsen J.S., Ebersbach T., Valentin-Hansen P., Sogaard-Andersen L., Kallipolitis B.H. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA. 2006;12:1383–1395. doi: 10.1261/rna.49706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pichon C., Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl Acad. Sci. USA. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Willkomm D.K., Minnerup J., Huttenhofer A., Hartmann R.K. Experimental RNomics in Aquifex aeolicus: identification of small non-coding RNAs and the putative 6S RNA homolog. Nucleic Acids Res. 2005;33:1949–1960. doi: 10.1093/nar/gki334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lenz D.H., Mok K.C., Lilley B.N., Kulkarni R.V., Wingreen N.S., Bassler B.L. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 84.Lease R.A., Smith D., McDonough K., Belfort M. The small noncoding DsrA RNA is an acid resistance regulator in Escherichia coli. J. Bacteriol. 2004;186:6179–6185. doi: 10.1128/JB.186.18.6179-6185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]