Abstract
Lsm proteins are ubiquitous, multifunctional proteins that are involved in the processing and/or turnover of many, if not all, RNAs in eukaryotes. They generally interact only transiently with their substrate RNAs, in keeping with their likely roles as RNA chaperones. The spliceosomal U6 snRNA is an exception, being stably associated with the Lsm2-8 complex. The U6 snRNA is generally considered to be intrinsically nuclear but the mechanism of its nuclear retention has not been demonstrated, although La protein has been implicated. We show here that the complete Lsm2-8 complex is required for nuclear accumulation of U6 snRNA in yeast. Therefore, just as Sm proteins effect nuclear localization of the other spliceosomal snRNPs, the Lsm proteins mediate U6 snRNP localization except that nuclear retention is the likely mechanism for the U6 snRNP. La protein, which binds only transiently to the nascent U6 transcript, has a smaller, apparently indirect, effect on U6 localization that is compatible with its proposed role as a chaperone in facilitating U6 snRNP assembly.
INTRODUCTION
In metazoa, the U1, U2, U4 and U5 spliceosomal snRNAs are produced by RNA polymerase II, acquire a 7-methylguanosine 5′ cap, like mRNA and are exported to the cytoplasm (1). Here they associate with a heteroheptameric complex of Sm proteins that promotes hypermethylation of the cap and 3′ end trimming (2). The Sm proteins also contribute to nuclear import of these core snRNPs (3–5). In contrast, U6 is produced by RNA polymerase III and acquires a γ-monomethyl phosphate cap (6). It is generally considered that U6 is nuclear restricted, with no cytoplasmic phase during its biogenesis (7–11), however, the mechanism of nuclear retention is unknown. The nuclear localization of U6 could be due to the lack of a nuclear export signal in the RNA, the presence of an active retention signal (such as the γ-monomethyl cap), or retention through interaction with proteins. Alternatively, U6 snRNA may exit the nucleus and be reimported by interaction with proteins. Olson and Siliciano (12) reported the apparent shuttling of U6 snRNA in a yeast heterokaryon assay, however, as every RNA they tested seemed to shuttle, they could not rule out the possibility that the nuclei may have been leaky.
Like other RNA polymerase III products, the nascent U6 transcript has an oligo(U) sequence at its 3′ end that is bound by La protein (Lhp1p in yeast) (13,14), and experiments with Xenopus oocytes led Boelens et al. (9) to propose that binding of La protein to this sequence might be responsible for its nuclear retention. However, La binds only transiently to newly synthesized U6 snRNA and it was subsequently shown that La/Lhp1p is replaced on the 3′ end of U6 by the heteroheptameric complex of Lsm2 to 8 proteins (15–18), that are structurally related to the Sm proteins. Also, Prp24p (p110 in humans) is another U6 associated protein (19) that could affect the U6 snRNP localization. We have therefore compared the effects of the Lsm proteins, Lhp1p, Prp24p and the γ-monomethyl cap on U6 localization in Saccharomyces cerevisiae.
MATERIALS AND METHODS
Yeast strains and plasmids
The yeast strains used in this study are listed in Table 1. Yeast strains were grown in YPDA, YPGalA or YMM medium as required. To create strains MPS3 and MPS5, genomic LSM8 was 13myc-tagged by homologous recombination, using PCR product amplified from the plasmid pFA6a-13myc-hphMX6 [modified from pFA6a-13myc-kanMX6 (20)]. Amplified DNA contained the last 45 base pairs of LSM8 coding sequence, 13 copies of the c-myc epitope, hygromycin resistance (Hph), and 45 base pairs of downstream sequence. DNA was introduced into BMA38a or mutant strain and transformants selected on YPDA containing Hygromycin at 300 ug/ml. To produce pMPS2 and pMPS8, that encode GFP fused to Lhp1p and Lsm8p respectively, the LHP1 or LSM8 coding sequence was inserted into pGFP-N-FUS (PMET-GFP- MCS-CYC1TERM, URA3, CEN1) (21) in frame with the GFP coding sequence. To create strain MPS70, plasmid pRS314-539H6-5′Δ11 (22), containing the SNR6 gene with the first 11 nucleotides deleted, was introduced into BSY557, and pBS1191, containing the wild-type SNR6 gene, was shuffled out by plating cells on 5-FOA medium. To visualize Lhp1p, LHP1 was cloned into plasmid pGFP-N-FUS (21), to produce pMPS2 that encodes GFP-tagged Lhp1p.
Table 1.
Yeast strains used in this work
| Strain | Genotype |
|---|---|
| AEMY20b | MATa, ade2-1, his3▵200, leu2-3, -112, trp1-1, ura3-1, can1-100, lsm6▵::HIS3 |
| AEMY21b | MATa, ade2-1, his3▵200, leu2-3, -112, trp1-1, ura3-1, can1-100, lsm7▵::HIS3 |
| AEMY25b | MATa, ade2-1, his3▵200, leu2-3, -112, trp1-1, ura3-1, can1-100, lsm1▵::TRP1 |
| AEMY31 (16) | MATα, ade2-1, his3-11,-15, leu2-3, -112, trp1▵, ura3-1, lsm3▵::TRP1 [pAEM64 (PGAL-HA-LSM3, URA, CEN4, ARS1)] |
| AEMY33 (16) | MATα, ade2-1, his3-11,-15, leu2-3, -112, trp1▵, ura3-1, lsm2▵::HIS3 [pAEM68 (PGAL-LSM2-HA, URA, CEN4, ARS1)] |
| AEMY45b | MATa, ade2-1, his3-11,-15, leu2-3, -112, trp1▵, ura3-1, lsm8▵::TRP1 [pAEM76 (PGAL-HA-LSM8, URA, CEN4, ARS1)] |
| AEMY48b | MATa, ade2-1, his3-11,-15, leu2-3, -112, trp1▵, ura3-1, lsm5▵::TRP1 [pAEM75 (PGAL-HA-LSM5, URA, CEN4, ARS1)] |
| BMA38a | MATa, trp1▵1, his3▵200, ura3-1, leu2-3,-112, ade2-1, can1-100 |
| BSY557 (42) | MATa, ade2, arg4, leu2-3,-112, trp1-289, ura3-52, snr6::ADE2 [pBS1191 (CEN-URA3-SNR6)] |
| MCY4 (43) | MATα, ade1-101, his3-1, leu2-3,-112, trp1-289, ura3-52, LEU2-PGAL1-LSM4 |
| MPS3a | MATa, trp1▵1, his3▵200, ura3-1, leu2-3,-112, ade2-1, can1-100, LSM8:13myc-HphMX6 |
| MPS5a | MATa, ade2-1, his3▵200, leu2-3, -112, trp1-1, ura3-1, can1-100, lsm6▵::HIS3, LSM8:13myc-HphMX6 |
| MPS30a | MATa, trp1▵1, his3▵200, ura3-1, leu2-3,-112, ade2-1, can1-100, PGAL-GST-PRP24-HIS3 |
| MPS70a | As BSY557 but with pBS1191 replaced by pRS314-539H6-5′Δ11 (22) |
| PSY871 (44) | MATa, rsl1-1/kap95, rnal-1, ade2, ade3, leu2, ura3 [pPS714(RNA1)] |
| PSY1102c | MATa, rsl1-3/kap95, ura3-52, leu2Δ1, trp1Δ63 |
| PSY580c | MATa, ura3–52, leu2Δ1, trp1Δ63 |
| YCA35 (28) | MATa, trp1, his3▵200, ura3-52, gal2, gal▵108, lhp1::Kl URA |
aThis work; bA.E. Mayes and Beggs, unpublished, UK; cP. Silver
Fluorescent in situ hybridization
The sequence of the U6 snRNA probe (U6A) was 5′GATCATCTCTGTATTGTTTCAAATTGACCAAATGTCCACGAAGGGTT, with a single fluorescein molecule at the 3′ end. Oligonucleotide probes for the U1 snRNA were labelled with a single fluorescein attached to the 5′ end and had the sequences 5′-ACACCAATTTGAATTTGGGTGTCAAACTTCTCCAGGCAGAAGAAACAAAGGGCCCCAAAAATCAGTTTAA (U1-1) and 5′-AATCTCCGTCAAAAACTAAAATGGCACGCTAGAGAAAAGTAGTCAAAAAGAATGCCTCTACAAAG (U1-2). The sequence of the U4 probe was GACGGTCTGGTTTATAATTAAATTTC, with a Cy3 label at its 3′ end. Preparation of cells for FISH was performed essentially as described (23,24). Briefly, yeast cells in logarithmic phase in liquid YPDA were fixed for 40 min in 4% formaldehyde and spheroplasted for 30 min with Lyticase in buffer B (0.1 M potassium phosphate pH 7.4, 1.2 M sorbitol). Spheroplasted cells were then probed with fluorescently-labelled oligonucleotide(s), stained with 4′,6′-diamidino-2-phenylindole (DAPI), and imaged as described below.
Immunofluorescence
Yeast cells grown to logarithmic phase in liquid YPDA were fixed in 4% formaldehyde and spheroplasted as above. Spheroplasted cells were incubated with anti-myc antibody (Santa Cruz Biotechnology) overnight in buffer B containing 5% milk, and then for 2 h with FITC-conjugated donkey anti-mouse antibody (Jackson Immuno-Research). Vectashield mounting medium with DAPI (Vector Labs) was added prior to imaging.
GFP imaging
GFP fluorescence was detected in log-phase yeast cells grown in liquid medium.
Image capture
Cells were viewed using a Leica FW400 microscope (Leica) and imaging was performed essentially as described previously (24,25). Images were captured using LeicaFW4000 software (Leica) with a CoolSNAP HQ cooled CCD camera (Roper Scientific). When images were restored, a three-dimensional data set, composed of 20 images separated by 200 nm in the axial direction, was acquired and deconvolved with an acquired point spread function (PSF) using Leica Deblur software (Leica).
RESULTS AND DISCUSSION
Lsm proteins are required for nuclear accumulation of U6 snRNA
Lsm proteins are involved in the processing and/or turnover of most, if not all, RNAs in eukaryotes (26,27). There are eight Lsm proteins in budding yeast. The heteroheptameric Lsm1-7 complex is cytoplasmic, interacts with proteins involved in mRNA turnover, and promotes mRNA decapping and decay (26). The Lsm2-8 complex is nuclear and, in addition to its effects on the U6 snRNA, it has been implicated in the processing of pre-tRNAs (28), pre-snoRNAs (29) and pre-rRNAs (30) and in the turnover of pre-mRNAs (31).
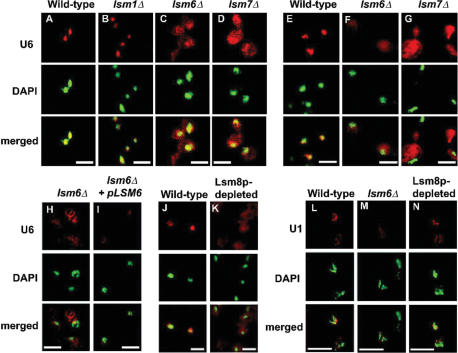
Saccharomyces cerevisiae strains lacking Lsm1p, Lsm6p or Lsm7p are viable at 30°C but not at 37°C. Therefore, fluorescent in situ hybridization (FISH) was carried out on strains carrying an lsm1Δ, lsm6Δ or lsm7Δ mutation (AEMY25, AEMY20 and AEMY21, respectively), as well as the wild-type control (BMA38a) grown at 30°C. In wild-type and lsm1Δ cells, U6 snRNA was exclusively nuclear (Figure 1A and B). In contrast, U6 snRNA was detected in both the nucleus and cytoplasm in lsm6Δ and lsm7Δ cells (Figure 1C and D). The U6 localization defect was more severe when the lsm6Δ and lsm7Δ cells were shifted to 37°C for 2.5 h prior to performing FISH (Figure 1E–G). Thus Lsm6p and Lsm7p, but not the cytoplasmic Lsm1p, are required for normal nuclear accumulation of the U6 snRNA. To confirm that the U6 mis-localization phenotype in lsm6Δ cells was due to the absence of Lsm6p, the lsm6Δ strain AEMY20 was transformed with plasmid pAEM61, which contains LSM6 expressed under control of the GAL1 promoter, or with empty vector as a control. Growth of these strains on galactose-containing medium confirmed that production of Lsm6p from pAEM61 suppressed the lsm6Δ growth defect (data not shown) as well as the U6 mis-localization (Figure 1H and I).
Figure 1.
U6 snRNA is delocalized in Lsm6-, Lsm7- and Lsm8-deficient cells. (A–G) U6 snRNA localization was examined in several lsm knockout strains. (A) wild-type cells (BMA38a), (B) lsm1Δ cells (AEMY25), (C) lsm6Δ cells (AEMY20) and (D) lsm7Δ cells (AEMY21) were grown at 30°C, fixed and hybridized with fluorescein-labelled U6A oligonucleotide at 37°C. Nuclei were visualized with DAPI. (E) wild-type (BMA38a), (F) lsm6Δ (AEMY20) and (G) lsm7Δ (AEMY21) cells were incubated at 37°C for 2.5 h prior to fixation and staining as above. (H–I) Production of Lsm6p rescues the lsm6Δ-associated defect in U6 localization. lsm6Δ cells carrying (H) YCpIF16 (empty vector) or (I) pAEM61 (plasmid carrying LSM6 under control of the GAL1 promoter) were grown in galactose medium to mid-log phase and prepared for in situ hybridization with the U6 probe as above. (J and K) U6 snRNA is delocalized upon depletion of Lsm8p. U6 localization was examined in (J) wild-type (BMA38a) and (K) AEMY45, in which LSM8 is under the control of the GAL1 promoter, enabling depletion of Lsm8p in glucose medium. Cells were grown in YPGalA and then shifted to YPDA for 12 h. (L–N) U1 localization in Lsm8-depleted strains. U1 localization was investigated using U1-1 and U1-2 probes simultaneously. (L) wild type (BMA38a), (M) lsm6Δ (AEMY20) and (N) Lsm8p-depleted cells (AEMY45). To improve the clarity of the Lsm8-depletion results, brightness was slightly increased in Photoshop. Scale bar, 10 μm.
To determine whether the essential Lsm8p was required for U6 localization, strain AEMY45 was used, in which LSM8 is under the control of a galactose-inducible promoter. AEMY45 cells were grown in galactose medium (YPGalA) then shifted to repressing medium (YPDA) for 12 h, which is long enough to deplete Lsm8p but not long enough to cause major degradation of U6 snRNA (ref 16 and data not shown). The U6 signal was detected throughout the Lsm8-depleted cells (Figure 1J and K). Furthermore, similar depletion of the essential Lsm2, Lsm3, Lsm4 or Lsm5 proteins also resulted in delocalization of the U6 snRNA (Supplementary data Figure S1). Thus, although Lsm8p, as the only Lsm protein that is nuclear restricted might conceivably have been responsible by itself for U6 nuclear accumulation, we conclude that the complete Lsm2-8 complex is required. These results also show that the nuclear accumulation of U6 snRNA is not due to the lack of a nuclear export signal in the RNA.
To test whether the defect in U6 localization seen in the absence of Lsm proteins is specific or reflects the mis-localization of small RNAs in general, the localization of two other snRNAs, U1 and U4, was investigated. The U1 snRNA has been shown to pass through the cytoplasm during its biogenesis in Xenopus laevis oocytes, although whether this nuclear export/re-import cycle is conserved in yeast is not currently known. Mature U1 does not interact with the Lsm proteins, rather it binds the Sm protein complex, which is required for U1 nuclear import in metazoa. Indeed, FISH showed that the nuclear localization of U1 snRNA was unaffected in Lsm-deficient strains, (Figure 1L–N). Mature U4 snRNA is also Sm-associated and under normal conditions is almost entirely associated with the U6 snRNA, either in the U4/U6 di-snRNP, or the U4/U6.U5 tri-snRNP. No difference in U4 localization was observed in wild-type and lsm6Δ cells (data not shown, the FISH signal for U4 was weak), indicating that the cytoplasmic U6 observed in Lsm-deficient strains is probably not associated with U4 snRNA. Thus the U6 localization defect observed in Lsm-deficient strains is specific to U6 and is not due to a general ‘leakage’ of RNAs out of the nucleus.
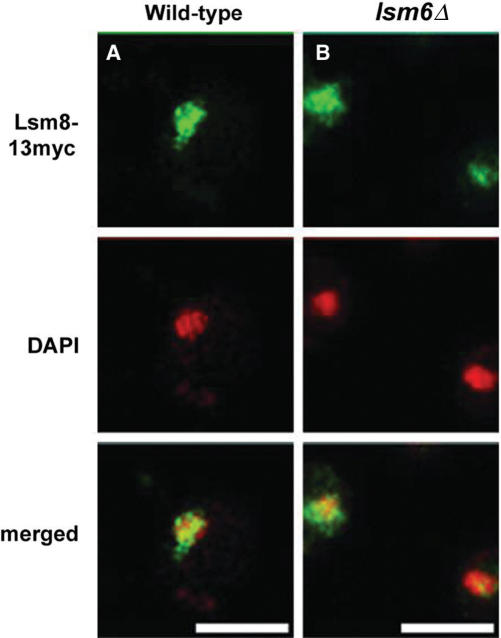
Lsm4p has been shown to co-precipitate some U6 snRNA from splicing extracts prepared from lsm6Δ or lsm7Δ cells, indicating that not all members of the Lsm2-8 complex are absolutely required for binding to U6 snRNA (32), but that its affinity for RNA in the absence of one of these subunits may be reduced. However, it is not known whether this Lsm4-associated U6 snRNA is of nuclear or cytoplasmic origin. To address the question of whether in lsm6Δ cells mis-localized U6 is bound by the remaining members of the Lsm2-8 complex, the localization of 13myc-tagged Lsm8p was analysed in an lsm6Δ strain (MPS5) and a wild-type control (MPS3) (Figure 2A and B). In lsm6Δ cells, Lsm8p remained nuclear, as in wild-type cells, indicating that there is not a significant amount of Lsm8p associated with the cytoplasmic U6. This result indicates that even though Lsm8p is able to accumulate in the nucleus in the absence of Lsm6p, the weak or transient interaction of a partial Lsm complex lacking Lsm6p does not support efficient U6 snRNA accumulation in the nucleus although it does support cell viability. This also points more towards inefficient nuclear retention than inefficient nuclear uptake of U6 snRNA bound by incomplete Lsm complex. Similarly, the nuclear localization of Lhp1p was unaffected by lsm6Δ (data not shown).
Figure 2.
Localization of Lsm8p in an Lsm6-deficient strain. Localization of Lsm8p in (A) wild-type (MPS3) and (B) lsm6Δ (MPS5) cells. Lsm8p was tagged with the 13-myc epitope, and was detected with mouse anti-myc antibodies. DNA was stained with DAPI. Scale bar, 10 μm.
La/Lhp1p has a smaller, probably indirect effect on U6 localization
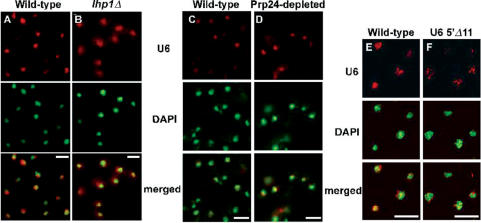
La/Lhp1 protein associates with nascent RNA polymerase III transcripts. It binds transiently to the uridine-rich 3′end of newly transcribed U6 snRNA that has a 3′ OH but, unlike the Lsm2-8 proteins, it does not bind to the mature U6 snRNA that has a 3′ terminal phosphate (33). La/Lhp1 is thought to have a chaperone-like function, facilitating correct RNA folding and/or RNP formation. It has been proposed (14) that Lhp1p hands the U6 RNA on to the Lsm2-8p complex. Despite the many functions of the LHP1 gene, its deletion in S. cerevisiae is not lethal, and causes no growth defect at any temperature. However, lhp1Δ is synthetic lethal with the temperature-sensitive mutation lsm8-1 (14), suggesting a functional relationship between Lhp1p and Lsm8p. FISH analysis of an lhp1Δ strain showed that U6 snRNA was mainly nuclear (Figure 3B), although there was increased cytoplasmic signal compared to wild-type cells (Figure 3A). This apparent low level of cytoplasmic U6 in the lhp1Δ strain could be explained by inefficient association of newly transcribed U6 RNA with the Lsm proteins in the absence of Lhp1p, as the fraction of total U6 RNA that co-precipitated with Lsm8p in an lhp1Δ cell extract was reduced compared to wild-type extract (data not shown). This suggests that Lhp1p affects U6 localization only indirectly and supports the proposal that La/Lhp1p might function as a chaperone, facilitating U6 snRNP assembly (9,14). Recently, Fok et al. (34) reported that human La, which is a nuclear phosphoprotein, undergoes nucleocytoplasmic shuttling. Therefore, conceivably La/Lhp1p could hand on U6 snRNA to the Lsm proteins either in the nucleus or in the cytoplasm. However, in Xenopus oocytes, the nuclear import of U6 RNA injected into the cytoplasm was not dependent on La (35); thus La most likely facilitates U6 snRNP assembly in the nucleus.
Figure 3.
U6 localization in Lhp1p-depleted or Prp24p-depleted yeast and localization of 5′ truncated U6 RNA. U6 localization was investigated in (A) wild-type (BMA38a) and (B) lhp1Δ (YCA35) cells grown at 30°C in YPDA; (C) wild-type (BMA38a) and (D) Prp24p-depleted cells (MPS30; PRP24 under control of the GAL1 promoter; grown in YPDA for 10 h); (E) wild-type (strain BSY557) and (F) U6 with 5′Δ 11 mutation (MPS70) cells. Scale bar, 10 μm.
Prp24p does not affect U6 localization
Prp24p is another U6-associated factor that might directly or indirectly affect U6 localization. Prp24p interacts with the U4 and U6 snRNAs and has been proposed to promote their annealing to form the U4/U6 di-snRNP (36). FISH was performed with a strain containing a PGAL-regulated PRP24 after incubation in YPDA for 10 h to deplete Prp24p, and with a temperature-sensitive prp24-1 strain following a shift to 37°C for 2 h. No change in U6 localization was observed in either case, indicating that Prp24p is not involved in the localization of the U6 snRNA (Figure 3C and D and data not shown).
The U6 snRNA cap does not affect its localization
The spliceosomal snRNAs that are produced by RNA polymerase II acquire a 7-methylguanosine 5′ cap co-transcriptionally. Although the cytoplasmic trafficking of snRNAs has not been demonstrated in yeast, in metazoan cells, the 7-methylguanosine cap contributes to their nuclear export (1). Further methylation in the cytoplasm produces the characteristic tri-methylguanosine cap which is important for their re-import (2). In contrast, the U6 snRNA has a single methyl group added to the 5′ γ−phosphate, which might contribute to its nuclear retention. Intriguingly, Kwan et al. (22) showed that an eleven nucleotide deletion at the 5′ end of the U6 gene results in the transcript receiving a tri-methylguanosine cap like the other snRNAs, despite still being transcribed by RNA polymerase III. This ‘5′Δ 11’ deletion does not have any effect on cell growth, showing this mutant U6 to be functional, but it is not known whether the Lsm proteins bind the truncated U6 RNA. The localization of U6 5′Δ 11 RNA was investigated in yeast strain MPS70 that lacks the genomic SNR6 and has the mutant 5′Δ11 snr6 gene and flanking sequences on a plasmid (22). The results (Figure 3E and F) show that deletion of the 5′ end of the U6 snRNA does not affect its localization. Thus, it appears that the cap structure is not a specific determinant of U6 nuclear localization in yeast, as seems also to be the case in Xenopus oocytes (8).
Nuclear accumulation of Lsm8p and U6 snRNA is importin β-dependent
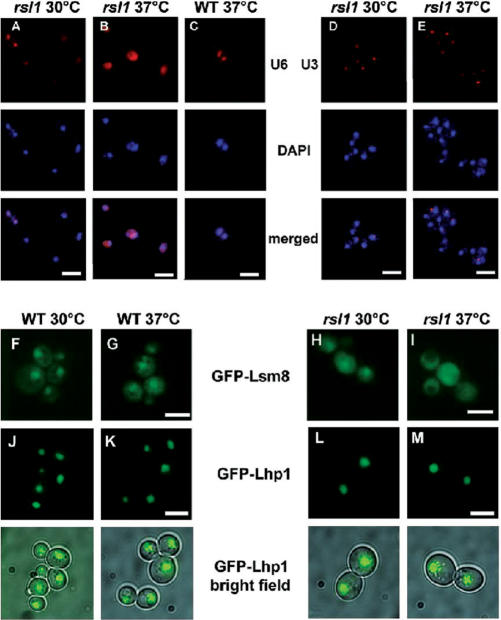
RSL1 (also known as KAP95) encodes the yeast homologue of importin (karyopherin) β (37), which is required for nuclear import of many nuclear proteins and, at least in metazoa, also mediates nuclear import of the tri-methylguanosine-capped snRNPs (38). To investigate whether it is also essential for nuclear accumulation of the U6 snRNP, wild-type and temperature-sensitive rsl1 cells were grown at 30°C and then shifted to 37°C for 2 h, and U6 localization was analysed by FISH (Figure 4). In comparison to the nuclear localization of U6 in wild-type yeast and in the karyopherin mutant at 30°C, the mutant cells at 37°C showed significant delocalization of U6 (Figure 4A–C), whereas localization of the nucleolar U3 snRNA was unaffected (Figure 4D and E). To test the dependence of Lsm8p localization on Rsl1p, a GFP-Lsm8 fusion protein was used. In wild-type cells, GFP-Lsm8 showed nuclear accumulation at 30 and 37°C (Figure 4F and G), whereas its nuclear accumulation was significantly reduced in rsl1 cells at 30°C, and absent after 1 h at 37°C (Figure 4H and I). In contrast, the rsl1 mutation did not affect the nuclear localization of a GFP-Lhp1 fusion protein (Figure 4J–M). This is compatible with the report (39) that in yeast the Sxm1/Kap108p karyopherin is responsible for the nuclear import of Lhp1p. Thus the dependence of U6 snRNA nuclear accumulation on the Rsl1-import pathway most likely reflects the nuclear uptake of the Lsm2-8 proteins by this pathway.
Figure 4.
U6 snRNA and Lsm8p are delocalized in rsl nuclear import mutant cells. (A–C) U6 localization was investigated in rsl1-1ts (PSY871) cells at (A) 30°C or (B) shifted to the restrictive temperature (37°C) for 2 h or (C) in wild-type (WT) cells grown at 37°C. (D) rsl1-1ts cells were grown at permissive temperature (30°C) or (E) shifted to the restrictive temperature (37°C) for 2 h and U3 localization was tested to control for nuclear integrity. (F–I) GFP-Lsm8p localization was investigated in live rsl1-3ts (PSY1102) or wild-type (PSY580) cells carrying pMPS8 (encodes GFP-Lsm8) at (F,H) 30°C or (G,I) shifted to the 37°C for 1 h. (J–M) GFP-Lhp1p localization was investigated in live rsl1-3ts (PSY1102) or wild-type (PSY580) cells carrying pMPS2 (encodes GFP-Lhp1) grown at (J,L) 30°C or (K, M) shifted to the 37°C for 2 h. In the case of GFP-Lhp1p, a bright field view is presented to show the outline of the cells. Scale bar, 10 μm.
The results presented here show a role for all seven components of the Lsm2-8 complex in the nuclear accumulation of U6 snRNA, and indicate the involvement of the full Lsm2-8 complex in this process. In contrast, Lsm8p nuclear localization appears to be Lsm6p-independent and therefore not dependent on the formation of a complete Lsm2-8 complex, and Lsm8p does not accumulate in the cytoplasm with U6 snRNA in the absence of Lsm6p. As the Lsm2 to 8 proteins are also involved in the processing of many other RNAs in the nucleus (28–31), presumably these proteins can enter the nucleus when not bound to RNA. Thus, the newly transcribed U6 snRNA is most likely handed on by Lhp1 to the Lsm2-8 complex within the nucleus and retained there.
If the 3′ end of U6 snRNA that is bound by Lsm proteins is effectively a nuclear retention sequence, its deletion should result in escape of U6 snRNA to the cytoplasm. Unfortunately, this cannot be directly tested for yeast U6 in vivo, as this uridine-rich sequence is important for correct termination of transcription by RNA polymerase III, and lack of association with the Lsm proteins results in instability of the U6 snRNA (16,17). Indeed, a 3′ end truncated U6 RNA that lacked this sequence was found to be highly unstable when injected into Xenopus oocyte nuclei. Nevertheless, a significant fraction of the residual RNA was observed to leave the nucleus, unlike full-length U6 RNA, suggesting that this sequence was responsible for its nuclear retention (9).
Although Lsm proteins interact with many different RNAs, these interactions are generally only transient, in keeping with their proposed roles as RNA chaperones. The fact that the spliceosomal U6 snRNA is an exception, being stably associated with Lsm proteins, might be explained, at least in part, by the need to retain the U6 snRNA in the nucleus in the absence of any other stably U6-associated proteins. Recently, Takano et al. (40) and Shaheen et al. (41) demonstrated that pre-tRNAs shuttle between cytoplasm and nucleus during their biogenesis. Thus, in S. cerevisiae there may be a general export of RNA-polymerase-III-transcribed RNAs that are not bound by nuclear retention factors.
Acknowledgements
We are grateful to Anita Hopper and Pam Silver for providing yeast strains and to David Tollervey and Joanna Kufel for helpful comments on this manuscript. This work was funded by the Darwin Trust of Edinburgh Studentships to M.P.S. and K-L.B., by the Wellcome Trust Studentship 71448 to M.A.M.R., and by the Wellcome Trust Grant 067311 to J.D.B. who is a Royal Society Darwin Trust Professor. ‘Funding to pay the Open Access publication charge was provided by the Wellcome Trust.’
Conflict of interest statement. None declared.
References
- 1.Hamm J, Mattaj IW. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 2.Kiss T. Biogenesis of small nuclear RNPs. J. Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- 3.Hamm J, Darzynkiewicz E, Tahara SM, Mattaj IW. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 1990;62:569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- 4.Fischer U, Lührmann R. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science. 1990;249:786–790. doi: 10.1126/science.2143847. [DOI] [PubMed] [Google Scholar]
- 5.Will CL, Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- 6.Singh R, Reddy R. Monomethyl phosphate: A cap structure in spliceosomal U6 small nuclear RNA. Proc. Natl. Acad. Sci. U.S.A. 1989;86:8280–8283. doi: 10.1073/pnas.86.21.8280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vankan P, Mcguigan C, Mattaj IW. Domains of U4 and U6 snRNAs required for snRNP assembly and splicing complementation in Xenopus oocytes. EMBO J. 1990;9:3397–3404. doi: 10.1002/j.1460-2075.1990.tb07541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terns MP, Dahlberg JE, Lund E. Multiple cis-acting signals for export of pre-U1 snRNA from the nucleus. Genes Dev. 1993;7:1898–1908. doi: 10.1101/gad.7.10.1898. [DOI] [PubMed] [Google Scholar]
- 9.Boelens WC, Palacios I, Mattaj IW. Nuclear retention of RNA as a mechanism for localization. RNA. 1995;1:273–283. [PMC free article] [PubMed] [Google Scholar]
- 10.Pante N, Jarmolowski A, Izaurralde E, Sauder U, Baschong W, Mattaj IW. Visualizing nuclear export of different classes of RNA by electron microscopy. RNA. 1997;3:498–513. [PMC free article] [PubMed] [Google Scholar]
- 11.Bertrand E, Bordonne R. Assembly and traffic of small nuclear RNPs. Prog. Mol. Subcell. Biol. 2004;35:79–97. doi: 10.1007/978-3-540-74266-1_4. [DOI] [PubMed] [Google Scholar]
- 12.Olson BL, Siliciano PG. A diverse set of nuclear RNAs transfer between nuclei of yeast heterokaryons. Yeast. 2003;20:893–903. doi: 10.1002/yea.1015. [DOI] [PubMed] [Google Scholar]
- 13.Rinke J, Steitz JA. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res. 1985;13:2617–2629. doi: 10.1093/nar/13.7.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pannone BK, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Lührmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3'-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayes AE, Verdone L, Legrain P, Beggs JD. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Séraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. EMBO J. 1999;18:3451–3462. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vidal VPI, Verdone L, Mayes AE, Beggs JD. Characterization of U6 snRNA-protein interactions. RNA. 1999;5:1470–1481. doi: 10.1017/s1355838299991355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shannon KW, Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev. 1991;5:773–785. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- 20.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1739–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 21.Niedenthal RK, Riles L, Johnston M, Hegemann JH. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 22.Kwan S, Gerlach VL, Brow DA. Disruption of the 5' stem-loop of yeast U6 RNA induces trimethylguanosine capping of this RNA polymerase III transcript in vivo. RNA. 2000;6:1859–1869. doi: 10.1017/s1355838200991325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long RM, Elliott DJ, Stutz F, Rosbash M, Singer RH. Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA. 1995;1:1071–1078. [PMC free article] [PubMed] [Google Scholar]
- 24.Samarsky DA, Fournier MJ, Singer RH, Bertrand E. The snoRNA box C/D motif directs nucleolar targeting and also couples snoRNA synthesis and localization. EMBO J. 1998;17:3747–3757. doi: 10.1093/emboj/17.13.3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carter KC, Bowman D, Carrington W, Fogarty K, McNeil JA, Fay FS, Lawrence JB. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- 26.He W, Parker R. Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol. 2000;12:350. doi: 10.1016/s0955-0674(00)00098-3. [DOI] [PubMed] [Google Scholar]
- 27.Beggs JD. RNA processing and the Lsm proteins. Novartis Medal Lecture. Biochem. Soc. Trans. 2005;33:433–438. doi: 10.1042/BST0330433. [DOI] [PubMed] [Google Scholar]
- 28.Kufel J, Allmang C, Verdone L, Beggs JD, Tollervey D. Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with La-homologous protein Lhp1p. Mol. Cell Biol. 2002;22:5248–5256. doi: 10.1128/MCB.22.14.5248-5256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kufel J, Allmang C, Verdone L, Beggs J, Tollervey D. A complex pathway for 3' processing of the yeast U3 snoRNA. Nucleic Acids Res. 2003;31:6788–6797. doi: 10.1093/nar/gkg904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kufel J, Allmang C, Petfalski E, Beggs J, Tollervey D. Lsm Proteins are required for normal processing and stability of ribosomal RNAs. J. Biol. Chem. 2003;278:2147–2156. doi: 10.1074/jbc.M208856200. [DOI] [PubMed] [Google Scholar]
- 31.Kufel J, Bousquet-Antonelli C, Beggs JD, Tollervey D. Nuclear pre-mRNA decapping and 5' degradation in yeast require the Lsm2-8p complex. Mol. Cell Biol. 2004;24:9646–9657. doi: 10.1128/MCB.24.21.9646-9657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verdone L, Galardi S, Page D, Beggs JD. Lsm proteins promote regeneration of pre-mRNA splicing activity. Curr. Biol. 2004;14:1487–1491. doi: 10.1016/j.cub.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 33.Terns MP, Lund E, Dahlberg JE. 3'-End-dependent formation of U6 small nuclear ribonucleoprotein particles in Xenopus laevis oocyte nuclei. Mol. Cell Biol. 1992;12:3032–3040. doi: 10.1128/mcb.12.7.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fok V, Friend K, Steitz JA. Epstein-Barr virus noncoding RNAs are confined to the nucleus, whereas their partner, the human La protein, undergoes nucleocytoplasmic shuttling. J. Cell Biol. 2006;173:319–325. doi: 10.1083/jcb.200601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grimm C, Lund E, Dahlberg JE. In vivo selection of RNAs that localize in the nucleus. EMBO J. 1997;16:793–806. doi: 10.1093/emboj/16.4.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raghunathan PL, Guthrie C. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–860. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- 37.Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J. Biol. Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- 38.Palacios I, Hetzer M, Adam SA, Mattaj IW. Nuclear import of U snRNPs requires importin beta. EMBO J. 1997;16:6783–6792. doi: 10.1093/emboj/16.22.6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenblum JS, Pemberton LF, Blobel G. A nuclear import pathway for a protein involved in tRNA maturation. J. Cell Biol. 1997;139:1655–1661. doi: 10.1083/jcb.139.7.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takano A, Endo T, Yoshihisa T. tRNA actively shuttles between the nucleus and cytosol in yeast. Science. 2005;309:140–142. doi: 10.1126/science.1113346. [DOI] [PubMed] [Google Scholar]
- 41.Shaheen HH, Hopper AK. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 2005;102:11290–11295. doi: 10.1073/pnas.0503836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luukkonen BG.M, Séraphin B. Construction of an in vivo-regulated U6 snRNA transcription unit as a tool to study U6 function. RNA. 1998;4:231–238. [PMC free article] [PubMed] [Google Scholar]
- 43.Cooper M, Parkes V, Johnston LH, Beggs JD. Identification and characterisation of Uss1p (Sdb23p): a novel U6 snRNA-associated protein with significant similarity to core proteins of small nuclear ribonucleoproteins. EMBO J. 1995;14:2066–2075. doi: 10.1002/j.1460-2075.1995.tb07198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koepp DM, Wong DH, Corbett AH, Silver PA. Dynamic localization of the nuclear import receptor and its interactions with transport factors. J. Cell Biol. 1996;133:1163–1176. doi: 10.1083/jcb.133.6.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]