Abstract
Delta is a major transmembrane ligand for Notch receptor that mediates numerous cell fate decisions. The Notch signaling pathway has long been thought to be mono-directional, because ligands for Notch were generally believed to be unable to transmit signals into the cells expressing them. However, we showed here that Notch also supplies signals to neighboring mouse neural stem cells (NSCs). To investigate the Notch–Delta signaling pathway in a bi-directional manner, we analyzed functional roles of the intracellular domain of mouse Delta like protein 1 (Dll1IC). In developing mouse NSCs, Dll1IC, which is released from cell membrane by proteolysis, is present in the nucleus. Furthermore, we screened for transcription factors that bind to Dll1IC and demonstrated that Dll1IC binds specifically to transcription factors involved in TGF-β/Activin signaling—Smad2, Smad3 and Smad4—and enhances Smad-dependent transcription. In addition, the results of the present study indicated that over-expression of Dll1IC in embryonic carcinoma P19 cells induced neurons, and this induction was blocked by SB431542, which is a specific inhibitor of TGF-β/Activin signaling. These observations strongly suggested that Dll1IC mediates TGF-β/Activin signaling through binding to Smads and plays an important role for bi-directional Notch–Delta signaling pathway.
INTRODUCTION
Delta is a major transmembrane ligand for Notch receptor and plays an important role in Notch signaling, which mediates the fates of numerous cells in both invertebrates and vertebrates (1,2). The Notch signaling pathway has long been thought to be mono-directional, because ligands for Notch were generally considered unable to transmit signals into the cells expressing these ligands (3,4). Several lines of evidence support this idea. For example, it was thought that none of the intracellular domains of putative Notch ligands display any significant sequence similarity throughout evolution (3). Indeed, replacement of most of the intracellular domain of LAG-2, a C. elegans lin-12 (Notch) ligand, with a β-galactosidase fusion protein has no discernible effect on LAG-2 function (3). In contrast, however, Baker and Schubiger reported that the extracellular domain of Notch expressed in the mesoderm provided a positive signal to the overlaying ectoderm in Drosophila (5). Furthermore, it has been reported that the intracellular domain of Delta (X-Delta-1) is required for normal development in Xenopus (6). In Drosophila, it was also shown that intracellular domains of these ligands act as antagonists of Notch signaling (7). These findings suggest that signaling in the opposite direction also exists. Thus, the important and critical question is whether signaling events occur not only from ligand-expressing cells to Notch-expressing cells but also vice versa, i.e. in a bi-directional manner.
Evidence has recently been accumulating in support of a functional role of the intracellular domains of Notch ligands, which implies the existence of bi-directional signaling mechanisms. For example, Delta has been shown to be cleaved by ADAM protease and γ-secretase to release an intracellular domain (8–11). This processing followed a mechanism of regulated intramembrane proteolysis (RIP), which was first described for the proteolytic activation of Notch (12). The RIP mechanism requires sequential cleavage steps to occur within the juxtamembrane (JM) and transmembrane (TM) domain, which are carried out by an ADAM protease and Presenilin γ-secretase, respectively. In the case of Notch, RIP serves to release an intracellular domain that has activity in the nucleus through binding to transcription factors, Suppressor of Hairless (SU[H], RBP-jκ in mammals) (1,2). Indeed, several groups have reported evidence of the nuclear localization of the intracellular fragments of ligands for Notch (10,11,13). Recently, it was demonstrated that many single transmembrane-spanning proteins including Notch, Delta and amyloid precursor protein (APP) are substrates for γ-secretase (14). Thus, it is likely that the intracellular domains of these substrates are released from the cell membrane by RIP. Common enzymes modulate proteolysis and the turnover of possible signaling molecules has led to the attractive speculation that similar mechanisms widely contribute to proteolysis-regulated signaling pathways (12,14).
Recently, Delwig et al. have reported that Drosophila Delta proteolysis differs from the conventional RIP, because TM processing of Drosophila Delta is not sensitive to preseniline, and TM and JM cleavages occur independently of each other. Based on these observations, they concluded that Delta proteolysis can act to modulate Delta activity (15). However, mouse and rat Delta like protein1 (Dll1) are sequentially processed and preseniline is essential for these proteolysis (10,11). Thus, they adhere more closely to the RIP mechanism. Therefore, it is possible that the intracellular domain of Delta plays a role in bi-directional signaling.
Indeed, Delta homologs display significant sequence similarity, which is restricted to vertebrates, in their intracellular domain. We show here evidence that the development of neurons from mouse neural stem cells (NSCs) was enhanced by co-culture with Notch1-expressing cells. We further demonstrated that, in developing mouse NSCs, Dll1 is already cleaved and an intracellular domain of Dll1 (Dll1IC) is present in the nucleus. Additionally, Dll1 proteolysis can be upregulated through interaction with Notch1 and this upregulation was strongly inhibited by the γ-secretase inhibitor. We also demonstrated the possibility that Dll1IC mediates transforming growth factor-β (TGF-β)/Activin signaling through binding to Smads, and boosts transcription of specific genes. Finally, we showed that over-expression of Dll1IC in embryonic carcinoma P19 cells induced neurons, and this induction was blocked by SB431542, a specific inhibitor of TGF-β/Activin signaling. These observations strongly suggested that Dll1IC mediates TGF-β/Activin signaling through binding to Smads, and plays an important role in the bi-directional Notch–Delta signaling pathway.
MATERIALS AND METHODS
Preparation of anti-Dll1IC Antibody
Dll1IC (nucleotides 1702–2166) was cloned into the pGEX vector (Amersham Biosciences, New Jersey). GST fusion protein was induced in Escherichia coli strain BL21 with 1 mM IPTG and purified by glutathione affinity column chromatography (Amersham). Rabbit antisera were prepared by eight subcutaneous injections of 500 μg of GST fusion protein with Freund's adjuvant at weekly intervals. Recombinant Dll1IC protein was released from the GST fusion protein by digestion with thrombin and coupled to a HiTrap affinity column (Amersham). Anti-Dll1IC antibody was purified using this affinity column. As shown in Supplementary Figure 1S, this antibody is specific for Dll1.
Preparation of NSCs and subcellular protein fractions
Pregnant mice (Crj: CD-1) were purchased from Charles River Japan (Japan). The enzymatic method used for preparation of NSCs from mouse embryonic day-10 (E10) embryos and culture conditions were described previously (16,17). For immunocytochemical study, 6 × 104 NSCs were cultured on Poly-l-Lysin coated cell-disk (Sumitomo, Japan), in a 1:1 mixture of Ham's nutrient mixture F12 and Dulbecco's modified Eagle's medium (DME/F12, Invitrogen, California), supplemented with bFGF (5 ng/ml; Pepro Tech, United Kingdom) and N2 supplement (Invitrogen). N- [N-(3,5- Difluorophenacetyl-l- alanyl)]-S- phenylglycine t-Butyl Ester (DAPT) was purchased from Calbiochem (Germany) as a γ-secretase inhibitor (18,19). For inhibition experiments, 10 μM DAPT was added to culture medium.
Subcellular protein fractions were prepared using FractionPREP™ Cell Fraction System (BioVision, California) according to manufacture's protocol.
Culture of NSCs on monolayer of Notch1-expressing or Dll1-expressing COS7 cells
Eight hundred nanograms of each expression vector for full-length Notch1, full-length Dll1 or vector alone were transfected into 8 × 104 COS7 cells using Lipofectamine (Invitrogen) and cultured for 48 h on cell-disk (Sumitomo) as a monolayer in DME/F12 (Invitrogen), supplemented with bFGF (5 ng/ml; Pepro Tech, United Kingdom) and N2 supplement (Invitrogen). Then 2 × 104 NSCs were further cultured on the monolayer of COS7 cells for 2 or 4 days followed by immunocytochemistry.
Co-culture of Dll1-expressing cells with Notch1-expressing cells
COS7 cells were cultured in DMEM with 10% fetal calf serum (FCS). Six micrograms of each expression vector for full-length Notch1, full-length Dll1 or vector alone were transfected into 106 COS7 cells using FuGENE6 (Roche Diagnostics, Indiana, USA). Forty-eight hours after transfection, cells were peeled off the dish using 0.53 mM EDTA in phosphate buffer saline (PBS). Aliquots of 5 × 104 Dll1-expressing cells and 4 × 105 Notch1-expressing cells were mixed and cultured in 6-well plates. Then 4 × 105 cells carrying vector alone were also mixed with 5 × 104 Dll1-expressing cells and cultured as a control. Two days before transfection, 10 μM DAPT was added to culture medium as a γ-secretase inhibitor for inhibition experiments. Cells were disrupted and aliquots of 20 μg of cell lysates were subjected to western blotting analysis with anti-Dll1IC antibody.
Immunocytochemistry
Cells were cultured on cell-disk (Sumitomo). After fixation with 4% paraformaldehyde in PBS for 30 min, cells were treated with 0.1% TritonX-100 and 0.1% Tween20 in PBS for 10 min, followed by blocking with 10% FCS in PBS for 1 h. Then cells were incubated with primary antibody at 4°C overnight, followed by incubation with fluorescence labeled secondary antibody. Primary antibody concentrations used were as followed: rabbit anti-Dll1, 0.5 ng/ml, rabbit anti-Nestin (17), 1:1000, mouse Tuj1 (Covance, California), 1:1000. All secondary antibodies were purchased from Molecular Probes (Oregon) and the antibody concentrations used were as followed: Alexa 546 Goat anti-mouse IgG, 1:2000, Alexa 488 Goat anti-rabbit IgG, 1:2000, Alexa 488 Goat anti-mouse IgG, 1:2000. Nuclear counterstaining was performed with 4′, 6-diamidino-2-phenylindole (DAPI, Molecular Probes). Signals were visualized using Axioplan2 fluorescent microscope and AxioCam digital camera (Zeiss, Germany).
Screening for transcription factors that bind to Dll1IC
A total of 150 biotinylated double-stranded DNA oligonucleotides representing known transcription factor binding sequences and arrays that were spotted with the same set of oligo DNAs (TF–TF Interaction Array) were generously supplied by B-Bridge International Inc. (California, USA) and Panomics (California, USA) and used to screen for transcription factors that bind to the intracellular domain of Dll1 protein. Detail information about these arrays can be seen at http://panomics.com/MA5011.cfm. Lists of oligonucleotide sequences also can be seen at http://panomics.com/TF_TF1.cfm and http://panomics.com/TF_TF2.cfm. NSCs were isolated from mouse E11 embryos, and nuclear proteins were prepared from NSCs using the Dignam method (20). Aliquots of 5 μg of nuclear proteins from E11 NSCs were incubated with 150 biotinylated double-stranded DNA oligonucleotides. DNA/protein complexes including Dll1IC were immunoprecipitated by the addition of 1 μg of anti-Dll1IC antibody and protein A-Sepharose (Amersham). After immunoprecipitation, double-stranded DNAs were eluted from precipitated complexes by heat treatment at 99°C for 5 min, followed by hybridization to the array. Arrays were incubated with HRP-labeled Avidin and were visualized using ECL Plus detection reagent (Amersham). Signals were detected using a FAS1000 cooled CCD camera (Toyobo, Japan).
Immunoprecipitation
Aliquots of 8 × 105 COS7 cells were transiently co-transfected with 4 μg of each cDNA encoding 8 Smads and 4 μg of Dll1IC with or without 4 μg of constitutively activated receptor (caALK5-HA or caALK6-HA) using Lipofectamine 2000 (Invitrogen). Forty-eight hours after transfection, cells were disrupted and aliquots of 20 μg of cell lysates were incubated with 1 μg of rabbit anti-Dll1IC antibody or 1 μg of mouse monoclonal anti-FLAG M2 antibody (Sigma), followed by immunoprecipitation and western blotting as described (21).
Establishment of Dll1IC expressing P19 cell lines and neuronal induction
P19 cells were cultured in alpha-modified Eagle's essential medium (Sigma) containing 10% FCS and 2 mM l-glutamine. Dll1IC expression vectors were transfected into P19 cells with FuGENE6 (Roche). Cells were also transfected with pCDNA (Invitrogen) as a negative control. After selection of transfected P19 cells in culture medium containing 500 µg/ml of G418 for 4 weeks, approximately twenty G418-resistant clones were picked and analyzed for the expression of Dll1IC by western blotting. To induce differentiation of established P19 cell lines, cells were allowed to aggregate at 1 × 106 cells per 10-cm bacterial grade Petri dish in culture medium without all-trans-retinoic acid (RA) for 4 days. After aggregation, cells were plated onto poly-l-Lysine coated cell-disk (Sumitomo). The culture medium was changed to DME/F12 (Invitrogen) containing N2 supplement (Invitrogen) and 10 ng/ml bFGF (Pepro Tech). The cells were cultured for 3 days to allow differentiation. For inhibition experiments, Dll1IC-expressing P19 cells were aggregated and plated in the presence of 1 µM SB431542 (Tocris Bioscience, Missouri, USA).
Dual Luciferase reporter assay
Aliquots of 4 × 104 HepG2 cells were transiently transfected with the indicated amounts of expression plasmids for Dll1IC, 100 ng of 9 × CAGA-Luc promoter–reporter plasmid (22,23), which responds specifically to Smad3, and 2 ng of pRL-CMV (Promega, Wisconsin, USA) as an internal control using FuGENE6 (Roche). In each experiment, total amounts of plasmids were adjusted to 0.5 μg by the addition of pCDNA plasmid (Invitrogen). After overnight incubation, 5 ng/ml TGF-β (Pepro Tech) was added to the medium for stimulation. Aliquots of 6 × 103 Dll1IC/P19 cells or pCDNA/P19 cells were transiently transfected with 200 ng of 9 × CAGA-Luc promoter–reporter plasmid and 5 ng of pRL-CMV (Promega) using FuGENE6 (Roche). After overnight incubation, cells were stimulated with TGF-β (Pepro Tech) at various concentrations. Cell lysates were prepared 48 h after transfection, and luciferase activity was measured using a dual-luciferase reporter system (Promega) as described (24). Values were normalized relative to Renilla luciferase activity.
RESULTS
Delta homologs display significant sequence similarity in their intracellular domain through the evolution of vertebrates
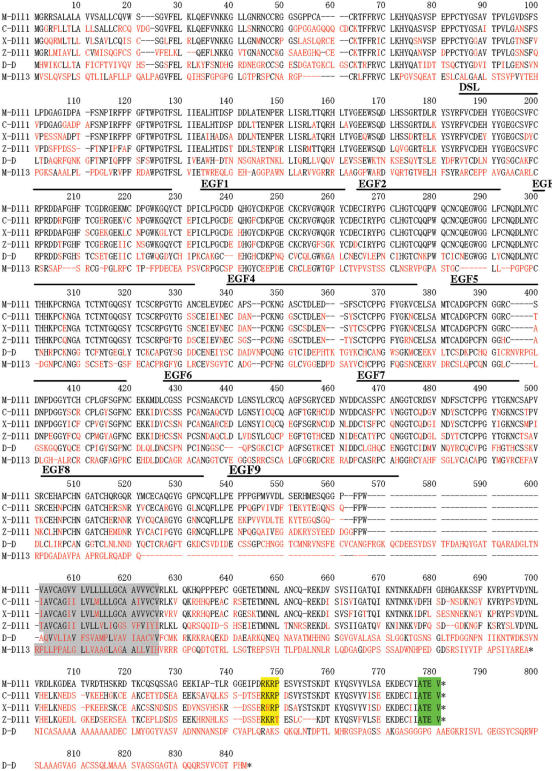
As shown in Figure 1, amino acid sequences in the extracellular domain of Delta are strongly conserved through evolution. For example, amino acid identities are 44.1% between mouse and Drosophila Delta. Remarkably, Delta homologs share up to 70% identity in individual EGF-like repeats. Significant homologies are still recognized within the intracellular domain of vertebrate Delta. For example, in the intracellular domain, amino acid sequence identities are 62.3% between mouse and chicken Delta, 55.8% between mouse and Xenopus, 51.0% between mouse and zebra fish. However, there is no homology between these vertebrate Deltas and Drosophila Delta. Thus, conservation of amino acid sequences in intracellular domain is restricted to vertebrate Delta. In addition, Dll3, a divergent type of Delta, does not show any homology to other Delta in the intracellular domain.
Figure 1.
Alignment of amino acid sequences of Delta homologs from various vertebrate species and Drosophila was generated using DNASIS program (Hitachi Software, Japan). Amino acids that are identical to those of mouse Dll1 are shown in black. Amino acids that differ from those of mouse Dll1 are shown in red. Gaps that were introduced for the alignment are shown as lines. The transmembrane region is shaded gray. RKRP predicted nuclear localization signal is shaded yellow, ATEV PDZ-binding motif is shaded green. M-Dll1: mouse Delta1, C-Dll1: chicken Delta1, X-Dll1: Xenopus Delta1, Z-Dll1: zebra fish DeltaD, D-D: Drosophila Delta, M-Dll3: mouse Delta3.
Three highly conserved regions are detected within vertebrate Delta; the first is located from amino acid position 643 to 680, the second is located from 690 to 701 and the last is located in the C-terminus (747–781). As has been reported, the RKRP nuclear localization signal (747–750) and ATEV consensus PDZ-binding motif (778–781) are present at the C-terminus region (25).
Emergence of neurons from NSCs was enhanced by the presence of Notch
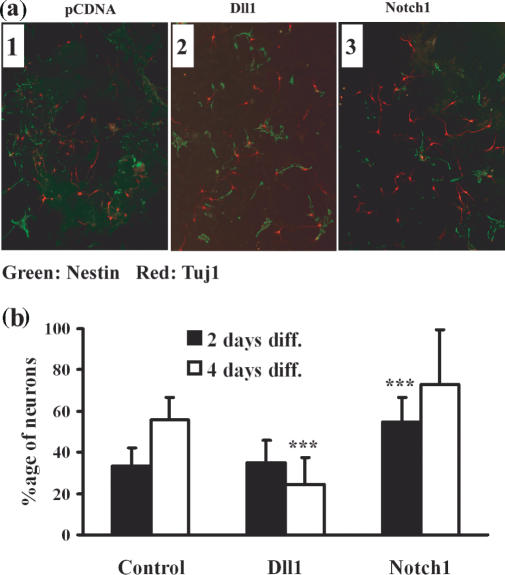
We isolated NSCs from mouse E10 embryos as described earlier (16,17). As shown in Figure 2a and b, when NSCs were co-cultured on a monolayer of mouse Notch1-expressing COS7 cells for 2 days, the rate of cells positive for Tuj1 antibody, which recognizes neuron-specific class III β-Tubulin and is a marker of young neurons (26), appearing from NSCs (55%) was higher than that in control cultures (33%). This difference was statistically significant (P < 0.001) using Student's t-test. After 4 days of culture, many NSCs had spontaneously differentiated into neurons (56%) in the control experiment. Although there was no significance by statistical analysis, more neurons (72%) were detected in the co-culture experiment with Notch1-expressing COS7 cells (Figure 2b). Conversely, when NSCs were co-cultured for 4 days on a monolayer of Dll1 over-expressing COS7 cells, the rate of emergence of Tuj1 positive neurons (25%) was lower than that in controls (56%) (Figure 2b). This difference was also statistically significant (P < 0.001).
Figure 2.
Co-culture of NSCs with Notch1-expressing cells or Dll1-expressing cells. (a) NSCs were cultured for 2 days on monolayers of Dll1 (2) or Notch1 (3) over-expressing COS7 cells and immunostained with Tuj1, a marker of neurons (red) and the NSC marker Nestin (green). (1): Control experiment with pCDNA-transfected COS7 cells. (b) NSCs were co-cultured for two (black bar: 2 days diff.) or four (white bar: 4 days diff.) days as described above. After immunostaining, Tuj1-positive neurons and Nestin-positive NSCs were counted. Values are means ± S.D. for 10 independent fields.***P < 0.001, Student's t-test (versus appropriate controls).
In developing NSCs, endogenous Dll1 is already cleaved and Dll1IC is present in the nucleus
To examine whether Delta was cleaved as described previously (8–11), NSCs isolated from E10 mouse embryos were subjected to western blotting analysis with anti-Dll1IC antibody. As shown in Figure 3a, at least four bands were detected on western blots. From the sizes of the bands, band 1 seemed to be the full-length Dll1, while the others appeared to be cleavage products of Dll1. The band with the lowest molecular weight (band 2) seemed like Dll1IC (18 kDa) released from the cell membrane by γ-secretase (8–11). To examine the cellular localization of Dll1IC, we prepared subcellular protein fractions from NSCs, followed by western blotting. Although full-length Dll1 was present in the membrane fraction, Dll1IC was present in nuclear fraction (Figure 3a). These results suggest that Dll1IC is localized in the nucleus. Developing NSCs were further stained with anti-Dll1IC antibody, a large proportion of the Dll1IC immunoreactivity was accumulated in the nucleus as shown in Figure 3b. These accumulations were observed in more than 35% of developing NSCs. However, inhibitor of γ-secretase strongly blocked these accumulations in almost all of NSCs (Figure 3b). These findings indicate that a large proportion of endogenous Dll1 is already cleaved by γ-secretase and activated in developing NSCs.
Figure 3.
Dll1 was cleaved and Dll1IC presented in the nucleus. (a) Aliquots of 20 μg of lysates from NSCs and aliquots of 30 μg of subcellular protein fractions were subjected to western blotting with anti-Dll1IC antibody. From the sizes of the bands, band 1 seemed to be the full-length mouse Delta (Dll1), while the others appeared to be cleavage products of Dll1. The band with the lowest molecular weight (band 2) seemed like Dll1IC (18 kDa) released from the cell membrane. S, lysates from NSCs; N, nuclear fraction; C, cytoplasm fraction; M, membrane fraction. (b) NSCs were stained with anti-Dll1IC antibody (1) and DAPI (2). Merged image (3) shows that immunoreactivity had accumulated in the nucleus. However, inhibitor of γ-secretase strongly blocked these accumulations; stained with anti-Dll1IC antibody (4), DAPI (5), merged image (6).
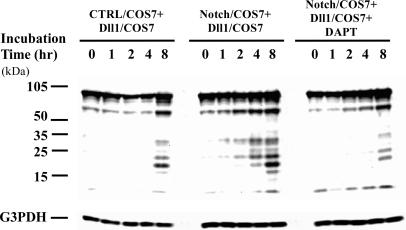
It has been shown that incubation of Drosophila Delta-expressing S2 cells with Notch-expressing S2 cells caused a transient accumulation of C-terminal fragments of Delta that had been cleaved by latent proteases (13,15). We therefore investigated whether mouse Dll1 processing could also be induced through interaction with Notch. Since COS7 cells express presenilin and have a γ-secretase activity (27,28), we employed COS7 cells for this experiment. Dll1-expressing COS7 cells were co-cultured with Notch1-expressing COS7 cells or control COS7 cells and Dll1 processing was detected by western blotting with anti-Dll1IC antibody. As shown in Figure 4, several bands were detected on western blots. From the size of band, 90 kDa protein seemed to be the full-length Dll1, while the others appeared to be the cleavage products of Dll1. When Dll1 expressing COS7 cells (Dll1/COS7) were co-cultured with COS7 cells carrying vector alone (CTRL/COS7) for 8 h, a small amount of cleavage products of Dll1 were initially detected. However, cleavage products of Dll1 were remarkably increased by co-culture with Notch1-expressing COS7 cells (Notch/COS7). For example, even after a 1 h incubation, weak bands were detected. Thereafter, amounts of cleaved products increased. As shown in Figure 4, this enhancement of Dll1processing was strongly inhibited by DAPT which is an inhibitor for γ-secretase.
Figure 4.
Co-culture of Dll1-expressing cells with Notch1-expressing cells. Each expression vector for full-length Notch1, full-length Dll1 or vector alone was transfected into COS7 cells. Forty-eight hours after transfection, Dll1-expressing cells (Dll1/COS7) were mixed with Notch-expressing cells (Notch/COS7) or control cells (CTRL/COS7) at a ratio of 1:8 and cultured for indicated times. DAPT was added to the culture medium as an inhibitor of γ-secretase. Dll1 processing was detected by western blotting with anti-Dll1IC antibody. From the size of the band, the 90 kDa protein (top band) seemed to be full-length Dll1, while the other appeared to be cleaved products of Dll1. Cleavage products of Dll1 were remarkably increased by co-culture with Notch1-expressing COS7 cells and these increases were strongly inhibited by DAPT. The same filter was also reacted with anti-G3PDH antibody as an internal control, showing that almost the same amount of cell lysates were processed for western blotting (bottom of the figure).
Dll1IC bound to Smads and enhanced Smad depending transcription
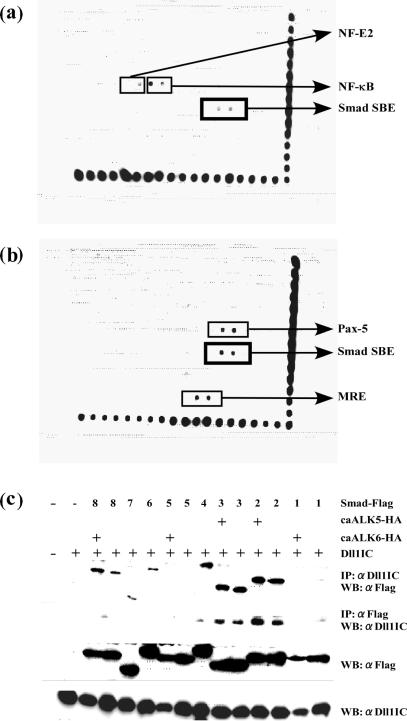
The nuclear localization of Dll1IC suggests that Dll1IC may have effects on the transcriptions of specific target genes. To examine this possibility, we searched for transcription factors capable of binding to Dll1IC using a new method. Briefly (see Supplementary Figure 2S), nuclear extracts from E11 NSCs were incubated with 150 double-stranded DNA oligonucleotides representing binding sequences of known transcription factors. After immunoprecipitation with an antibody against Dll1IC, double-stranded oligos were eluted from the precipitated complexes and hybridized to arrays that were spotted with the same set of oligos in duplicate. The hybridization signals obtained are shown in Figure 5a. The Smad binding sequences (5′-AGTATGTCTAGACTGA-3′) showed strong signals, which were enhanced by the addition of recombinant Dll1IC to nuclear extracts prior to immunoprecipitation (Figure 5b). Two spots, signals of which were enhanced by the addition of recombinant Dll1IC protein, were Pax-5 binding sequence and mineral corticoid response element (MRE). The other spots, signals of which were disappeared by the addition of recombinant Dll1IC protein, were NF-κB binding site and NF-E2 binding sequence.
Figure 5.
Dll1IC bound to Smad2, Smad3 and Smad4. (a) Result of screening for transcription factors that bind to Dll1IC. Note that Smad binding sequences showed strong signals (boxed). (b) Signal from Smad binding sequences (boxed) were enhanced by the addition of recombinant Dll1IC protein to the nuclear extract before immunoprecipitation. Signals from Pax-5 binding sequence and mineral corticoid response element (MRE) were also enhanced by the addition of recombinant Dll1IC protein. On the other hand, signals from NF-κB binding site and NF-E2 binding sequence were disappeared by the addition of recombinant Dll1IC protein. Spots along the right and bottom side of arrays are markers for alignment. (c) COS7 cells were transiently co-transfected with each expression vector for 8 Smads and Dll1IC expression vector with or without expression vectors for constitutive activated receptor (caALK5-HA or caALK6-HA). Forty-eight hours after transfection, lysates from co-transfected COS7 cells were subjected to immunoprecipitation (IP) with the indicated antibodies, followed by western blotting (WB). αDll1IC, rabbit anti-Dll1IC antibody; αFlag, mouse monoclonal anti-FLAG M2 antibody. Levels of expression of Smads and Dll1IC were determined by western blotting and are shown in the bottom two panels.
These findings suggest that Dll1IC forms complexes with Smads in the nucleus of NSCs. Thus, co-immunoprecipitation experiments were performed to confirm these results. FLAG-tagged Smad cDNAs were transfected along with Dll1IC into COS7 cells and cell extracts were subjected to immunoprecipitation with anti-FLAG or anti-Dll1IC antibody followed by western blotting (Figure 5c). Eight vertebrate Smads, designated Smad1 to Smad8, have been identified to date (29,30). Although Smad1 and Smad5 did not bind to Dll1IC, Smad2 and Smad3 showed strong binding. Co-transfection with caALK5-HA or caALK6-HA, an expression vector for the constitutively active receptor that phosphorylates Smad2, Smad3 and Smad1, Smad 5, Smad 8, respectively did not show any significant effects on binding. Smad4 also showed strong binding to Dll1IC. Smad6, Smad7 and Smad8 showed weak binding to Dll1IC.
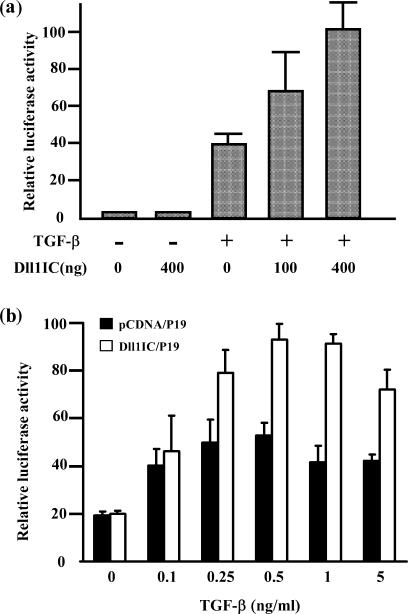
Next, we examined the effects of Dll1IC on Smad-dependent transcription. We performed dual luciferase assay using 9 × CAGA-Luc promoter–reporter plasmids that respond specifically to Smad3 (22,23). These promoter–reporter plasmids were transfected into HepG2 cells with or without Dll1IC expression vector. After transfection, the cells were stimulated with or without TGF-β, and luciferase activities were measured as an indicator of transcriptional activity. As shown in Figure 6a, only a little activity was detected without TGF-β stimulation, regardless of whether Dll1IC expression vectors were co-transfected. Stimulation of HepG2 cells with TGF-β significantly induced transcriptional activity from this promoter, and expression of Dll1IC further enhanced the transcriptional activity in a dose-dependent manner. For example, co-transfection of 100 and 400 ng of Dll1IC expression vectors were associated with a 175 and 250% enhancement of luciferase activity respectively as compared with TGF-β stimulation alone (Figure 6a).
Figure 6.
Dll1IC enhanced Smad-dependent transcription. (a) HepG2 cells were transiently transfected with the indicated amounts of Dll1IC expression vector and 9 × CAGA-Luc promoter–reporter plasmid, which responds specifically to Smad3. Cells were stimulated by the addition of TGF-β to the medium. Luciferase activities were normalized using the pRL-CMV vector that was always co-transfected as an internal control. Values are shown as the means ± S.D. of four independent experiments. (b) 9 × CAGA-Luc promoter–reporter plasmid was transfected into P19 cells stably over-expressing Dll1IC (Dll1IC/P19). P19 cells, which carry the vector alone (pCDNA/P19), were used as control. After transfection, cells were stimulated with TGF-β at various concentrations. Luciferase activities were normalized using the pRL-CMV vector that was always co-transfected as an internal control. Values are shown as the means ± S.D. of four independent experiments.
We established P19 cells stably over-expressing Dll1IC (Dll1IC/P19). P19 cells, which carry the vector alone (pCDNA/P19), were also established and used as control. 9 × CAGA-Luc promoter–reporter plasmids were transfected into Dll1IC/P19 or pCDNA/P19. After transfection, cells were stimulated with TGF-β at various concentrations and luciferase activities were measured. No significant difference of luciferase activity was detected between Dll1IC/P19 and control without TGF-β stimulation (Figure 6b). However, stimulation of these cells with TGF-β made significant difference of luciferase activity between Dll1IC/P19 and control cells. In brief, stimulation of Dll1IC/P19 with 0.5 and 1 ng/ml TGF-β were associated with 180 and 220% enhancement of luciferase activity respectively as compared with control cells (Figure 6b).
Over-expression of Dll1IC in embryonic carcinoma P19 cells induced neurons
Dll1IC may be involved in neuronal differentiation through binding to Smads. To examine this possibility, we employed Dll1IC/P19 as well as pCDNA/P19 as controls.
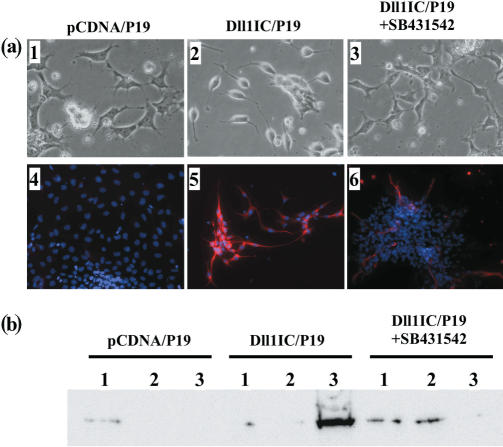
It is well known that stimulation with retinoic acid (RA) is essential for the induction of neurons from P19 cells (31–34). Thus, Dll1IC/P19 and pCDNA/P19 were cultured as aggregates for 4 days without RA, and then plated onto coverslips where the cells were allowed to differentiate for 3 days. Although pCDNA/P19 retained epithelial cell-like morphology, Dll1IC/P19 cells became round and showed a bipolar morphology with neurite extension (Figure 7a). pCDNA/P19 cells were negative for Tuj1, a marker of neurons, while almost all Dll1IC/P19 cells were Tuj1-positive (Figure 7a). These results indicate that over-expression of Dll1IC in P19 cells induced neurons. However, when 1 μM SB431542, a specific inhibitor of the TGF-β type1 receptor that activates Smad2 and Smad3 by phosphorylation (35–37), was added to Dll1IC/P19 cultures, the appearance of neurons was strongly inhibited. For example, Dll1IC/P19 treated with SB431542 retained epithelial cell-like morphology similar to pCDNA/P19 (Figure 7a). Although a few cells had long neurites, less than 10% of the cells were positive for Tuj1 (Figure 7a). These findings were confirmed by western blotting. As described above, Dll1IC/P19 and pCDNA/P19 were cultured without RA, allowed to differentiate, and lysates were prepared from subcultured, aggregated and re-plated cells, then subjected to western blotting with Tuj1 antibody. As shown in Figure 7b, expression of Tuj1 protein was induced after differentiation of Dll1IC/P19. Moreover, SB431542 strongly inhibited this induction and there were no changes detected in pCDNA/P19 throughout the culture period (Figure 7b).
Figure 7.
Over-expression of Dll1IC in P19 cells induced neurons. (a) Dll1IC-expressing P19 cells (Dll1IC/P19) and those carrying vector alone (pCDNA/P19) were cultured without RA. For inhibition experiments, Dll1IC/P19 cells were cultured with 1 μM SB431542. (1)–(3) are phase-contrast micrographs, and (4)–(6) show the results of staining with Tuj1, a marker of neurons (red) and DAPI (blue). (b) Cells were cultured as described and lysates were prepared from subcultured cells (1), aggregated cells (2) and re-plated cells (3). Aliquots of 10 μg of lysates were subjected to western blotting with Tuj1.
DISCUSSION
As indicated above, the important question is whether signaling events occur not only from ligand-expressing cells to Notch-expressing cells but also vice versa, i.e. in a bi-directional manner. As shown in Figure 1, up to 50% homologies are still recognized within the intracellular domain through the evolution of vertebrates. Thus, it is highly possible that conservation of these amino acid sequences reflect the functional importance of the intracellular domain. Indeed, we show here evidence that Dll1IC plays an important role in a bi-directional signaling pathway. Since the conservation of these sequences is restricted to vertebrate Delta, it seems that these functions, which are discussed below, would be common to vertebrates, but not invertebrates. Invertebrate DeltaIC may have a different function from that in vertebrates.
As shown in Figure 2, when NSCs were co-cultured on a monolayer of Dll1-expressing COS7 cells, the rate of emergence of neurons was lower than that in controls. It is well known that Notch signaling helps maintain the undifferentiated state of NSCs and inhibits the determination of neurons (1,2). Our findings therefore suggest that Dll1 on COS7 cells generates signals to neighboring NSCs that express Notch and thus activates Notch signaling. Conversely, when NSCs were co-cultured on a monolayer of mouse Notch1-expressing COS7 cells for 2 days, the rate of neurons developing from NSCs (55%) was significantly (P < 0.001) higher than that in control cultures (33%) (Figure 2a and b). These observations indicate that Notch1 on COS7 cells may also generate signals to neighboring NSCs and these ligands, probably Delta, may then transmit signals into the cells expressing them to mediate neuronal differentiation. Thus, signaling events may occur not only from Delta-expressing cells to Notch-expressing cells but also vice versa, that is, in a bi-directional manner, during differentiation of NSCs. The Drosophila mosaic experiment supports this idea. Baker and Schubiger published some data from a mosaic experiment in Drosophila, showing that expression of Notch in the mesoderm of Notch mutant could suppress the ectodermal defects of the mutant (5). This effect was inferred to be due to the extracellular domain of the protein and not its signaling function, since activated Notch failed to produce non-autonomous suppression (5). This suggested that the extracellular domain of Notch expressed in mesoderm was acting as a positive signal to the overlaying ectoderm in Drosophila.
It has been reported that Delta is cleaved sequentially by proteases, including ADAM and γ-secretase (8–11) and processing of Drosophila Delta was up-regulated in co-cultures with Notch-expressing S2 cells (13,15). It has also been reported that a truncated intracellular isoform of Delta shows prominent nuclear localization (10,11,13). Thus, we examined whether the same events occurred in mouse NSCs. Indeed, in developing NSCs, a large proportion of endogenous Dll1 is already cleaved and Dll1IC was localized in the nucleus (Figure 3a). Immunoreactivity against Dll1IC was accumulated in the nucleus and inhibitor of γ-secretase strongly blocked this accumulation (Figure 3b). Moreover, co-culture experiment showed that Dll1 processing could be enhanced through interaction with Notch1 (Figure 4). This enhancement was strongly inhibited by γ-secretase inhibitor (Figure 4). Although, as indicated in the introduction, Delwig et al. reported that Drosophila Delta proteolysis differs from the conventional RIP (15), mouse and rat Dll1 adhere more closely to RIP mechanism. Because, mouse and rat Dll1 are sequentially processed and preseniline is essential for these proteolysis (10,11). Thus, these observations further support the idea that signaling event also occurred from Notch-expressing cells to Delta-expressing cells.
The nuclear localization of Dll1IC suggests that Dll1IC may have effects on the transcriptions of specific target genes. To examine this possibility, we searched for transcriptional factors capable of binding to Dll1IC using a new method (see Supplementary Figure 2S). Since Smad binding sequence showed a strong signal (Figure 5a and b), co-immunoprecipitation experiments were performed to confirm these findings. Smads have been shown to act as mediators of signaling by the TGF-β superfamily. Eight vertebrate Smads, designated Smad1 to Smad8, have been identified to date (29,30). Smad2 and Smad3 are activated by TGF-β and activin (21,38,39), while Smad1 and Smad5 are major components that are activated by bone morphogenic proteins (BMPs) (40–42). Although Smad1 and Smad5 did not bind to Dll1IC, Smad2 and Smad3 showed strong binding (Figure 5c). These findings suggest that Dll1IC mediates TGF-β/Activin signaling through binding to Smad2 and/or Smad3. Phosphorylation of Smad2 and Smad3 was not required for binding to Dll1IC, as co-transfection with caALK5-HA, an expression vector for the constitutively active receptor that phosphorylates Smad2 and Smad3, did not show any significant effects on binding (Figure 5c). However, BMP signaling, which is known to inhibit neurogenesis and to enhance the appearance of astrocytes (43,44), may not be affected by Dll1IC, as Dll1IC did not bind to Smad1 or Smad5. Although Smad8 is thought to be involved in BMP signaling, it showed only weak binding to Dll1IC (Figure 5c). The common Smad, Smad4, which ubiquitously forms heterotrimeric complexes with two other Smads (41,45), also showed strong binding to Dll1IC (Figure 5c). Thus, binding of Dll1IC to not only Smad2 and Smad3 but also to Smad4 may stabilize Dll1IC–Smad complexes. Smad6 and Smad7 are thought to be inhibitory Smads. Smad7 inhibits both TGF-β/Activin signaling and BMP signaling, whereas Smad6 preferentially inhibits BMP signaling (30). Although interactions between Dll1IC and these inhibitory Smads were weak, it is interesting if these interaction can inhibit BMP signaling, which is known to inhibit neurogenesis and to enhance the appearance of astrocytes (43,44).
Although we have yet to determine the actual target genes for the Dll1IC complex with Smad2 or Smad3, we performed dual luciferase assay using 9 × CAGA-Luc promoter–reporter plasmids that respond specifically to Smad3 (22,23) as a model system. As shown in Figure 6a, stimulation of HepG2 cells with TGF-β significantly induced transcriptional activity from this promoter and expression of Dll1IC further enhanced the activity in a dose-dependent manner. These findings strongly suggest that Dll1IC mediates transcription of certain genes, which are targets of TGF-β/Activin signaling, through binding to Smads. As shown in Figure 6b, similar transfection experiments using P19 cells stably over-expressing Dll1IC (Dll1IC/P19) also showed that Dll1IC enhanced transcriptional activity and support this conclusion. Interestingly, Pfister et al. identified the PDZ protein, Acvrinp1 as a binding protein to mouse Dll1IC (25). Acvrinp1 also interacts with Activin Type II receptors and Smad3 (46). Thus, it is likely that Dll1IC can already join in assembling signaling molecules at a subcellular site, then translocate to the nucleus with Smads.
Dll1IC may be involved in neuronal differentiation through binding to Smads. To examine this possibility, we employed Dll1IC/p19 again. Although P19 cells have been shown to be induced to differentiate into neurons (31–34), RA stimulation is essential for the induction of neurons from P19 cells. However, neurons could be induced from P19 cells stably over-expressing Dll1IC without RA stimulation and this induction was strongly inhibited by SB431542, a specific inhibitor of TGF-β type1 receptor that activates Smad2 and Smad3 (Figure 7a and b). Expressions of TGF-βs and their receptors were detected through the differentiation of P19 cells (Supplementary Figure 3S). These results indicate that over-expression of Dll1IC in P19 cells induced neurons through binding to Smad2 and/or Smad3, suggesting that Dll1IC plays an important role in neuronal differentiation. Recently, it has been reported that TGF-β inhibits proliferation and accelerates differentiation of hippocampal granule neuron (47). This observation also supports our hypothesis.
These observations lead us to conclude (see schematic model in Supplementary Figure 4S) that the Notch receptor also supplies signals to Delta that are expressed on the surface of neighboring NSCs. Delta is then cleaved sequentially by proteases, probably including ADAM and γ-secretase (8–11), and finally Dll1IC is released from the cell membrane and translocates to the nucleus, where it mediates TGF-β/Activin signaling through binding to Smads and enhances transcription of specific genes leading to neuronal differentiation. This conclusion strongly implies the existence of Delta signaling, which means that the Notch-Delta signaling pathway is bi-directional. It is well known that the intracellular domain of Notch is also released from the cell membrane by the RIP mechanism (1,2,12,14) similar to those involved in the cleaving of Delta and translocates to the nucleus to modulate gene expression through binding to the transcription factor, Suppressor of Hairless (SU[H], RBP-jκ in mammals) (1,2). Therefore, similar mechanisms are involved in both directions of the Notch–Delta signaling pathway.
In the view of a bidirectional signaling pathway, it would be interesting to see if Delta pathway antagonizes Notch pathway. As a pilot experiment, we have examined if Dll1IC is able to regulate the Notch target genes by luciferase assay. However, we could not detect any antagonized or agonized activity of Dll1IC against expression of Hes1 or Hes5 genes as shown in Supplementary Figure 5S.
BMPs, another group belonging to the TGF-β superfamily, have recently been shown to inhibit neurogenesis and to enhance the generation of astrocytes from NSCs (43,44). It is thus likely that the TGF-β superfamily mediates both neurogenesis and gliogenesis from neural stem cells.
ACKNOWLEDGEMENTS
A total of 150 biotinylated double-stranded DNA oligonucleotides representing known transcription factor binding sequences and arrays that were spotted with the same set of oligo DNAs were generously supplied by B-Bridge International Inc. (California) and Panomics (California). We are grateful to T. Imamura (The cancer institute of JFCR) and K. Miyazono (University of Tokyo) for generously donating Smads expression vectors, R. Kageyama (Kyoto University) for Hes1 and Hes5-Luc promoter–reporter plasmids, J.S. Nye (Johnson and Johnson Pharmaceutical Research and Development) for Notch1 expression vector and Y. Tomooka (Tokyo University of Science) for anti-Nestin antibody. This work was supported by Special Coordination Funds from the Ministry of Education, Culture, Sports, Science and Technology of Japan and Japan Health Sciences Foundation.
REFERENCES
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Justice NJ, Jan YN. Variations on the Notch pathway in neural development. Curr. Opin. Neurobiol. 2002;12:64–70. doi: 10.1016/s0959-4388(02)00291-x. [DOI] [PubMed] [Google Scholar]
- 3.Henderson ST, Gao D, Lambie EJ, Kimble J. Lag-2 may encode a signaling ligand for the GLP-1 and LIN-12 receptors of C. elegans. Development. 1994;120:2913–2924. doi: 10.1242/dev.120.10.2913. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald K, Greenwald I. Interchangeability of Caenorhabditis elegans DSL proteins and intrinsic signalling activity of their extracellular domains in vivo. Development. 1995;121:4275–4282. doi: 10.1242/dev.121.12.4275. [DOI] [PubMed] [Google Scholar]
- 5.Baker R, Schubiger G. Autonomous and nonautonomous Notch functions for embryonic muscle and epidermis development in Drosophila. Development. 1996;122:617–626. doi: 10.1242/dev.122.2.617. [DOI] [PubMed] [Google Scholar]
- 6.Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- 7.Sun X, Artavanis-Tsakonas S. The intracellular deletions of DELTA and SERRATE define dominant negative forms of the Drosophila Notch ligands. Development. 1996;122:2465–2474. doi: 10.1242/dev.122.8.2465. [DOI] [PubMed] [Google Scholar]
- 8.Qi H, Rand MD, Wu X, Sestan N, Wang W, Rakic P, Xu T, Artavanis-Tsakonas S. Processing of the Notch ligand Delta by the metalloprotease Kuzbanian. Science. 1999;283:91–94. doi: 10.1126/science.283.5398.91. [DOI] [PubMed] [Google Scholar]
- 9.Ikeuchi T, Sisodia SS. The Notch ligands, Delta1 and Jagged2, are substrates for presenilin-dependent “γ-secretase” cleavage. J. Biol. Chem. 2003;278:7751–7754. doi: 10.1074/jbc.C200711200. [DOI] [PubMed] [Google Scholar]
- 10.LaVoie MJ, Selkoe DJ. The Notch ligands, Jagged and Delta, are sequentially processed by α-secretase and presenilin/γ-secretase and release signaling fragments. J. Biol. Chem. 2003;278:34427–34437. doi: 10.1074/jbc.M302659200. [DOI] [PubMed] [Google Scholar]
- 11.Six E, Ndiaye D, Laabi Y, Brou C, Gupta-Rossi N, Israel A, Logeat F. The Notch ligand Delta1 is sequentially cleaved by an ADAM protease and γ-secretase. Proc. Natl. Acad. Sci. U.S.A. 2003;100:7638–7643. doi: 10.1073/pnas.1230693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu. Rev. Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- 13.Bland CE, Kimberly P, Rand MD. Notch-induced proteolysis and nuclear localization of the Delta ligand. J. Biol. Chem. 2003;278:13607–13610. doi: 10.1074/jbc.C300016200. [DOI] [PubMed] [Google Scholar]
- 14.Koo EH, Kopan R. Potential role of presenilin-regulated signaling pathways in sporadic neurodegeneration. Nat. Med. 2004;10:S26–S33. doi: 10.1038/nm1065. [DOI] [PubMed] [Google Scholar]
- 15.Delwig A, Bland C, Beem-Miller M, Kimberly P, Rand MD. Endocytosis-independent mechanisms of Delta ligand proteolysis. Exp. Cell Res. 2006;312:1345–1360. doi: 10.1016/j.yexcr.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 16.Kitani H, Shiurba R, Sakakura T, Tomooka Y. Isolation and characterization of mouse neural precursor cells in primary culture. In Vitro Cell Dev. Biol. 1991;27:615–624. doi: 10.1007/BF02631104. [DOI] [PubMed] [Google Scholar]
- 17.Tomooka Y, Kitani H, Jing N, Matsushima M, Sakakura T. Reconstruction of neural tube-like structures in vitro from primary neural precursor cells. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9683–9687. doi: 10.1073/pnas.90.20.9683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A-γ-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3:688–694. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olivier A, Lauret E, Gonin P, Galy A. The Notch ligand delta-1 is a hematopoietic development cofactor for plasmacytoid dendritic cells. Blood. 2006;107:2694–2701. doi: 10.1182/blood-2005-03-0970. [DOI] [PubMed] [Google Scholar]
- 20.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription inhibition by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakao A, Imamura T, Souchelnytskyi S, Kawabata M, Ishisaki A, Oeda E, Tamaki K, Hanai J, Heldin C-H, Miyazono K, et al. TGF-β receptor-mediated signalling through Smad2, Smad3 and Smad4. EMBO J. 1997;16:5353–5362. doi: 10.1093/emboj/16.17.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier JM. Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–3100. doi: 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jonk LJC, Itoh S, Heldin C.-H, ten Dijke P, Kruijer W. Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin, and bone morphogenetic protein-inducible enhancer. J. Biol. Chem. 1998;273:21145–21152. doi: 10.1074/jbc.273.33.21145. [DOI] [PubMed] [Google Scholar]
- 24.Nakayama K, Nagase K, Tokutake Y, Koh C-S, Hiratochi M, Ohkawara T, Nakayama N. Multiple POU-binding motifs, recognized by tissue-specific nuclear factors, are important for Dll gene expression in neural stem cells. Biochem. Biophys. Res. Commun. 2004;325:991–996. doi: 10.1016/j.bbrc.2004.10.138. [DOI] [PubMed] [Google Scholar]
- 25.Pfister S, Przemeck GKH, Gerber JK, Beckers J, Adamski J, Hrabe de Angelis M. Interaction of the MAGUK family member Acvrinp1 and the cytoplasmic domain of the Notch ligand Delta1. J. Mol. Biol. 2003;333:229–235. doi: 10.1016/j.jmb.2003.08.043. [DOI] [PubMed] [Google Scholar]
- 26.Easter SS, Jr, Ross LS, Frankfurter A. Initial tract formation in the mouse brain. J. Neurosci. 1993;13:285–299. doi: 10.1523/JNEUROSCI.13-01-00285.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lichtenthaler SF, Ida N, Multhaup G, Masters CL, Beyreuther K. Mutations in the transmembrane domain of APP altering γ-secretase specificity. Biochemistry. 1997;36:15396–15403. doi: 10.1021/bi971071m. [DOI] [PubMed] [Google Scholar]
- 28.Diehmann A, Ida N, Weggen S, Grunberg J, Haass C, Masters CL, Bayer TA, Beyreuther K. Analysis of Presenilin 1 and Presenilin 2 expression and processing by newly developed monoclonal antibodies. J. Neurosci. Res. 1999;56:405–419. doi: 10.1002/(SICI)1097-4547(19990515)56:4<405::AID-JNR8>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 29.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 30.Miyazawa K, Shinozaki M, Hara T, Furuya T, Miyazono K. Two major Smad pathways in TGF-β superfamily signalling. Genes Cells. 2002;7:1191–1204. doi: 10.1046/j.1365-2443.2002.00599.x. [DOI] [PubMed] [Google Scholar]
- 31.McBurney MW, Jones-Villeneuve EM, Edwards MK, Anderson PJ. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 1982;299:165–167. doi: 10.1038/299165a0. [DOI] [PubMed] [Google Scholar]
- 32.Jones-Villeneuve EM, Rudnicki MA, Harris JF, McBurney MW. Retinoic acid-induced neural differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 1983;3:2271–2279. doi: 10.1128/mcb.3.12.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nye JS, Kopan R, Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 34.Gao X, Bian W, Yang J, Tang K, Kitani H, Atsumi T, Jing N. A role of N-Cadherin in neuronal differentiation of embryonic carcinoma P19 cells. Biochem. Biophys. Res. Commun. 2001;284:1098–1103. doi: 10.1006/bbrc.2001.5089. [DOI] [PubMed] [Google Scholar]
- 35.Laping NJ, Grygielko E, Mathur A, Butter S, Bomberger J, Tweed C, Martin W, Fornwald J, Lehr R, Harling J, et al. Inhibition of transforming growth factor (TGF)-β1-induced extracellular matrix with a novel inhibitor of the TGF-β type I receptor kinase activity: SB-431542. Mol. Pharmacol. 2002;62:58–64. doi: 10.1124/mol.62.1.58. [DOI] [PubMed] [Google Scholar]
- 36.Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-β superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 37.Watabe T, Nishihara A, Mishima K, Yamashita J, Shimizu K, Miyazawa K, Nishikawa S, Miyazono K. TGF-β receptor kinase inhibitor enhances growth and integrity of embryonic stem cell-derived endothelial cells. J. Cell Biol. 2003;163:1303–1311. doi: 10.1083/jcb.200305147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eppert K, Scherer SW, Ozcelik H, Pirone R, Hoodless P, Kim H, Tsui LC, Bapat B, Gallinger S, Andrulis IL, et al. MADR2 maps to 18q21 and encodes a TGFβ-regulated MAD-related protein that is functionally mutated in colorectal carcinoma. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y, Feng X, We R, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature. 1996;383:168–172. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
- 40.Hoodless PA, Haerry T, Abdollah S, Stapleton M, O'Connor MB, Attisano L, Wrana JL. MADR1, a MAD-related protein that functions in BMP2 signaling pathways. Cell. 1996;85:489–500. doi: 10.1016/s0092-8674(00)81250-7. [DOI] [PubMed] [Google Scholar]
- 41.Kretzschmar M, Liu F, Hata A, Doody J, Massague J. The TGF-β family mediator Smad1 is phosphorylated directly and activated functionally by the BMP receptor kinase. Genes Dev. 1997;11:984–995. doi: 10.1101/gad.11.8.984. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki A, Chang C, Yingling JM, Wang XF, Hemmati-Brivanlou A. Smad5 induces ventral fates in Xenopus embryo. Dev. Biol. 1997;184:402–405. doi: 10.1006/dbio.1997.8548. [DOI] [PubMed] [Google Scholar]
- 43.Nakashima K, Takizawa T, Ochiai W, Yanagisawa M, Hisatsune T, Nakafuku M, Miyazono K, Kishimoto T, Kageyama R, Taga T. BMP2-mediated alteration in the developmental pathway of fetal mouse brain cells from neurogenesis to astrocytogenesis. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5868–5873. doi: 10.1073/pnas.101109698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takizawa T, Ochiai W, Nakashima K, Taga T. Enhanced gene activation by Notch and BMP signaling cross-talk. Nucleic Acids Res. 2003;31:5723–5731. doi: 10.1093/nar/gkg778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lagna G, Hata A, Hemmati-Brivanlou A, Massague J. Partnership between DPC4 and SMAD proteins in TGF-β signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- 46.Shoji H, Tsuchida K, Kishi H, Yamakawa N, Matsuzaki T, Liu Z, Nakamura T, Sugino H. Identification and characterization of a PDZ protein that interacts with activin type II receptors. J. Biol. Chem. 2000;275:5485–5492. doi: 10.1074/jbc.275.8.5485. [DOI] [PubMed] [Google Scholar]
- 47.Lu J, Wu Y, Sousa N, Almeida OFX. SMAD pathway mediation of BDNF and TGFβ2 regulation of proliferation and differentiation of hippocampal granule neurons. Development. 2005;132:3231–3242. doi: 10.1242/dev.01893. [DOI] [PubMed] [Google Scholar]