Abstract
Recent work has demonstrated an unexpected prevalence of copy number variation in the human genome, and has highlighted the part this variation may play in predisposition to common phenotypes. Some important genes vary in number over a high range (e.g. DEFB4, which commonly varies between two and seven copies), and have posed formidable technical challenges for accurate copy number typing, so that there are no simple, cheap, high-throughput approaches suitable for large-scale screening. We have developed a simple comparative PCR method based on dispersed repeat sequences, using a single pair of precisely designed primers to amplify products simultaneously from both test and reference loci, which are subsequently distinguished and quantified via internal sequence differences. We have validated the method for the measurement of copy number at DEFB4 by comparison of results from >800 DNA samples with copy number measurements by MAPH/REDVR, MLPA and array-CGH. The new Paralogue Ratio Test (PRT) method can require as little as 10 ng genomic DNA, appears to be comparable in accuracy to the other methods, and for the first time provides a rapid, simple and inexpensive method for copy number analysis, suitable for application to typing thousands of samples in large case-control association studies.
INTRODUCTION
Several recent studies have demonstrated that some genes or groups of genes can show variation in copy number, and that this variation can have important functional consequences (1–6). For example, the genes CCL3L1, CCL4L1 and TBC1D3 are present on a segmental duplication that can vary between 0 and 10 copies per person (7); this variation appears to be a determinant of individual susceptibility to, and progression of, infection with HIV-1 (8). Similarly, a group of beta-defensin genes including DEFB4 commonly varies between two and seven copies per person, with occasional extremely expanded alleles containing 8–11 repeats visible as ‘euchromatic variants’ of 8p23.1 (9). These beta-defensin genes, as well as the independently variable alpha-defensins DEFA1A3 (10–12), are candidate genes for variation in susceptibility to infectious disease, as well as autoimmune and inflammatory disorders (10), and low copy number of the DEFB4 segmental duplication has been associated with Crohn's disease of the colon (13).
Where frequent copy number variation encompasses the 0–3 copy number range, many established technologies can be used to provide rapid, cheap and accurate measurement of DNA copy number. In contrast, where the copy number range is higher, such as for CCL3L1 or the DEFB4 cluster, accurately distinguishing a count of five copies from six requires a precision not available from easily implemented methods. For example, combining MAPH with determination of Restriction Enzyme Digest Variant Ratios (MAPH/REDVR) has been capable of high reproducibility in determining copy number at DEFB4 (10) and DEFA1A3 (11), but uses large amounts of genomic DNA (>1 μg) and is labour-intensive. Real-time PCR is gaining in popularity as a method of determining copy number (8,13), but requires careful set-up and does not easily provide the highest throughput required for large association studies.
An ideal high-throughput method for measuring copy number in large-scale association studies would be accurate, inexpensive, robust and use only small amounts of genomic DNA, and the pressing need for methods with these properties is increasingly recognized (14). In assessing pathological deletions or duplications of single-copy genes, relatively simple multiplex fluorescent PCR methods (15–17) have delivered an acceptable level of accuracy in this range. Multiplex fluorescent PCR methods compare the amount of PCR product made from a test amplicon with the yield from a reference locus in the same multiplex reaction. These approaches have the advantage of simplicity, but are prone to variability. The experimental variability of multiplex PCR is presumably due to the different amplification properties of the test and reference loci, and the differential sensitivity of the yield of each amplification reaction to the precise conditions. To obtain reliable results in multiplex PCR, great care needs to be taken with a number of experimental factors, including DNA quality and (as far as possible) matching the amplification properties of test and reference amplicons. Some of this experimental variation has been reduced by the design of short amplicons, combined with careful attention to primer design and PCR conditions, in the QMPSF technology (15,16).
In this study we have adapted the quantitative multiplex PCR approach by using primers designed to amplify from repeated DNA elements. The primers are precisely designed to amplify from a copy of the element within the variable repeat unit, plus exactly one other unlinked reference locus. We have applied this method to the copy-variable DEFB4 repeat unit, and compare the results from this approach with results on the same samples from three independent alternative methods. This new PRT method is comparable in accuracy to these alternative methods, and its simple format, and requirement for only small (10–20 ng) amounts of genomic DNA, should allow accurate, inexpensive and rapid copy number typing of large cohorts of samples in association studies.
MATERIALS AND METHODS
Samples and formats
Genomic DNA from Dutch and HapMap samples was used at concentrations of 5–10 ng/μl, and for most experiments samples were arrayed in 96-well microtitre plates and processed in batches of 96. Dutch genomic DNA samples included those from the Nijmegen Biomedical Study (18). All liquid-handling operations could be carried out using multi-channel pipettes.
MLPA and array-CGH data
MLPA was carried out as described (19) using 250 ng genomic DNA and the SALSA MLPA kit P139 Defensin from MRC-Holland, and data were analyzed as described (20). Array-CGH data were downloaded from the Wellcome Trust Sanger Institute website www.sanger.ac.uk/humgen/cnv/data. Details on methods used to collect the data, downloading from the website and clones used are also available on the site.
PRT assay
PCR was carried out using 5 ng input genomic DNA, 0.5 μM primer HSPD5.8F (CCAGATGAGACCAGTGTCC) and 0.5 μM FAM- or HEX-labelled primer HSPD5.8R (TTTTAAGTTCAGCAATTACAGC) (Figure 1), in a buffer [modified from (21)] containing final concentrations of 50 mM Tris–HCl (pH 8.8), 12 mM ammonium sulphate, 5 mM magnesium chloride, 125 μg/ml BSA (non-acetylated, Ambion Inc), 7.4 mM 2-mercaptoethanol and 1.1 mM each dNTP (sodium salts), with 0.5U Taq DNA polymerase in a total volume of 10 μl. Products were amplified using 30 cycles of 95°C for 30 s, 53°C for 30 s and 70°C for 30 s, followed by a single ‘chase’ phase of 53°C for 1 min/70°C for 20 min to enhance complete extension in the final round and hence reduce levels of single-stranded DNA products. The cycle number of 30 was chosen after empirical tests to determine conditions that yielded quantifiable (i.e. not saturating) amounts of product; the annealing temperature was also chosen after empirical comparisons, and the choice of 53°C is near the maximum annealing temperature that can be used, probably associated with a reduction in the efficiency of amplification.
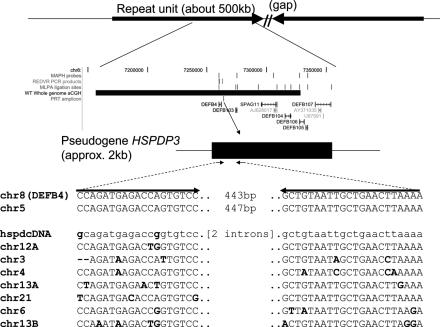
Figure 1.
Principle of the PRT assay at DEFB4. The top line shows the general structure of the repeat unit containing DEFB4 (which has two inverted rather than tandem repeats in the March 2006 assembly). The middle panel shows the locations of the genes SPAG11, DEFB4 and DEFB103–107, together with the locations of probes or amplicons used in MAPH, REDVR, MLPA and PRT, and the extent of the clone (10C3) used in array-CGH ('WT Whole Genome aCGH'). In the detailed display at the bottom, the primers amplify products from the HSPDP3 pseudogene upstream of DEFB4 on chromosome 8, and from a reference copy on chromosome 5, but have multiple mismatches (bold) with other copies of the element. In this way a single primer pair can be used to amplify two very similar products, one from near DEFB4, the other from chromosome 5.
Two amplifications were carried out for each sample, one with a fluorescent FAM label, the other with a fluorescent HEX label; 1 μl of each PCR product was added, without further purification, to a 10 μl digestion containing 1× ReAct 2 buffer [50 mM Tris–HCl (pH8.0), 10 mM MgCl2, 50 mM NaCl] (Invitrogen) and 5U HaeIII (New England Biolabs). After incubation at 37°C for 4–16 h, 2 μl was added to 10 μl HiDi formamide with ROX-500 marker (Applied Biosystems), and analyzed by electrophoresis on an ABI3100 36 cm capillary using POP4 polymer and an injection time of 30 s.
Data analysis
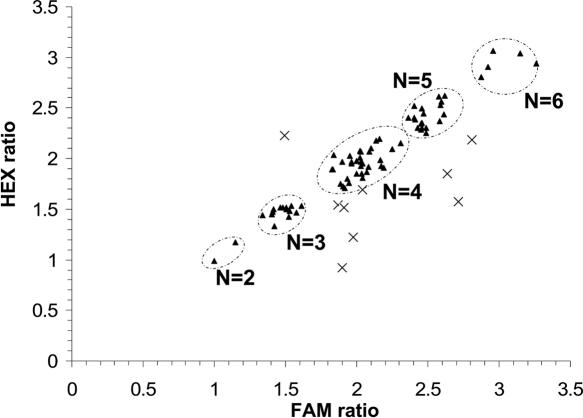
Peak areas corresponding to the 302 bp HaeIII fragment from near DEFB4 and the 315 bp fragment from chromosome 5 were recorded for both FAM- and HEX-labelled products using GeneScan and Genotyper software (Applied Biosystems). The ratio 302/315 bp was compared between FAM- and HEX-labelled products, and results were accepted if the difference between the ratios was <15% of their mean; this criterion led to the rejection of ∼10% of tests (Figure 2).
Figure 2.
Comparison of ratios (chromosome 8: chromosome 5) from FAM- and HEX-labelled products in a single experiment. Pairs of ratios (triangles) meeting the quality-control criteria (73 samples) cluster around groups corresponding to copy numbers of 2, 3, 4, 5 and 6. Crosses show results rejected for having too great a difference between FAM and HEX ratios (10 samples).
If accepted, the mean of the ratios of the FAM- and HEX-labelled products was used in further analysis. Although naïve inference of a copy number equal to double the mean ratio would lead to reasonably accurate results, there were small but definite shifts between experiments in the relative amplification of the test and reference products. Mean ratios were therefore used in conjunction with reference standards to calibrate each experiment, and the resulting (least-squares) linear regression used to infer the copy numbers for unknown samples. In most experiments MAPH/REDVR copy numbers were used to calibrate PRT assays; subsequently, DNA samples giving reproducible results from several PRT assays have also been successfully used as calibration standards.
RESULTS AND DISCUSSION
We reasoned that many of the problems of accuracy and reproducibility associated with multiplex PCR might be avoided if the test and reference amplicons were as similar as possible. This principle has recently been successfully exploited in an innovative approach to the diagnosis of trisomy; because some sequences present on chromosome 21 (for example) have nearly identical paralogues at another site in the genome, a single pair of primers can be used to amplify both test and reference loci, distinguishing them via minor differences of internal sequence (22). Although copy-variable loci are very unlikely to contain extensive regions with nearly identical counterparts at other locations, we were able to exploit the same advantages of amplifying both test and reference loci with a single primer pair by designing precisely placed primers in a diverged (low copy number) repetitive sequence, thereby allowing a Paralogue Ratio Test (PRT).
A heat-shock protein pseudogene of ∼2 kb (HSPDP3) is found ∼2 kb upstream of the DEFB4 gene (Figure 1), and at 10 locations elsewhere in the genome. We were able to design primers that matched the copy near DEFB4 and one other copy on chromosome 5 exactly, but which had multiple mismatches to copies at other chromosomal locations. The (test) chromosome 8 (DEFB4) and (reference) chromosome 5 copies give PCR products too close in size (443 and 447 bp respectively) to separate reliably by capillary electrophoresis, but they could be easily distinguished and quantified after digestion with the restriction enzyme HaeIII to give products of 302 and 315 bp. Because products from other copies of the pseudogene were predicted to have characteristic alternative fragment lengths following HaeIII digestion, the absence of detectable fragments of these lengths confirmed that under the conditions used, the products detected came exclusively from the chromosome 5 and chromosome 8 loci. Measuring the ratios of products from the test and reference loci (see ‘Materials and Methods’) allowed inference of DEFB4 copy number, which was linearly related to product ratio.
In the absence of ‘gold standard’, error-free methods to count DEFB4 copy numbers for reference samples, we validated the method by comparisons with results from three other established techniques. PRT was used to type 591 genomic DNA samples which had already been typed for DEFB4 copy number by the MAPH/REDVR method. The MAPH/REDVR approach uses a primary copy number estimate by MAPH (23), refined by examining ratios of sequence variants from the repeat unit (10,11). For 486 samples (82%), a single PRT assay gave the same integer copy number as MAPH/REDVR; for a further 64 samples (11%), the MAPH/REDVR copy number was confirmed by PRT on repeat testing. Thus for 93% of samples, the MAPH/REDVR value was confirmed by PRT either on first-pass testing or after a single repeat test. For 25 samples (4.2%) PRT consistently gave a value different from MAPH/REDVR, and for these samples MAPH/REDVR was assumed to have been in error. No consistent results were obtained for 16 samples (2.7%).
A subset of 135 of these samples was also typed by MLPA (19), so that this smaller sample set was typed by three independent methods: MAPH/REDVR, PRT and MLPA. For samples which had undergone more than one PRT assay, only the first test result was included in this analysis. These three methods appear to be comparable in their accuracy; all three methods agreed on the integer copy number for 113 out of 135 samples (84%), and of the 22 remaining samples, the result from one method disagreed with the other two—first-pass PRT gave the discordant result for seven samples, MAPH/REDVR for 6 samples, and MLPA for nine samples. From the 135 MLPA results, 119 (88%) agreed with the integral copy number measured by a single PRT assay. Comparisons between unrounded MLPA copy number estimates and unrounded copy number values from first-pass PRT are shown in Figure 3. As expected, clusters of values corresponding to integral copy numbers are seen, and the spread of measured values is higher at higher copy number.
Figure 3.
Comparison between unrounded copy number estimates from PRT, with unrounded data from MLPA for the same 135 samples.
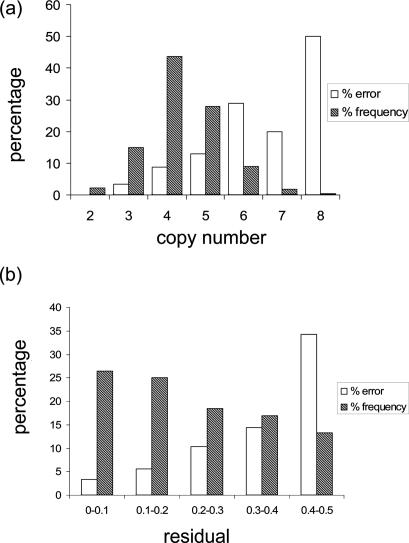
We estimated rates of error for single-pass PRT typing from a larger data set, combining results from MAPH/REDVR and PRT data, plus the MLPA results for the subset of 135 samples, to define a consensus copy number for all 575 samples (after removing the 16 samples for which no consistent data were obtained). First-pass PRT results disagreeing with this consensus were scored as measurement errors. There were two striking patterns in the distribution of error for single-pass PRT typing. First, the highest copy numbers had the greatest error, as expected for a method that relies on ratios of measurements. Estimated error rates were <10% for copy numbers of 2, 3 and 4, but rose to 20% or higher for samples with copy numbers of six or more (Figure 4a). Second, unrounded PRT values close to an integer value were more likely to give the correct integer value; values within 0.2 of an integer had estimated error rates <10%, but if the unrounded value was as much as 0.4 from an integer value, the error rate was >30% (Figure 4b). Overall, the analyses suggested that a single PRT test yielded the correct integral value in 89% of samples (511 out of 575), a figure that can be further improved by confirmatory and repeat testing as recommended below.
Figure 4.
Correlation between error attributable to first-pass PRT and (a) copy number or (b) absolute difference between the unrounded and integer values measured (‘residual’). In each case the inferred percentage error in each subcategory is shown by the open columns. Shaded columns show the relative frequencies of each copy number or residual category in the data set.
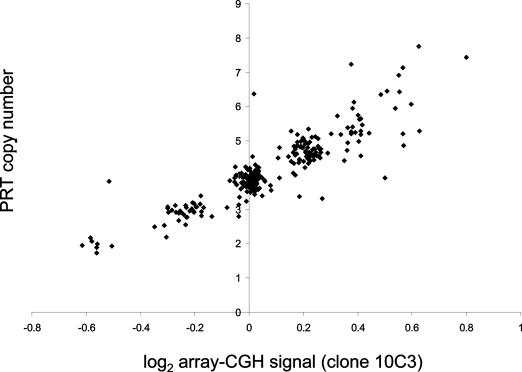
As a further comparison of PRT with established methodology, PRT was used to type 261 samples from the HapMap collection (24). These data were then compared with array-CGH data from a clone (10C3) within the DEFB4 repeat unit. Figure 5 shows the comparison between unrounded PRT results and normalized signal from array-CGH of clone 10C3, demonstrating clusters of data-points corresponding to integral copy numbers of 2, 3, 4 and 5, with a greater spread at copy numbers of 6 and 7. Integral copy numbers from array-CGH data were assigned to these samples based on ranges defined by the clusters, and agreed with the PRT-derived integer copy number for 202 samples (77%). This relatively low figure combines error attributable to PRT with an unknown but presumably higher rate of error from array-CGH. Analysis of flanking SNP haplotypes in the HapMap samples showed very weak evidence for association with flanking haplotypes, and no SNPs or haplotypes were found that could reliably be used as a surrogate for copy number typing.
Figure 5.
Comparison between first-pass PRT typing (unrounded values) and normalized signal from a clone from the DEFB4 region in array-CGH for 261 samples from the HapMap study.
Finally, a total of 99 samples underwent duplicate testing by PRT without any prior selection for concordance or disagreement with expected values, and 83 (83.8%) gave the same integer values on both tests. Because the frequency of samples with discordant duplicate tests is expected to be approximately twice the rate of error for a single test, this suggests that the error rate for integer values from a single PRT test is ∼8.1% (95% confidence interval 5.6–11.8%).
All these analyses together suggest that a single PRT test would on its own return the correct integer value in ∼85–95% of tests. In typing large numbers of samples for case-control association studies using any error-prone measurement or genotyping method, a compromise has to be made between throughput and the cost and replication work required to obtain accurate data. The accuracy of PRT testing for copy number could clearly be improved by requiring concordant duplicate testing of all samples, but for many large-scale studies such a doubling of the genotyping cost and effort may not be justified. The patterns discovered in our analyses suggest that a low error rate can be achieved with selective re-testing of samples. For example, if re-testing was triggered by integral copy numbers of five or more with unrounded PRT values >0.3 from the nearest integer, then the data predict that only 15% of samples would require re-testing, and of the 85% of results accepted by these criteria, ∼92% would return the correct integral value. Depending on the balance required between throughput, cost, labour and accuracy, alternative criteria for acceptance or re-testing can be formulated.
In the presence of somatic mosaicism, the true copy number in a DNA sample may not be an integer, but instead an intermediate value reflecting the mean copy number of the cell population. In such a case, if the correct measure of copy number is frequently non-integral, it is predicted that multiple measurements of the same samples (typed by several methods, or in duplicate by a single method) should have a tendency to cluster around non-integer values. In our data set, common mosaicism should therefore be detectable as a significant correlation between residuals (i.e. the unrounded measurement minus the rounded integer value) between duplicate PRT tests or between different methods. No such correlation was detected in our data (not shown), suggesting that determination of a whole number of repeats is the biologically relevant measurement result, and in turn that differences between the measured values and integer values are the results of measurement error.
The analysis assumes that the reference locus (in this case on chromosome 5) does not itself vary in copy number, and that substitutional mismatches at either the test or reference locus primer binding sites do not lead to `drop-out' of one or more copies. In addition to their absence from all available sequence databases and sequence trace archives, substitutional variants do not appear to pose a problem at the chromosome 5 reference locus for DEFB4 measurement; drop-out of one copy at the reference locus should lead to apparent doubling of the copy number of the test locus, and no PRT results showed a discrepancy of exactly double the consensus value. Drop-out at the test locus may lead to subtler anomalies, including failure of only a single copy to amplify, reducing the apparent copy number by one. In particular, gene conversion among diverged members of a copy-variable array (3) could act to propagate even quite rare substitutional variants, leading to errors in the estimation of copy number. It is unlikely that this is a significant source of error at DEFB4; in addition to the close agreement of results from different methods, where the methods disagreed, there was no tendency for PRT to record systematically lower values than MAPH/REDVR or MLPA. Further reassurance on both these sources of drop-out may be obtained by use of a second PRT system with a different reference locus, and using a different part of the repeat element; at DEFB4 a second PRT system can be generated using a reference locus on chromosome 4. Although in practice the assay does not frequently give rise to partial digestion products, incomplete action of HaeIII might in principle distort the product ratios. However, one would predict that (unless incomplete digestion affected the test and reference loci differentially) the correct product ratios would nevertheless be preserved in the population of completely digested molecules. Furthermore, there is no absolute need to use restriction digestion as the method to discriminate test and reference products; the assay could easily be modified to determine the ratios using other methods such as pyrosequencing.
How general is the applicability of PRT to copy number measurement? Although many genes and regions have been defined as copy variable, the true extent of the variable region has been accurately determined for only a few. Consequently, the precise extent of the DNA to be measured cannot be defined for most loci. Furthermore, even when there are appropriately diverged dispersed repeats, without extensive individual examination it is not easy to assess the critical requirement for a pair of primers specific to precisely two loci. Compounding this problem is the uncertainty surrounding the extent to which primer mismatches can be relied upon to discriminate against additional, alternative amplification products. Finally, we have not yet determined whether using two or more co-amplified reference loci may be as accurate as using a single reference locus; if so, then the scope for developing accurate PRT systems may be even wider than we suggest here.
Where there are closely linked paralogous sequences, as may frequently happen in the evolution of gene family clusters, paralogues assumed to be present at constant copy number may be used as specific reference loci. Thus, for example, the CYP2D8P pseudogene has been used as a reference locus in the accurate measurement of gene copy number for CYP2D6 (25), and the RHCE gene, usually present at two copies, could be used as an effective reference locus for RHD. However, it is very difficult to be sure that gene conversion does not exchange test and reference sequences in such local tandem arrays, and at other loci such as DEFA1A3 it is known that the paralogous variants can interchange locations (11).
For this reason an unlinked reference locus is desirable. To examine how frequently a copy-variable locus would harbour a sufficiently diverged repeat element to allow a PRT assay, we examined 20 well-characterized examples of gene-containing copy-variable regions in addition to DEFB4. We found 18 loci at which primers could be designed to amplify a test product plus exactly one reference product, with mismatches to other genomic loci (Table 1). In many cases, the two products differ in size, so that no restriction enzyme digestion or other sequence-specific processing is required to distinguish them. Some of the systems proposed involve diverged members of sequence families present in the genome at very high copy number (Alu, FLAM-C, L1) for which it may be difficult to generate the locus-specificity required. Nevertheless, although they remain to be tested in practice, at many of these loci it is clear that the primers are very likely to form the basis of a specific and accurate PRT assay for copy number. We may therefore conservatively suggest that useful PRT systems could be designed for at least 50% of copy-variable genes.
Table 1.
Potential PRT systems for DEFB4 and 20 further copy variable genes
| Gene/locus | Citation | Closely linked reference paralogue? | Unlinked reference amplicon | |
|---|---|---|---|---|
| Element | Coordinates (March 2006 assembly) | |||
| DEFB4 | (9) | — | HSPDP3 | chr5:21918479–21918925 |
| AMY1A | (26) | AMY2A | ERV1 LTR | chr12:49600721–49601110 |
| C4A | (27) | (C4B) | HERVKC4 | chr19:7769331–7769661 |
| CCL3L1 | (7) | CCL3 | LTR61 | chr10:82018627–82019006 |
| CFHR1 | (28) | CFH | AluSp | chr2:116372603–116372838 |
| CNTNAP3 | (29) | — | LTR8A | chr1:32677895–32678213 |
| CYP2D6 | (25) | CYP2D7P/8P | — | — |
| DEFA1 | (30) | DEFA3 | MLT1A0 LTR | chr7:152825444–152825655 |
| DNMT1 | (31) | — | MLT1E2 | chr7:76691729–76691822 |
| FCGR3 | (32) | — | MRE11 | chr1:47139266–47139433 |
| GSTM1 | (33) | GSTM5 | — | — |
| GSTT1 | (34) | — | FLAM-C | chr1:211834045–211834128 |
| IGL | (35) | — | (DUP) | chr22:30937538–30938261 |
| NPHP1 | (36) | — | (DUP) | chr2:88842054–88842506 |
| OPN1MW | (37) | OPN1LW | MER73 | chr15:91444669–91444891 |
| PGA5 | (38) | PGA3 | (DUP) | chr1:173259630–173259694 |
| PRAMEF8/F9 | (39) | — | L1MA4 | chr11:84670650–84670997 |
| RHD | (40) | RHCE | THE1D | chr11:96087596–96087729 |
| SMN2 | (41) | (SMN1) | LTR19C | chr17:64241628–64241737 |
| PRB1 | (42) | PRB2 | L1PB4 | chr1:96304675–96305394 |
| UGT2B17 | (43) | UGT2B15 | THE1D | chr16:13662429–13662603 |
The final ‘coordinates’ column specifies the reference locus and the placement of primers that should discriminate against other copies in the genome. In column 4, ‘(DUP)’ indicates a duplicon not associated with an identifiable dispersed repeat element. Where there is a potentially useful reference locus closely linked to the test locus, this is shown in the third column; in the specific cases of C4B and SMN2, the paralogues are not distinguished by substitutional divergence.
Acknowledgments
Diana Olthuis and Gys de Jongh are acknowledged for technical assistance, and Evelien Hoenselaar and Gaby Schobers for technical assistance with the MLPA assays. We thank Mark Jobling for access to HapMap DNA samples, Matt Hurles for helpful discussions, and Jess Tyson and Tamsin Majerus for helpful comments on the manuscript. This work was supported by a grant to JS from the Netherlands Organization for Scientific Research (NWO-Genomics 050-10-102), and a Wellcome Trust Bioarchaeology Postdoctoral Fellowship to EJH (no.071024). Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Tuzun E., Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nature Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 2.Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fredman D., White S.J., Potter S., Eichler E.E., Den Dunnen J.T., Brookes A.J. Complex SNP-related sequence variation in segmental genome duplications. Nature Genet. 2004;36:861–866. doi: 10.1038/ng1401. [DOI] [PubMed] [Google Scholar]
- 4.Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nature Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 5.Feuk L., Carson A.R., Scherer S.W. Structural variation in the human genome. Nature Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 6.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Maner S., Massa H., Walker M., Chi M.Y., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 7.Townson J.R., Barcellos L.F., Nibbs R.J.B. Gene copy number regulates the production of the human chemokine CCL3-L1. Eur. J. Immunol. 2002;32:3016–3026. doi: 10.1002/1521-4141(2002010)32:10<3016::AID-IMMU3016>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez E., Kulkarni H., Bolivar H., Mangano A., Sanchez R., Catano G., Nibbs R.J., Freedman B.I., Quinones M.P., Bamshad M.J., et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1340. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 9.Hollox E.J., Armour J.A., Barber J.C. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am. J. Hum. Genet. 2003;73:591–600. doi: 10.1086/378157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hollox E.J., Davies J., Griesenbach U., Burgess J., Alton E.W., Armour J.A. Beta-defensin genomic copy number is not a modifier locus for cystic fibrosis. J. Negat. Results Biomed. 2005;4:9. doi: 10.1186/1477-5751-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aldred P.M.R., Hollox E.J., Armour J.A.L. Copy number polymorphism and expression level variation of the human alpha-defensin genes DEFA1 and DEFA3. Hum. Mol. Genet. 2005;14:2045–2052. doi: 10.1093/hmg/ddi209. [DOI] [PubMed] [Google Scholar]
- 12.Linzmeier R.M., Ganz T. Human defensin gene copy number polymorphisms: comprehensive analysis of independent variation in alpha- and beta-defensin regions at 8p22-p23. Genomics. 2005;86:423–430. doi: 10.1016/j.ygeno.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Fellermann K., Stange D.E., Schaeffeler E., Schmalzl H., Wehkamp J., Bevins C.L., Reinisch W., Teml A., Schwab M., Lichter P., et al. A Chromosome 8 Gene-Cluster Polymorphism with Low Human Beta-Defensin 2 Gene Copy Number Predisposes to Crohn Disease of the Colon. Am. J. Hum. Genet. 2006;79:439–448. doi: 10.1086/505915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Todd J.A. Statistical false positive or true disease pathway? Nature Genet. 2006;38:731–733. doi: 10.1038/ng0706-731. [DOI] [PubMed] [Google Scholar]
- 15.Casilli F., Di Rocco Z.C., Gad S., Tournier I., Stoppa-Lyonnet D., Frebourg T., Tosi M. Rapid detection of noval BRCA1 rearrangements in high-risk breast-ovarian cancer families using multiplex PCR of short fluorescent fragments. Hum. Mutat. 2002;20:218–226. doi: 10.1002/humu.10108. [DOI] [PubMed] [Google Scholar]
- 16.Charbonnier F., Raux G., Wang Q., Drouot N., Cordier F., Limacher J.-M., Saurin J.-C., Puisieux A., Olschwang S., Frebourg T. Detection of exon deletions and duplications of the mismatch repair genes in hereditary nonpolyposis colorectal cancer families using multiplex polymerase chain reaction of short fluorescent fragments. Cancer Res. 2000;60:2760–2763. [PubMed] [Google Scholar]
- 17.Vaurs-Barriere C., Bonnet-Dupeyron M.N., Combes P., Gauthier-Barichard F., Reveles X.T., Schiffmann R., Bertini E., Rodriguez D., Vago P., Armour J.A.L., et al. Golli-MBP copy number analysis by FISH, QMPSF and MAPH in 195 patients with hypomyelinating leukodystrophies. Ann. Hum. Genet. 2006;70:66–77. doi: 10.1111/j.1529-8817.2005.00208.x. [DOI] [PubMed] [Google Scholar]
- 18.Hoogendoorn E.H., Hermus A.R., de Vegt F., Ross H.A., Verbeek A.L., Kiemeney L.A., Swinkels D.W., Sweep F.C., den Heijer M. Thyroid function and prevalence of anti-thyroperoxidase antibodies in a population with borderline sufficient iodine intake: influences of age and sex. Clin. Chem. 2006;52:104–111. doi: 10.1373/clinchem.2005.055194. [DOI] [PubMed] [Google Scholar]
- 19.Schouten J.P., McElgunn C.J., Waaijer R., Zwijnenburg D., Diepvens F., Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koolen D.A., Nillesen W.M., Versteeg M.H.A., Merkx G.F.M., Knoers N., Kets M., Vermeer S., van Ravenswaaij C.M.A., de Kovel C.G., Brunner H.G., et al. Screening for subtelomeric rearrangements in 210 patients with unexplained mental retardation using multiplex ligation dependent probe amplification (MLPA) J. Med. Genet. 2004;41:892–899. doi: 10.1136/jmg.2004.023671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeffreys A.J., Neumann R., Wilson V. Repeat unit sequence variation in minisatellites—a novel source of DNA polymorphism for studying variation and mutation by single molecule analysis. Cell. 1990;60:473–485. doi: 10.1016/0092-8674(90)90598-9. [DOI] [PubMed] [Google Scholar]
- 22.Deutsch S., Choudhury U., Merla G., Howald C., Sylvan A., Antonarakis S.E. Detection of aneuploidies by paralogous sequence quantification. J. Med. Genet. 2004;41:908–915. doi: 10.1136/jmg.2004.023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armour J.A., Sismani C., Patsalis P.C., Cross G. Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res. 2000;28:605–609. doi: 10.1093/nar/28.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altshuler D., Brooks L.D., Chakravarti A., Collins F.S., Daly M.J., Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soderback E., Zackrisson A.L., Lindblom B., Alderborn A. Determination of CYP2D6 gene copy number by pyrosequencing. Clin. Chem. 2005;51:522–531. doi: 10.1373/clinchem.2004.043182. [DOI] [PubMed] [Google Scholar]
- 26.Groot P.C., Bleeker M.J., Pronk J.C., Arwert F., Mager W.H., Planta R.J., Eriksson A.W., Frants R.R. The human alpha-amylase multigene family consists of haplotypes with variable numbers of genes. Genomics. 1989;5:29–42. doi: 10.1016/0888-7543(89)90083-9. [DOI] [PubMed] [Google Scholar]
- 27.Collier S., Sinnott P.J., Dyer P.A., Price D.A., Harris R., Strachan T. Pulsed field gel-electrophoresis identifies a high degree of variability in the number of tandem 21-hydroxylase and complement C-4 gene repeats in 21-hydroxylase deficiency haplotypes. EMBO J. 1989;8:1393–1402. doi: 10.1002/j.1460-2075.1989.tb03520.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes A.E., Orr N., Esfandiary H., Diaz-Torres M., Goodship T., Chakravarthy U. A common CFH haplotype, with deletion of CFHR1 and CFHR3, is associated with lower risk of age-related macular degeneration. Nature Genet. 2006;38:1173–1177. doi: 10.1038/ng1890. [DOI] [PubMed] [Google Scholar]
- 29.Lecce R., Murdolo M., Gelli G., Steindl K., Coppola L., Cupelli A.R.E., Neri G., Zollino M. The euchromatic 9p+polymorphism is a locus-specific amplification caused by repeated copies of a small DNA segment mapping within 9p12. Hum. Genet. 2006;118:760–766. doi: 10.1007/s00439-005-0085-x. [DOI] [PubMed] [Google Scholar]
- 30.Mars W.M., Patmasiriwat P., Maity T., Huff V., Weil M.M., Saunders G.F. Inheritance of unequal numbers of the genes encoding the human neutrophil defensins HP-1 and HP-3. J. Biol. Chem. 1995;270:30371–30376. doi: 10.1074/jbc.270.51.30371. [DOI] [PubMed] [Google Scholar]
- 31.Franchina M., Kay P.H. Allele-specific variation in the gene copy number of human cytosine 5-methyltransferase. Hum. Hered. 2000;50:112–117. doi: 10.1159/000022898. [DOI] [PubMed] [Google Scholar]
- 32.Aitman T.J., Dong R., Vyse T.J., Norsworthy P.J., Johnson M.D., Smith J., Mangion J., Roberton-Lowe C., Marshall A.J., Petretto E., et al. Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature. 2006;439:851–855. doi: 10.1038/nature04489. [DOI] [PubMed] [Google Scholar]
- 33.Board P., Coggan M., Johnston P., Ross V., Suzuki T., Webb G. Genetic-heterogeneity of the human glutathione transferases—a complex of gene families. Pharm. Therapeut. 1990;48:357–369. doi: 10.1016/0163-7258(90)90054-6. [DOI] [PubMed] [Google Scholar]
- 34.Brockmoller J., Gross D., Kerb R., Drakoulis N., Roots I. Correlation between trans-Stilbene oxide-glutathione conjugation activity and the deletion mutation in the glutathione-S-transferase class Mu gene detected by polymerase chain-reaction. Biochem. Pharmacol. 1992;43:647–650. doi: 10.1016/0006-2952(92)90591-6. [DOI] [PubMed] [Google Scholar]
- 35.van der Burg M., Barendregt B.H., van Gastel-Mol E.J., Tumkaya T., Langerak A.W., van Dongen J.J.M. Unraveling of the polymorphic C lambda 2-C lambda 3 amplification and the Ke(+)Oz(−) polymorphism in the human Ig lambda locus. J. Immunol. 2002;169:271–276. doi: 10.4049/jimmunol.169.1.271. [DOI] [PubMed] [Google Scholar]
- 36.Baris H., Bejjani B.A., Tan W.H., Coulter D.L., Martin J.A., Storm A.L., Burton B.K., Saitta S.C., Gajecka M., Ballif B.C., et al. Identification of a novel polymorphism - The duplication of the NPHP1 (nephronophthisis 1) gene. Am. J. Med. Genet. Part A. 2006;140A:1876–1879. doi: 10.1002/ajmg.a.31390. [DOI] [PubMed] [Google Scholar]
- 37.Deeb S.S., Alvarez A., Malkki M., Motulsky A.G. Molecular-patterns and sequence polymorphisms in the red and green visual pigment genes of Japanese men. Hum. Genet. 1995;95:501–506. doi: 10.1007/BF00223860. [DOI] [PubMed] [Google Scholar]
- 38.Evers M.P.J., Zelle B., Bebelman J.P., Vanbeusechem V., Kraakman L., Hoffer M.J.V., Pronk J.C., Mager W.H., Planta R.J., Eriksson A.W., et al. Nucleotide-sequence comparison of 5 human pepsinogen-a (Pga) genes—evolution of the Pga multigene family. Genomics. 1989;4:232–239. doi: 10.1016/0888-7543(89)90325-x. [DOI] [PubMed] [Google Scholar]
- 39.Birtle Z., Goodstadt L., Ponting C. Duplication and positive selection among hominin-specific PRAME genes. BMC Genomics. 2005;6:120. doi: 10.1186/1471-2164-6-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colin Y., Cherifzahar B., Vankim C.L., Raynal V., Vanhuffel V., Cartron J.P. Genetic-basis of the Rhd-positive and Rhd-negative blood-group polymorphism as determined by southern analysis. Blood. 1991;78:2747–2752. [PubMed] [Google Scholar]
- 41.Feldkotter M., Schwarzer V., Wirth R., Wienker T.F., Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time LightCycler PCR: Fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am. J. Hum. Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lyons K.M., Stein J.H., Smithies O. Length polymorphisms in human proline-rich protein genes generated by intragenic unequal crossing over. Genetics. 1988;120:267–278. doi: 10.1093/genetics/120.1.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson W., III, Pardo-Manuel de Villena F., Lyn-Cook B.D., Chatterjee P.K., Bell T.A., Detwiler D.A., Gilmore R.C., Valladeras I.C., Wright C.C., Threadgill D.W., et al. Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics. 2004;84:707–714. doi: 10.1016/j.ygeno.2004.06.011. [DOI] [PubMed] [Google Scholar]