Abstract
Controlling the size and shape of DNA condensates is important in vivo and for the improvement of nonviral gene delivery. Here, we demonstrate that the morphology of DNA condensates, formed under a variety of conditions, is shifted completely from toroids to rods if the bacterial protein HU is present during condensation. HU is a non-sequence-specific DNA binding protein that sharply bends DNA, but alone does not condense DNA into densely packed particles. Less than one HU dimer per 225 bp of DNA is sufficient to completely control condensate morphology when DNA is condensed by spermidine. We propose that rods are favored in the presence of HU because rods contain sharply bent DNA, whereas toroids contain only smoothly bent DNA. The results presented illustrate the utility of naturally derived proteins for controlling the shape of DNA condensates formed in vitro. HU is a highly conserved protein in bacteria that is implicated in the compaction and shaping of nucleoid structure. However, the exact role of HU in chromosome compaction is not well understood. Our demonstration that HU governs DNA condensation in vitro also suggests a mechanism by which HU could act as an architectural protein for bacterial chromosome compaction and organization in vivo.
INTRODUCTION
Multivalent cations and molecular crowding agents can cause DNA to collapse from solution into well-defined nanometer-scale particles (1–5). This phenomenon of DNA condensation has been studied for many years as a model of high-density DNA packing in living systems, particularly in sperm cells and viruses (6–8). More recently, efforts to enhance artificial gene delivery for the improvement of gene therapies have generated substantial interest in the development of methods to control the size and shape of DNA condensates (9–11). The principal morphologies of DNA condensates formed in vitro are toroids, rods and spheroids. Toroidal condensates are the predominant morphology observed under many experimental conditions, and have historically received the most attention.
We have recently demonstrated that the size of toroidal DNA condensates can be controlled by solution conditions and by static DNA loops that act as nucleation sites for toroid formation. Such loops can significantly decrease the average diameter of toroidal condensates (12–14). Monovalent and divalent salt concentrations have also been shown to affect the size and thickness of DNA toroids (13,14). In contrast, controlling the morphology of DNA condensates between toroids and rods is largely an unexplored problem. Bloomfield and co-workers have demonstrated that condensation of DNA with me8-spermidine (a derivative of spermidine with methylated amines), or with other condensing agents in water–alcohol mixtures, increases the population of rod-like condensates (4,15–17). However, a general method for obtaining complete control over condensate morphology for a range of condensing agent structure and solution conditions has not been previously reported.
We have hypothesized that some natural proteins active in DNA condensation could be adapted to gain further control over DNA condensation in vitro. Protamines, which package DNA in vertebrate sperm cells, are obvious candidates because sperm cell chromatin represents one of the most highly condensed forms of DNA found in nature (18–21). The DNA-condensing properties of protamines have been the subject of numerous investigations (7,21–24), and have been used for artificial gene delivery (25–27). Our laboratory has recently demonstrated that cysteine-rich mammalian protamines readily condense DNA into spherical particles that are salt-stable (28). Nevertheless, sperm cell proteins represent only one possible class of proteins to aid the control of DNA condensation in vitro.
Prokaryotic cells contain a set of proteins that bind DNA and are associated with the nucleoid. Among these proteins, HU has been characterized as the most important protein for structural organization of the bacterial chromosome. For example, the lack of HU in the hupAhupB mutant results in decondensed nucleoids and anucleate cells (29–32). In E. coli, HU exits predominantly as a 20-kDa heterodimer composed of two subunits, HUα and HUβ, encoded by the hupA and HUpB genes. While HU was initially characterized as the bacterial equivalent of histones, more recent studies have revealed that the main role of HU in a number of cellular processes is that of a DNA architectural protein that bends DNA in a non-sequence-specific manner (33–37). Thus, HU appears to be more similar in function to the HMG proteins of eukaryotes than it is to histones (32, 38–40). The structure of Anabaena HU in a co-crystal with DNA revealed that conserved prolines of HU intercalate between the bases of DNA from the minor groove to induce pronounced kinks in the double helix (41). The crystal structure reveals that the HU-induced bend angle in DNA is as great as 105° to 140° (41). However, these HU–DNA co-crystals contained DNA with both mismatched and unpaired bases. Solution state studies with un-nicked DNA support a bend angle of approximately 60° (42,43). Measurements of DNA bending by HU also vary because HU–DNA complexes form flexible hinges that can accommodate a range of different bend angles (41,44,45).
In the present study, we have investigated the effects of HU on the process of DNA condensation in vitro. HU does not, by itself, cause DNA to condense into high-density particles. Furthermore, HU has been shown previously by in vitro assays to only moderately decrease the concentration of macromolecular crowding agents required to condense DNA (46,47). These previous experiments did not explore the potential role of HU in shaping the morphology of condensed DNA. Here, we report that HU causes a substantial change in the preferred morphology of DNA condensates formed upon the addition of molecular crowding agents or polyamines. The results presented provide additional support that local alterations in nucleic acid structure can be used to control both the size and morphology of DNA condensates (12,13,48). To the best of our knowledge, this is the first report of controlling DNA condensation with a protein that does not by itself condense DNA into high-density condensates. The results presented here are also suggestive of how HU may work in some capacity as an architectural protein in the compaction of the bacterial chromosome in vivo.
MATERIALS AND METHODS
DNA preparation
Bluescript II SK- (Stratagene) plasmid was isolated from the E. coli strain DH5α (Life Technologies) using the Qiagen maxi-prep kit (Valencia, CA), and linearized by digestion with the restriction endonuclease HindIII (New England Biolabs). Following enzymatic digestion, the DNA was rinsed at least five times with 1× TE (10 mM Tris, 1 mM EDTA, pH 7.8) using a Microcon-YM 30 spin column (Millipore) to remove salts and buffers from the restriction digest. The DNA was finally resuspended from the spin column in 1× TE. DNA concentration was verified spectrophotometrically. Bluescript II SK− plasmid is abbreviated as “linear DNA” throughout the text. Supercoiled Bluescript II SK− plasmid, obtained directly from the plasmid isolation procedure, was determined to be more than 90% supercoiled based on agarose gel electrophoresis analysis. Supercoiled DNA was also rinsed at least five times with 1× TE to ensure that the buffer conditions of all DNA stock samples were identical.
HU protein
HU protein was isolated and purified from the E. coli strain RLM1078 following a procedure previously described by Wojtuszewski et al. (49). To eliminate a contaminating nuclease, isolated HU protein was further purified on an FPLC MonoS 5/5 or 10/10 cation exchange column (Amersham-Pharmacia). The column was developed with a 0.05 to 1.0 M linear NaCl gradient and HU eluted at 0.35 M NaCl. The lack of nuclease activity was verified by the absence of digested products after incubating plasmid DNA with the protein. The extinction coefficient at 230 nm of 37.5 mM−1cm−1 was used to calculate HU protein concentration (49).
Preparation of DNA condensates
PEG-induced DNA condensates were prepared by mixing solutions of DNA and PEG 8000 to yield a final reaction mixture 5 µM DNA bp (given in units of base pair throughout), 125 mg/ml PEG 8000, 50 mM Tris-HCl (pH 7.8), 1 mM EDTA, 100 mM NaCl. The condensate reaction mixtures were allowed to equilibrate at room temperature for 20 min before depositing on grids. For PEG-induced condensates prepared in the presence of HU, HU was introduced to the DNA at three different points in the condensation process: (1) DNA was condensed with solutions of PEG containing HU for 20 min (HU during condensation); (2) DNA was incubated with HU for 10 min and then condensed with PEG for 10 min (HU before condensation); (3) HU was incubated for 10 min after DNA had been condensed by PEG for 10 min (HU after condensation). In all cases, the reaction mixtures were allowed to equilibrate at room temperature for 20 min before depositing on grids, and final condensate solutions were all 5 µM DNA bp, 125 mg/ml PEG 8000, 50 mM Tris-HCl (pH 7.8), 1 mM EDTA, 100 mM NaCl.
Spermidine-induced condensates were prepared by mixing solutions of DNA and spermidine to yield a condensation reaction mixture of 5 µM DNA, 700 µM spermidine, 0.33 × TE (pH 7.8), 15 mM NaCl. The same protocol was followed for the preparation of spermine-induced condensates, in which DNA was mixed with spermine to yield a condensation reaction mixture of 5 µM DNA, 15 µM spermine, 0.33 × TE (pH 7.8), 15 mM NaCl. For spermidine and spermine DNA condensation in the presence of HU, the same three protocols were followed as described above for the condensation of DNA by PEG in the presence of HU (i.e. HU before, during and after condensation).
Electron microscopy and analysis of DNA condensates
A 5 µl aliquot of each DNA condensate reaction mixture was deposited directly onto a carbon-coated electron microscopy grid (Ted Pella, Redding, CA). After allowing condensates to settle onto the grid for 15 min, 2% uranyl acetate was added to the grid for 2 min, and then the grid was rinsed in 95% ethanol and air-dried. The condensates prepared with PEG 8000 were rinsed in 20% ethanol to reduce the deposition of PEG aggregates. The size and morphology of DNA condensates were examined using a JEOL-100C transmission electron microscope (TEM). To obtain the relative toroid and rod populations in each sample, the grid surface was randomly scanned and the number of unaggregated toroids and rods visible on the viewing screen were counted. Several hundred structures were counted for each grid. Each measurement reported is the average of the counts from three different grid preparations. Images were acquired at 100, 000× magnification, and film negatives were scanned into digital format at 300 pixels/in. A computer graphics program was used to measure the dimensions of individual condensates for each sample.
RESULTS
HU governs the morphology of condensates formed under molecular crowding conditions
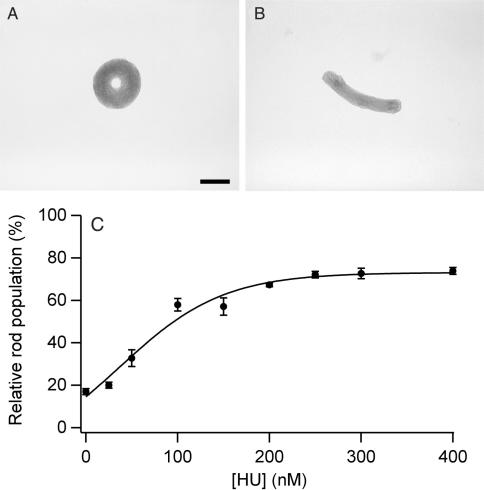
The addition of PEG 8000 to a sample of linear DNA at physiological ionic strength causes the condensation of DNA into toroidal and rod-like particles (Figure 1A, B). The mean outer diameter of toroids was 199 nm (σ, ±94 nm) with a mean thickness of 71 nm (σ, ±39 nm). The average length of rods was 386 nm (σ, ±81 nm) with a mean thickness of 95 nm (σ, ±27 nm). The relative populations of rods and toroids measured under the PEG-induced condensation conditions were 83% toroids and 17% rods.
Figure 1.
PEG-induced DNA condensate morphologies and morphology statistics as a function of HU concentration. (A) TEM image of a representative condensate of linear DNA condensed by PEG 8000 (no HU present). (B) TEM image of a representative condensate produced under identical conditions as in A, except in the presence of 200 nM HU. Scale bar is 100 nm. (C) Relative rod populations versus HU concentration for linear DNA condensed by PEG. Samples were 5 µM DNA bp, 125 mg/ml PEG 8000, 50 mM Tris-HCl, 1 mM EDTA (pH 7.8), 100 mM NaCl, and indicated concentrations of HU dimer. Each rod population measurement reported is the average of counts from three different EM grid preparations.
When the protein HU was added along with PEG to DNA, a definitive shift was observed in condensate morphology from toroids to rods as a function of HU concentration (Figure 1C). A plateau in relative rod population was reached at around 250 nM HU (in units of HU dimer). At this HU concentration, and up to at least 400 nM HU, rods represent 75% of the condensates tallied. The midpoint between the initial rod population and that of the plateau occurs at an HU concentration of 80 nM. Given that the DNA concentration for these experiments was 5 µM in bp, and that the binding site for HU is one HU dimer per 9 DNA bp, the concentration of HU required to cause the observed change in condensate morphology is less than what would be required to fully load the DNA (about 560 nM HU). The number of HU molecules necessary to control DNA condensate morphology was actually found to be significantly less than full loading (vide infra).
The addition of HU alone to DNA (i.e. in the absence of PEG) was not observed to condense DNA into densely packed particles, even up to an HU concentration of 400 nM. Furthermore, the DNA toroids and rods produced by PEG in the presence of HU are similar in size to those produced by PEG alone. Thus, HU apparently functions primarily as an architectural protein in condensation reactions rather than as either a strong protagonist or an antagonist of DNA condensation, at least for the range of HU concentrations studied here.
We note that PEG 8000 (without DNA or HU) was observed to form globule structures under the same solution conditions and EM grid preparation procedure (Materials and Methods). PEG 8000 did not produce particles with toroidal or rod-like morphologies. Thus, the statistics reported for PEG-induced DNA condensate morphology accurately represent the relative populations of toroids and rods. However, it cannot be ruled out that the globule structures observed when DNA is condensed by PEG do not contain any condensed DNA. This caveat does not apply to the other condensation protocols presented below, because the polyamine condensing agents did not produce any particles on the EM grids when DNA was not present.
HU governs the morphology of spermidine-DNA condensates
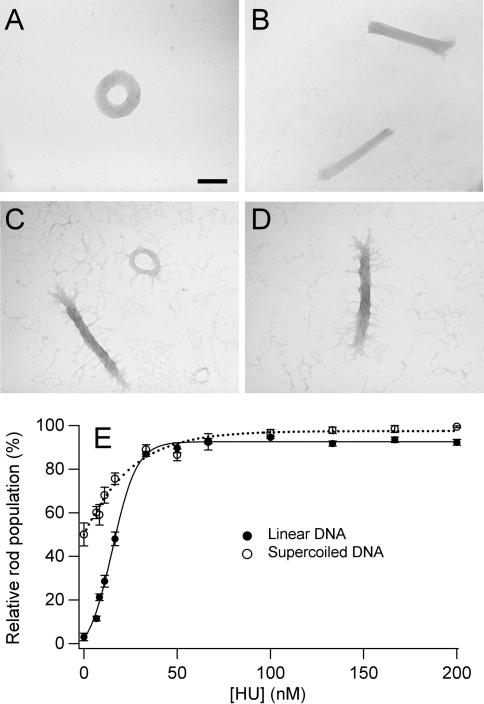
When linear DNA of 3 kb or greater length is condensed from solution by a wide variety of cationic molecules (e.g. polyamines, poly-lysine, protamines), the resulting condensate particles are mostly toroids, with the remaining particles being almost entirely rods (1,2,4,5,23,50,51). Under our experimental conditions, spermidine condensed linear 3 kb DNA into 97% toroids and only 3% rods. To elucidate the effect of HU on DNA condensation by polyamines, the morphology of spermidine-induced condensates was examined as a function of HU concentration. As shown in Figure 2, HU also causes a significant increase in the population of rod-like condensates for linear DNA (5 µM bp) when condensed by spermidine (700 µM). Specifically, the relative population of rod-like condensates increases from 3% rods (in the absence of HU) to greater than 90% rods in the presence of 50 nM HU, with the half maximum rod population being observed at 15 nM HU (Figure 2C).
Figure 2.
Spermidine-induced DNA condensate morphologies and morphology statistics as a function of HU concentration. (A) TEM image of a representative condensate of linear DNA condensed by spermidine (no HU present). (B) TEM image of representative condensates produced under identical conditions as in A, except in the presence of 50 nM HU. (C) TEM image of representative condensates of supercoiled DNA condensed by spermidine (no HU present). (D) TEM image of representative condensates produced under identical conditions as in C, except in the presence of 50 nM HU. Scale bar is 100 nm. (E) Relative rod populations versus HU concentration for linear and supercoiled DNA condensed by spermidine. Samples were 5 µM DNA bp, 700 µM spermidine chloride, 0.33 × TE (pH 7.8), 15 mM NaCl, and indicated concentrations of HU dimer.
DNA toroids produced by spermidine-induced condensation were of similar dimensions as those produced by PEG-induced condensation, with a mean outside diameter of 250 nm (σ, ±22 nm) and a mean thickness of 82 nm (σ, ±12 nm), but with smaller standard deviations in these dimensions. DNA rods produced by spermidine-induced condensation in the presence of HU exhibited an overall mean length of 450 nm and a mean width of 78 nm for all HU concentrations investigated. Rod length and thickness proved to be relatively insensitive to HU concentration. For example, rods formed in the presence 100 nM HU and 200 nM HU were the same size within experimental variation, i.e. mean rod length of 458 nm (σ, ±46 nm) vs. 450 nm (σ, ±61 nm), and mean rod thickness of 76 nm (σ, ± 4 nm) vs. 82 nm (σ, ±16 nm), respectively. We note that these mean thicknesses are also the same, within experimental error, for toroids and rods observed in absence and presence of HU. These observations demonstrate that HU does not significantly affect the dimensions of condensates produced by spermidine, only the relative population of rods. Thus, HU can act as a guide for DNA condensate morphology for both crowding-induced and polyamine-induced condensations. We note that HU causes a similar morphology shift for condensates formed by another commonly used tri-cationic DNA condensing agent, hexammine cobalt (III) (data not shown).
HU and supercoiling work together to promote the formation of rod-like condensates
It is known that supercoiling of DNA can provide high-affinity binding sites for HU (52). To explore the possibility that HU can work in conjunction with supercoiling to govern DNA condensate morphology, we investigated the condensation of supercoiled DNA as a function of HU concentration. For these particular studies, we chose to use spermidine-induced condensation because, as mentioned above, the statistics for DNA condensate morphology were potentially more accurate than those obtained for PEG-induced condensation. Additionally, the dimensions of rods and toroids were more uniform for the spermidine condensation reaction, which arguably makes rod versus toroid population statistics more relevant with regards to the amount of DNA condensed into each of these two morphologies.
The condensation of supercoiled DNA by spermidine (in the absence of HU) produces a greater population of rods than linear DNA, i.e. 50% vs. 3%, respectively (Figure 2). The addition of HU along with spermidine to supercoiled DNA again resulted in a concentration-dependent increase in the population of rods (Figure 2C). We observed that this combination of HU and superhelical stress increases the population of DNA rods to at least 99% at 100 nM HU (5 µM DNA bp; 700 µM spermidine), which is greater than the maximum rod population observed with linear DNA. The half maximum rod population was observed at 15 nM HU dimer, which is similar to that observed for linear DNA condensed by spermidine. Thus, HU and superhelical stress apparently work together to increase the population of rod-like condensates, and the effect of HU on DNA condensation is not fundamentally different for linear versus supercoiled DNA.
The role of HU in controlling DNA condensate morphology, rather than as a true condensing agent, is again illustrated by the similar condensate structures observed for supercoiled DNA in the absence and presence of HU. Under the conditions of our study, the rod-like condensates of supercoiled DNA condensed by spermidine exhibited thin fibril structures extending out from the main mass of the condensate (Figure 2C, D). We have previously shown that such structures indicate the existence of partially condensed DNA that collapses into fibrils upon preparation of EM grids (14). Slight changes in condensation conditions (e.g. ionic strength, temperature, sample dilution) can cause the appearance or disappearance of such fibrils (14). Thus, the coexistence of these fibrils on DNA condensates prepared in samples that only differ by the presence of HU is another strong indication that HU does not significantly promote or hinder DNA condensation at the protein concentrations used in this study. We note that fibrils extending from DNA condensates are not particular to supercoiled DNA, but are also observed for condensed linear DNA, depending on specific sample conditions (14).
HU governs the morphology of spermine–DNA condensates
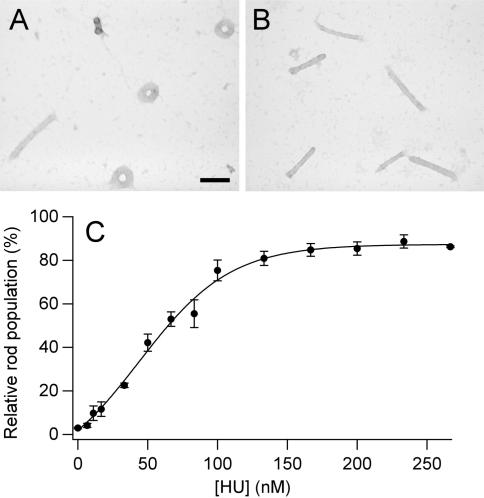
Condensation reactions similar to those described above were also performed with linear DNA and the tetracation spermine to further explore the possibility that guiding DNA condensation is a general property of HU, regardless of the condensing agent. Similar to spermidine, when linear DNA was condensed by spermine, the majority of particles formed were well-defined toroids with a minor population of rods (97% toroids, 3% rods) (Figure 3).
Figure 3.
Spermine-induced DNA condensate morphologies and morphology statistics as a function of HU concentration. (A) TEM image of representative condensates of linear DNA condensed by spermine (no HU present). (B) TEM image of representative condensates produced under identical conditions as that shown in A, except in the presence of 100 nM HU. Scale bar is 100 nm. (C) Relative rod populations plotted as a function of HU concentration for linear DNA condensed by spermine. Samples were 5 µM DNA bp, 15 µM spermine chloride, 0.33 × TE (pH 7.8), 15 mM NaCl and indicated concentrations of HU dimer.
DNA condensed by spermine also exhibited a gradual increase in the percentage of rod-like condensates as a function of HU (Figure 3C). A maximum plateau for rod population of approximately 90% was observed near 200 nM HU, with the half maximum population observed at 60 nM HU. Thus, HU is somewhat less effective in controlling the condensates formed by spermine in comparison to spermidine (a polyamine with one less charge).
In contrast to the condensates formed in the presence of PEG or spermidine, the condensates formed by spermine are considerably smaller (Figure 3A, B). Toroids had a mean outside diameter of 102 nm (σ, ±12 nm) and a mean thickness of 34 nm (σ, ±6 nm). The overall mean length and thickness of spermine–DNA rods, 215 nm and 28 nm, respectively, for all samples were also considerably lesser than those produced by PEG or spermidine. These condensate dimensions did not change significantly with increasing HU concentration. For example, at 100 nM HU the mean rod length was 221 nm (σ, ±32 nm) and the mean rod thickness was 26 nm (σ, ±5 nm), compared to 212 nm (σ, ±23 nm) and 29 nm (σ, ±5 nm), respectively, at 200 nM HU. The similar dimensions for spermine–DNA condensates formed in the absence and presence of HU again demonstrates the general ability of HU to guide DNA condensate morphology without significantly altering condensate size. The small change observed in condensate size with increasing HU concentration is even less significant when one considers the much greater difference in rod lengths that are associated with different condensing agents.
How many HU proteins are necessary to guide the condensation of a DNA molecule?
DNA condensed by spermidine appeared to be the most practical system among those presented here from which to determine the minimum number of HU proteins necessary to guide DNA condensation. A series of condensation experiments was conducted, in which the concentration of linear DNA was increased above 5 µM bp (i.e. the concentration used in all other experiments). For these experiments, HU concentration was fixed at 67 nM and spermidine concentration at 700 µM. This concentration of HU was chosen because it represents a point at which the rod population was within the plateau region (Figure 2C). At the initial concentrations of 5 µM DNA bp and 67 nM HU, there is the potential to bind at most one HU dimer per 75 bp. Our analysis by EM did not reveal any reduction in rod population when DNA concentration was increased to 15 µM bp, at which point there could be no more than one HU dimer bound per 225 bp. In a condensation reaction where the DNA concentration was increased to 20 µM bp (i.e. 1 HU dimer per 300 bp), the rod population exhibited the first appreciable decrease to approximately 80%. However, at this concentration of DNA, the condensates began to aggregate, which did not allow for the collection of precise morphology statistics. At DNA concentrations higher than 20 µM bp, condensate aggregation became even worse. Thus, within the limits of these experiments, our results demonstrate that at most one HU dimer is required per 225 bp to guide spermidine–DNA condensates into rod-like structures. This ratio could be smaller, as this estimate assumes that all HU is bound to DNA that is condensed in the rod-like condensates.
For DNA rods 450 nm in length, one 180° bend occurs in the DNA helix approximately every 1300 bp. It is known that HU binds more tightly to bent DNA (42,53,54), so it is reasonable to hypothesize that HU proteins would be localized at the end regions of rods. As mentioned above, HU can stabilize DNA bend angles that range from 60 to 140° (41–45). Thus, at most three HU dimers would be expected to localize at each DNA bend within a spermidine–DNA rod. This estimate translates to a maximum of one HU dimer per 430 bp. Our determination that not more than one HU dimer is required per 225 bp is within a factor of two of this simple theoretical estimate. Our observation that rod populations begin to decrease at a ratio of 1 HU per 300 bp (at 67 nM HU) is also in reasonable agreement with our estimate for the maximum number of HU dimers bound per rod.
Our determination that 67 nM HU will guide the condensation of DNA by spermidine into almost exclusively rod-like structures at a ratio of 1 HU dimer per 225 DNA bp also has important implications regarding the observed initial increase in rod population as a function of HU concentrations below 50 nM HU (Figure 2C). In particular, the same ratio of HU to DNA bp does not result in the formation of more than 66% rods for a DNA concentration of 5 µM bp and an HU concentration of 22 nM (Figure 2C). Thus, for DNA samples 5 µM in bp and HU concentrations below approximately 50 nM, the ability for HU to guide DNA condensation is not simply limited by the number of HU molecules present in the sample but apparently by the number of HU molecules actually bound to DNA (i.e. association constant limited). This conclusion is also supported by a set of experiments in which relative rod populations were measured for HU concentrations from 0 to 50 nM with a reduced DNA concentration of 2.5 µM bp. Rod populations were found to be the same as those measured for DNA at 5 µM bp (data not shown), which also indicates that rod populations are governed by the HU–DNA association constant for HU concentrations below 50 nM, rather than simply the ratio of HU to DNA present in the samples.
The HU–DNA disassociation constant (Kd) at the ionic strength of the condensation experiments carried out with spermidine was previously determined to be 400 nM (55). However, it is not currently possible to establish a complete equilibrium description of HU association with DNA in the presence of spermidine, as the possibility exists that HU binds more tightly to the sharp bends within a rod-like condensate than free DNA in solution.
Order of HU addition affects condensation morphology statistics
We have previously shown that DNA condensate size and morphology is affected differently by chemical agents that alter DNA structure if these agents are added to DNA before, during (i.e. coincident) or after the addition of a condensing agent (14). For example, order-of-addition studies provided important clues regarding how Mg2+ (which promotes helix–helix contacts) influences the different stages of DNA condensation (i.e. nucleation, proto-toroid formation and equilibrium growth) (14). The results presented above that demonstrate the effects of HU on DNA condensation were from experiments in which HU was added to DNA coincident with the condensing agents. As a means to gain insight into the stages at which HU controls the morphology of DNA condensates, we have also performed experiments in which HU was added before or after the addition of each condensing agent.
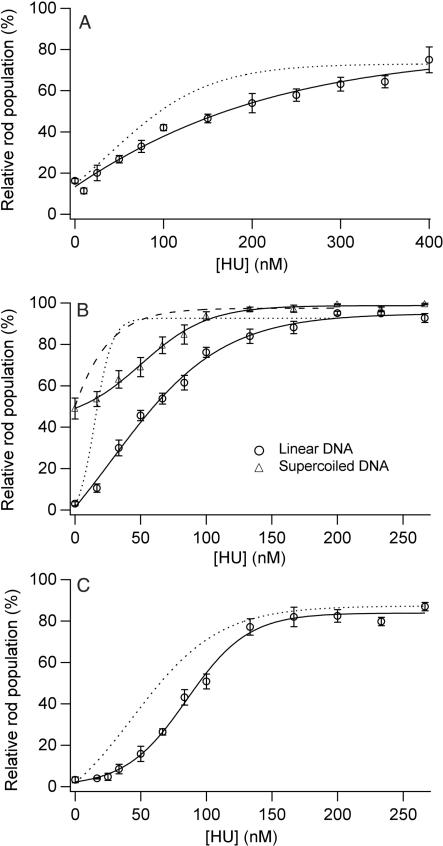
When HU was incubated with DNA prior to condensing agent addition, an increase in rod population was also observed for condensation by PEG/NaCl, spermidine and spermine (Figure 4). However, for all three condensing conditions, and for supercoiled DNA, a higher concentration of HU was required to achieve the same rod population measured when HU was added coincident with the condensing agent (Figure 4). The addition of HU to a PEG–DNA solution after condensation resulted in an even lower percentage of rod-like condensates. For example, an HU concentration of 400 nM resulted in an increase in rod population to 44% (data not shown), as compared to the 70% rod population observed when 400 nM HU was added before or during condensation by PEG (Figure 4). For condensates prepared with spermidine or spermine in the absence of HU, no apparent increase in rod population was observed when HU was added up to a concentration of 400 nM to solutions containing the pre-formed DNA–polyamine condensates.
Figure 4.
Condensate morphology statistics versus HU concentration for reactions with HU added to DNA before condensation. (A) Relative rod populations versus HU concentration for linear DNA condensed by PEG. Samples were 5 µM DNA bp, 125 mg/ml PEG 8000, 100 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA (pH 7.8), and indicated concentration of HU dimer. (B) Relative rod populations versus HU concentration for linear (circle) and supercoiled DNA (triangle) condensed by spermidine. Samples were 5 µM DNA bp, 700 µM spermidine chloride, 0.33 × TE (pH 7.8), 15 mM NaCl and indicated concentrations of HU dimer. (C) Relative rod populations versus HU concentration for linear DNA condensed by spermine. Samples were 5 µM DNA bp, 15 µM spermine chloride, 0.33 × TE (pH 7.8), 15 mM NaCl and indicated concentrations of HU dimer. Dashed curves are best-fit curves from rod populations measured for corresponding experiments in which HU was added coincident with the condensing agent (see Figures 1–3 for details).
The difference in rod populations observed for experiments in which the same concentration of HU was added before or coincident with condensation versus after condensation clearly demonstrates that HU influences condensate morphology during the process of DNA particle formation (i.e. nucleation and initial growth). On the other hand, the ability of HU to significantly increase the population of rods formed in the presence of PEG after condensation has occurred demonstrates that the ability of HU to increase the relative populations of rod-like condensates is also thermodynamic in nature. We note that the addition of HU to DNA condensed by spermidine and spermine caused a substantial increase in condensate aggregation, which may have limited DNA rearrangement into rods (data not shown).
DISCUSSION
A model for how HU guides DNA condensation in vitro
The coexistence of rods and toroids as products of in vitro DNA condensation reactions reflects the nearly equivalent energetics of DNA packing within these two distinct morphologies, including the energy required for the smooth bending of DNA within toroids versus that required to produce sharp bends at the ends of rods (16,56,57). When DNA is condensed in the presence of high concentrations of alcohol, or condensing agents with hydrophobic groups (e.g. permethylated spermidine), rod populations increase with respect to toroid populations (15,17,58–60). Under such conditions, hydrophobic groups of the solvent or condensing agent interact favorably with unstacked DNA bases. These interactions lower the free energy penalty associated with sharp bending at the ends of rods, which renders rod formation more energetically favorable (15,17,61). The higher population of rods observed with supercoiled DNA versus linear DNA can be attributed to torsional strain, which also makes DNA more prone to the formation of sharp bends (62,63). These correlations between the promotion of sharp bends and increased rod populations inspired us to investigate the possibility that DNA-bending proteins could be used to control the morphology of DNA condensates.
To understand the origin of the increase in rod populations observed in the presence of HU, we must consider both what is known about HU binding to DNA and the state of DNA within rods and toroids. DNA condensed into a rod exists in two states, in a sharply bent state in the end regions, and presumably unbent state in the linear region between the two ends. HU has been shown to preferentially bind pre-bent DNA, and to bend linear DNA (42,43,53,54). Therefore, HU binding is expected at the ends of rods, where binding would increase rod stability, and not in the linear regions, at least not at the HU concentrations used in the present study. DNA condensed within toroids is smoothly bent over a radius of curvature that is much greater than that of the sharp bends induced by HU (64). Thus, HU would not be expected to stabilize toroids, and may even suppress toroid formation, by introducing bends that are incompatible with DNA packing within a toroid.
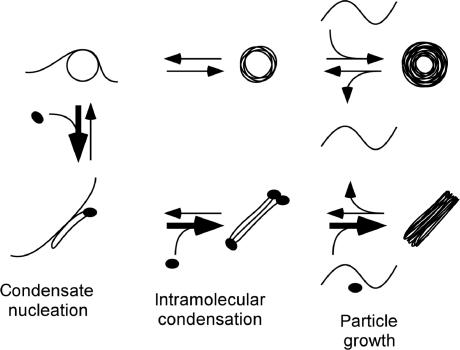
DNA condensation in vitro is a nucleation–growth process that includes rod/toroid nucleation, proto-rod/proto-toroid formation (i.e. intramolecular condensation) and particle growth by intermolecular condensation (57). In Figure 5, we present a schematic diagram of this process for DNA condensed in the presence of HU. Steps involving rod nucleation and growth are indicated by bold arrows as being more favorable in the presence of HU, as our data indicates that DNA condensation steps under both kinetic and thermodynamic control are more favorable towards rod formation in the presence of HU.
Figure 5.
A model for how HU affects the process of DNA condensation in which rods and toroids are formed. The three stages of DNA condensation in vitro, as described in text, are: rod/toroid nucleation; proto-rod/proto-toroid formation (intramolecular condensation); and condensate growth (intermolecular condensation), which includes strand exchange between condensates (under some conditions). Bold arrows indicate steps that apparently become more favorable in the presence of HU. Black ellipsoids represent HU molecules.
The apparent kinetic advantage provided to rods by HU can be understood in terms of how HU binding would preferentially promote rod nucleation. The nucleation structure for rods has not been investigated by experiment as it has been for toroids (12,13). However, Langevin dynamics simulations of DNA condensation indicate that rods are also nucleated by DNA loops, with toroids being nucleated by loops with an obtuse internal contact angle and rods being nucleated by loops with an acute internal contact angle (56). We propose that the binding of HU to either type of DNA loop would increase the probability for the loop to collapse on itself (Figure 5), forming a condensed oval structure that would nucleate rod formation. This proposed path to rod nucleation would simultaneously increase the probability of rod nucleation and reduce the probability of toroid nucleation. Creation of rod nucleation sites, by this or an alternative route, can also be viewed as a reduction in the activation energy required for rod nucleation as a result of DNA binding and bending by HU.
The intramolecular condensation of a single 3 kb DNA molecule gives rise to a proto-rod or a proto-toroid, depending on the nucleation structure (Figure 5). HU binding is again expected to be most favored at the ends of a proto-rod, which would increase the stability of the proto-rod. HU-stabilized proto-rods would then be more likely to grow into full-size rods by the addition of DNA from solution (Figure 5), versus proto-rods in the absence of HU. Finally, the binding of HU to DNA within a rod at any stage of growth would also be expected to provide additional stability to the rod structure versus DNA condensed into a toroid, and thereby provide a thermodynamic advantage to rods under equilibrium conditions (i.e. after the kinetically controlled stages of condensation).
While the model described above explains our observation that rod populations increase in the presence of HU, this model does not explain the reduced effectiveness of HU in promoting rod formation when HU is added prior to a DNA-condensing agent, as compared to coincident addition. However, this observation can be understood considering what is known about the bound lifetime of HU on DNA versus the timescale of DNA condensation. HU–DNA complexes have a dissociation half-life of 0.6 min and 6.3 min in 50 mM NaCl and 5 mM NaCl, respectively (65). The initial stages of DNA condensation, including nucleation and intramolecular collapse of a DNA strand, are complete within milliseconds (66,67). Thus, HU molecules preassociated with DNA may not be able to completely redistribute within the time frame of condensate formation to maximize their influence on the nucleation of rods or the stabilization of proto-rods. In contrast, HU added coincident with a condensing agent would be more efficient in guiding condensate structure if the on-rate of binding to DNA is faster for bent DNA, in which case HU would preferentially bind at the ends of nucleated rods rather than in the linear region of a rod or along a DNA strand that has nucleated a toroid.
We have observed that the relative rod population of DNA condensates formed in the presence of PEG/NaCl increases when HU is added even after condensation has taken place. In contrast, HU did not significantly increase rod populations when added after DNA was condensed by spermidine or spermine. We have previously presented evidence that the conversion between fully formed toroids and rods takes place through the exchange of condensed DNA with solution (57). The ability for HU to cause the conversion of toroids formed in the presence of PEG/NaCl into rods suggests that appreciable strand exchange occurs between toroids and solution after condensation is complete. In contrast, our observation that HU does not promote rod formation after condensation by spermidine or spermine suggests that strand exchange in these preparations is minimal, and that the observed effects of HU on spermidine and spermine condensate morphology occurs during the earlier stages of condensation (i.e. nucleation, proto-structure formation and initial growth). Nevertheless, it is likely that HU can alter the morphology of condensates formed by polyamines under equilibrium conditions if conditions are used that allow DNA strand exchange with solution after initial condensation (e.g. higher monovalent ionic strength). This possibility is currently being explored. We also note that the amount of HU bound to DNA within HU-induced rods has not been determined directly for any of the condensate preparations reported here. It is possible that less HU may be necessary to maintain the rod morphology than is initially required to guide the condensation of DNA into rods. Experimental conditions may even exist for which it is possible to completely remove HU from rods without reverting to alternative condensate morphologies.
Implications regarding the functionality of HU in DNA condensation
Our present study of controlling DNA condensation with the protein HU also suggests a possible functionality of this nucleoid-associated protein within bacteria cells. Chromosomal DNA in bacteria is condensed into the nucleoid by the combined effects of DNA supercoiling, nucleoid-associated proteins, cellular macromolecular crowding effects and polyamines (68–70). Although HU is one of the most abundant nucleoid-associated proteins and has often been referred to as a ‘histone-like’ protein, the role of HU in condensing the bacterial chromosome is not understood (33,43,45,71–74). Our use of spermidine (one of the major polyamines in bacteria) and PEG (a macromolecular crowding agent) is clearly far too simplistic to be considered a reasonable model of the bacterial cytoplasm. Nevertheless, we have demonstrated that HU guides the condensation of DNA into structures with linear bundles (i.e. rods) when DNA is condensed by two very different solution conditions, polyamines and a crowding environment. Thus, the ability to control DNA condensate morphology appears to be an intrinsic property of HU. These combined results indicate that HU is much more effective in controlling the morphology of DNA condensation than it is in promoting DNA condensation, which may reflect how HU plays an architectural role in the condensation of bacterial chromosomal DNA.
Azam et al. have estimated that 30,000 HU dimers exist per E. coli cell during the exponential growth phase (75). If HU is evenly distributed throughout the chromosomal DNA of an E. coli cell in this phase, then HU loading of DNA would be on average only 1 HU dimer per ∼300 to 400 bp (75). Our results demonstrate that HU can act as an architectural protein for guiding DNA condensing at such low loading levels (≤1 HU dimer per 225 bp). As HU is believed to be somewhat evenly distributed throughout the bacterial chromosome, our experimental evidence of the architectural role of HU in guiding DNA condensation is definitely of physiological relevance (76).
Mutational and biochemical analyses have implicated HU as a determinant protein in packaging the bacterial chromosome (29–32,77). Zimmerman and coworkers demonstrated that HU reduces the concentration of crowding agents (i.e. PEG 8000, albumin) required to condense DNA in vitro (46,47). However, more recent studies question the exact role of HU in chromosome condensation. Single molecule investigations of HU binding to DNA have revealed that HU has a dual mode of binding. At low HU-to-DNA ratios, (e.g. less than 1 HU dimer per 150 bp), HU-induced bends decrease the persistence length of DNA (43,45,73). In contrast, at high HU-to-DNA ratios (e.g. 1 HU dimer per 9 bp), HU actually increases the stiffness of DNA (43,45,72,73). While the high-loading mode of HU is intriguing, it is still unknown why such a mode exists for a protein that is believed to facilitate DNA compaction. In the present study, we have observed a dramatic effect of HU on DNA condensation at much lower HU concentrations and at much lower DNA loading levels than those of the above-mentioned studies.
We propose that HU can function in vivo as an architectural protein during chromosome condensation. Over the range of HU concentrations we have investigated, HU primarily functions as an architectural protein but not as an antagonist to DNA condensation, as has been suggested in recent reports (45,68,72,73,78). We hypothesize that HU could locally organize bacterial chromosome DNA in the presence of polyamines and a crowded environment to facilitate DNA condensation into a more ordered, bundle-like state. We emphasize that the rod-like DNA structures observed with HU and DNA condensing agents are not likely the same dimensions as condensed domains of DNA within bacterial cells, which will be restricted by higher levels of chromosome structure and domain supercoiling. Nevertheless, the morphology of DNA condensates produced in vitro appears to be useful for monitoring the conditions under which HU affects DNA condensation, which could prove valuable for further exploration of HU-controlled condensation within more sophisticated models of the bacterial cytoplasm.
ACKNOWLEDGMENTS
We thank Prof. Roger McMacken for the gift of the RLM1078 E. coli strain, Dr Christine C. Conwell and Prof. James Maher for helpful discussions, the Georgia Tech EM Center for use of the JEOL-100C and Yolande Berta for her technical assistance. This work was supported by the NIH (GM62873) (NVH), the NSF (MCB 0316625) (IM) and the Patrick and Catherine Weldon Donaghue Medical Research Foundation (IM).
REFERENCES
- 1.Laemmli UK. Characteriation of DNA condensates induced by poly(ethylene oxide) and polylysine. Proc. Natl. Acad. Sci. U.S.A. 1975;72:4288–4292. doi: 10.1073/pnas.72.11.4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gosule LC, Schellman JA. Compact form of DNA induced by spermidine. Nature. 1976;259:333–335. doi: 10.1038/259333a0. [DOI] [PubMed] [Google Scholar]
- 3.Bloomfield V. DNA Condensation. Curr. Opin. Struct. Biol. 1996;6:334–341. doi: 10.1016/s0959-440x(96)80052-2. [DOI] [PubMed] [Google Scholar]
- 4.Bloomfield V. DNA condensation by multivalent cations. Biopolymers. 1997;44:269–282. doi: 10.1002/(SICI)1097-0282(1997)44:3<269::AID-BIP6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 5.Hud NV, Vilfan ID. Toroidal DNA condensates: unraveling the fine structure and the role of nucleation in determining size in vitro. Annu. Rev. Biophys. Biomol. Struct. 2005;34:295–318. doi: 10.1146/annurev.biophys.34.040204.144500. [DOI] [PubMed] [Google Scholar]
- 6.Klimenko S, Tikchonenko T, Andreev V. Packing of DNA in the head of bacteriophage T2. J. Mol. Biol. 1967;23:523–533. doi: 10.1016/s0022-2836(67)80122-0. [DOI] [PubMed] [Google Scholar]
- 7.Hud NV, Allen MJ, Downing KH, Lee J, Balhorn R. Identification of the elemental packing unit of DNA in mammalian sperm cells by atomic-force microscopy. Biochem. Biophys. Res. Commun. 1993;193:1347–1354. doi: 10.1006/bbrc.1993.1773. [DOI] [PubMed] [Google Scholar]
- 8.Hud NV. Double-stranded DNA organization in bacteriophage heads - an alternative toroid-based model. Biophys. J. 1995;69:1355–1362. doi: 10.1016/S0006-3495(95)80002-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rolland A. From genes to gene medicines: recent advances in nonviral gene delivery. Crit. Rev. Ther. Drug Carrier Syst. 1998;15:143–198. doi: 10.1615/critrevtherdrugcarriersyst.v15.i2.20. [DOI] [PubMed] [Google Scholar]
- 10.Mahato RI, Smith LC, Rolland A. Pharmaceutical perspectives of nonviral gene therapy. Adv. Genet. 1999;41:95–156. doi: 10.1016/s0065-2660(08)60152-2. [DOI] [PubMed] [Google Scholar]
- 11.Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat. Biotechnol. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 12.Shen M, Downing K, Balhorn R, Hud N. Nucleation of DNA condensation by static loops: formation of DNA toroids with reduced dimensions. J. Am. Chem. Soc. 2000;122:4833–4834. [Google Scholar]
- 13.Conwell CC, Vilfan ID, Hud NV. Controlling the size of nanoscale toroidal DNA condensates with static curvature and ionic strength. Proc. Natl. Acad. Sci. U.S.A. 2003;100:9296–9301. doi: 10.1073/pnas.1533135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conwell CC, Hud NV. Evidence that both kinetic and thermodynamic factors govern DNA toroid dimensions: effects of magnesium(II) on DNA condensation by hexammine cobalt(III) Biochemistry. 2004;43:5380–5387. doi: 10.1021/bi049872u. [DOI] [PubMed] [Google Scholar]
- 15.Plum GE, Arscott PG, Bloomfield VA. Condensation of DNA by trivalent cations. 2. Effects of cation structure. Biopolymers. 1990;30:631–643. doi: 10.1002/bip.360300515. [DOI] [PubMed] [Google Scholar]
- 16.Bloomfield VA. Condensation of DNA by multivalent cations - considerations on mechanism. Biopolymers. 1991;31:1471–1481. doi: 10.1002/bip.360311305. [DOI] [PubMed] [Google Scholar]
- 17.Arscott PG, Ma CL, Wenner JR, Bloomfield VA. DNA condensation by cobalt hexaammine(III) in alcohol-water mixtures: dielectric constant and other solvent effects. Biopolymers. 1995;36:345–364. doi: 10.1002/bip.360360309. [DOI] [PubMed] [Google Scholar]
- 18.Koehler JK. Fine structure observations in frozen-etched bovine spermatozoa. J. Ultrastruct. Res. 1966;16:359–375. doi: 10.1016/s0022-5320(66)80068-0. [DOI] [PubMed] [Google Scholar]
- 19.Koehler JK, Wurschmidt U, Larsen MP. Nuclear and chromatin structure in rat spermatozoa. Gamete Res. 1983;8:357–370. [Google Scholar]
- 20.Allen MJ, Lee C, Lee JD, Pogany GC, Balooch M, Siekhaus WJ, Balhorn R. Atomic-force microscopy of mammalian sperm chromatin. Chromosoma. 1993;102:623–630. doi: 10.1007/BF00352310. [DOI] [PubMed] [Google Scholar]
- 21.Allen MJ, Bradbury EM, Balhorn R. The natural subcellular surface-structure of the bovine sperm cell. J. Struct. Biol. 1995;114:197–208. doi: 10.1006/jsbi.1995.1019. [DOI] [PubMed] [Google Scholar]
- 22.Hud NV, Milanovich FP, Balhorn R. Evidence of novel secondary structure in DNA-bound protamine is revealed by Raman-spectroscopy. Biochemistry. 1994;33:7528–7535. doi: 10.1021/bi00190a005. [DOI] [PubMed] [Google Scholar]
- 23.Allen MJ, Bradbury EM, Balhorn R. AFM analysis of DNA-protamine complexes bound to mica. Nucleic Acids Res. 1997;25:2221–2226. doi: 10.1093/nar/25.11.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Balhorn R, Brewer L, Corzett M. DNA condensation by protamine and arginine-rich peptides: analysis of toroid stability using single DNA molecules. Mol. Reprod. Dev. 2000;56:230–234. doi: 10.1002/(SICI)1098-2795(200006)56:2+<230::AID-MRD3>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 25.Li S, Huang L. In vivo gene transfer via intravenous administration of cationic lipid-protamine-DNA (LPD) complexes. Gene Ther. 1997;4:891–900. doi: 10.1038/sj.gt.3300482. [DOI] [PubMed] [Google Scholar]
- 26.Faneca H, Simoes S, de Lima MCP. Association of albumin or protamine to lipoplexes: enhancement of transfection and resistance to serum. J. Gene Med. 2004;6:681–692. doi: 10.1002/jgm.550. [DOI] [PubMed] [Google Scholar]
- 27.Maruyama K, Iwasaki F, Takizawa T, Yanagie H, Niidome T, Yamada E, Ito T, Koyama Y. Novel receptor-mediated gene delivery system comprising plasmid/protamine/sugar-containing polyanion ternary complex. Biomaterials. 2004;25:3267–3273. doi: 10.1016/j.biomaterials.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 28.Vilfan ID, Conwell CC, Hud NV. Formation of native-like mammalian sperm cell chromatin with folded bull protamine. J. Biol. Chem. 2004;279:20088–20095. doi: 10.1074/jbc.M312777200. [DOI] [PubMed] [Google Scholar]
- 29.Huisman O, Faelen M, Girard D, Jaffe A, Toussaint A, Rouviere-Yaniv J. Multiple defects in Escherichia coli mutants lacking HU protein. J. Bacteriol. 1989;171:3704–3712. doi: 10.1128/jb.171.7.3704-3712.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hillyard DR, Edlund M, Hughes KT, Marsh M, Higgins NP. Subunit-specific phenotypes of Salmonella typhimurium HU mutants. J. Bacteriol. 1990;172:5402–5407. doi: 10.1128/jb.172.9.5402-5407.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dri AM, Rouviere-Yaniv J, Moreau PL. Inhibition of cell-division in hupa hupb mutant bacteria lacking HU Protein. J. Bacteriol. 1991;173:2852–2863. doi: 10.1128/jb.173.9.2852-2863.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paull TT, Johnson RC. DNA looping by Saccharomyces cerevisiae high-mobility group proteins NHP6A/B – consequences for nucleoprotein complex assembly and chromatin condensation. J. Biol. Chem. 1995;270:8744–8754. doi: 10.1074/jbc.270.15.8744. [DOI] [PubMed] [Google Scholar]
- 33.Drlica K, Rouviere-Yaniv J. Histone-like proteins of bacteria. Microbiol. Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodman SD, Nicholson SC, Nash HA. Deformation of DNA during site-specific recombination of bacteriophage-lambda - replacement of IHF protein by HU protein or sequence-directed bends. Proc. Natl. Acad. Sci. U.S.A. 1992;89:11910–11914. doi: 10.1073/pnas.89.24.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavoie BD, Chaconas G. Site-specific HU binding in the Mu-transpososome - conversion of a sequence-independent DNA-binding protein into a chemical nuclease. Genes Dev. 1993;7:2510–2519. doi: 10.1101/gad.7.12b.2510. [DOI] [PubMed] [Google Scholar]
- 36.Lavoie BD, Shaw GS, Millner A, Chaconas G. Anatomy of a flexer-DNA complex inside a higher-order transposition intermediate. Cell. 1996;85:761–771. doi: 10.1016/s0092-8674(00)81241-6. [DOI] [PubMed] [Google Scholar]
- 37.Aki T, Adhya S. Repressor induced site-specific binding of HU for transcriptional regulation. EMBO J. 1997;16:3666–3674. doi: 10.1093/emboj/16.12.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Megraw TL, Chae CB. Functional complementarity between the HMG1-like yeast mitochondrial histone HM and the bacterial histone-like protein HU. J. Biol. Chem. 1993;268:12758–12763. [PubMed] [Google Scholar]
- 39.Bianchi ME. Prokaryotic HU and eukaryotic HMG1-a kinked relationship. Mol. Microbiol. 1994;14:1–5. doi: 10.1111/j.1365-2958.1994.tb01261.x. [DOI] [PubMed] [Google Scholar]
- 40.Bewley CA, Gronenborn AM, Clore GM. Minor groove-binding architectural proteins: structure, function, and DNA recognition. Annu. Rev. Biophys. Biomol. Struct. 1998;27:105–131. doi: 10.1146/annurev.biophys.27.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swinger KK, Lemberg KM, Zhang Y, Rice PA. Flexible DNA bending in HU-DNA cocrystal structures. EMBO J. 2003;22:3749–3760. doi: 10.1093/emboj/cdg351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wojtuszewski K, Mukerji I. HU binding to bent DNA: A fluorescence resonance energy transfer and anisotropy study. Biochemistry. 2003;42:3096–3104. doi: 10.1021/bi0264014. [DOI] [PubMed] [Google Scholar]
- 43.Sagi D, Friedman N, Vorgias C, Oppenheim AB, Stavans J. Modulation of DNA conformations through the formation of alternative high-order HU-DNA complexes. J. Mol. Biol. 2004;341:419–428. doi: 10.1016/j.jmb.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 44.Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr. Opin. Struct. Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 45.van Noort J, Verbrugge S, Goosen N, Dekker C, Dame RT. Dual architectural roles of HU: formation of flexible hinges and rigid filaments. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6969–6974. doi: 10.1073/pnas.0308230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy LD, Zimmerman SB. Condensation and cohesion of lambda-DNA in cell extracts and other media - implications for the structure and function of DNA in prokaryotes. Biophys. Chem. 1995;57:71–92. doi: 10.1016/0301-4622(95)00047-2. [DOI] [PubMed] [Google Scholar]
- 47.Murphy LD, Zimmerman SB. Macromolecular crowding effects on the interaction of DNA with Escherichia coli DNA-binding proteins - a model for bacterial nucleoid stabilization. BBA-Gene Struct. Expr. 1994;1219:277–284. doi: 10.1016/0167-4781(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 48.Sarkar T, Conwell CC, Harvey LC, Santai CT, Hud NV. Condensation of oligonucleotides assembled into nicked and gapped duplexes: potential structures for oligonucleotide delivery. Nucleic Acids Res. 2005;33:143–151. doi: 10.1093/nar/gki156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wojtuszewski K, Hawkins ME, Cole JL, Mukerji I. HU binding to DNA: Evidence for multiple complex formation and DNA bending. Biochemistry. 2001;40:2588–2598. doi: 10.1021/bi002382r. [DOI] [PubMed] [Google Scholar]
- 50.Chattoraj DK, Gosule LC, Schellman JA. DNA condensation with polyamines. II. Electron microscopic studies. J. Mol. Biol. 1978;121:327–337. doi: 10.1016/0022-2836(78)90367-4. [DOI] [PubMed] [Google Scholar]
- 51.Hansma HG, Golan R, Hsieh W, Lollo CP, Mullen-Ley P, Kwoh D. DNA condensation for gene therapy as monitored by atomic force microscopy. Nucleic Acids Res. 1998;26:2481–2487. doi: 10.1093/nar/26.10.2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shindo H, Furubayashi A, Shimizu M, Miyake M, Imamoto F. Preferential binding of Escherichia Coli histone-like protein HU-alpha to negatively supercoiled DNA. Nucleic Acids Res. 1992;20:1553–1558. doi: 10.1093/nar/20.7.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tanaka H, Goshima N, Kohno K, Kano Y, Imamoto F. Properties of DNA-binding of HU heterotypic and homotypic dimers from. Escherichia coli. J. Biochem. (Tokyo) 1993;113:568–572. doi: 10.1093/oxfordjournals.jbchem.a124084. [DOI] [PubMed] [Google Scholar]
- 54.Shimizu M, Miyake M, Kanke F, Matsumoto U, Shindo H. Characterization of the binding of HU and IHF, homologous histone-like proteins of Escherichia coli, to curved and uncurved DNA. Biochim. Biophys. Acta. 1995;1264:330–336. doi: 10.1016/0167-4781(95)00173-5. [DOI] [PubMed] [Google Scholar]
- 55.Pinson V, Takahashi M, Rouviere-Yaniv J. Differential binding of the Escherichia coli HU, homodimeric forms and heterodimeric form to linear, gapped and cruciform DNA. J. Mol. Biol. 1999;287:485–497. doi: 10.1006/jmbi.1999.2631. [DOI] [PubMed] [Google Scholar]
- 56.Ou Z, Muthukumar M. Langevin dynamics of semiflexible polyelectrolytes: Rod-toroid-globule-coil structures and counterion distribution. J. Chem. Phys. 2005;123:074905–01–074905–09. doi: 10.1063/1.1940054. [DOI] [PubMed] [Google Scholar]
- 57.Vilfan ID, Conwell CC, Sarkar T, Hud NV. Time study of DNA condensate morphology: implications regarding the nucleation, growth, and equilibrium populations of toroids and rods. Biochemistry. 2006;45:8174–8183. doi: 10.1021/bi060396c. [DOI] [PubMed] [Google Scholar]
- 58.Lang D. Regular superstructures of purified DNA in ethanolic solutions. J. Mol. Biol. 1973;78:247–254. doi: 10.1016/0022-2836(73)90113-7. [DOI] [PubMed] [Google Scholar]
- 59.Eickbush TH, Moudrianakis EN. The compaction of DNA helices into either continuous supercoils or folded-fiber rods and toroids. Cell. 1978;13:295–306. doi: 10.1016/0092-8674(78)90198-8. [DOI] [PubMed] [Google Scholar]
- 60.Lang D, Taylor TN, Dobyan DC, Gray DM. Dehydrated circular DNA - electron microscopy of ethanol-condensed molecules. J. Mol. Biol. 1976;106:97–107. doi: 10.1016/0022-2836(76)90302-8. [DOI] [PubMed] [Google Scholar]
- 61.Herskovits TT, Singer SJ, Geiduschek EP. Nonaqueous solutions of DNA - denaturation in methanol and ethanol. Arch. Biochem. Biophys. 1961;94:99–114. doi: 10.1016/0003-9861(61)90016-9. [DOI] [PubMed] [Google Scholar]
- 62.Grosberg AY, Zhestkov AV. On the toroidal condensed state of closed circular DNA. J. Biomol. Struct. Dyn. 1985;3:515–520. doi: 10.1080/07391102.1985.10508438. [DOI] [PubMed] [Google Scholar]
- 63.Arscott PG, Li AZ, Bloomfield VA. Condensation of DNA by trivalent cations. 1. Effects of DNA length and topology on the size and shape of condensed particles. Biopolymers. 1990;30:619–630. doi: 10.1002/bip.360300514. [DOI] [PubMed] [Google Scholar]
- 64.Hud NV, Downing KH. Cryoelectron microscopy of lambda phage DNA condensates in vitreous ice: the fine structure of DNA toroids. Proc. Natl. Acad. Sci. U.S.A. 2001;98:14925–14930. doi: 10.1073/pnas.261560398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Broyles SS, Pettijohn DE. Interaction of the Escherichia Coli HU protein with DNA - evidence for formation of nucleosome-like structures with altered DNA helical pitch. J. Mol. Biol. 1986;187:47–60. doi: 10.1016/0022-2836(86)90405-5. [DOI] [PubMed] [Google Scholar]
- 66.Porschke D. Dynamics of DNA condensation. Biochemistry. 1984;23:4821–4828. doi: 10.1021/bi00316a002. [DOI] [PubMed] [Google Scholar]
- 67.Tecle M, Preuss M, Miller AD. Kinetic study of DNA condensation by cationic peptides used in nonviral gene therapy: analogy of DNA condensation to protein folding. Biochemistry. 2003;42:10343–10347. doi: 10.1021/bi034325e. [DOI] [PubMed] [Google Scholar]
- 68.Dame RT. The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 2005;56:858–870. doi: 10.1111/j.1365-2958.2005.04598.x. [DOI] [PubMed] [Google Scholar]
- 69.Travers A, Muskhelishvili G. Bacterial chromatin. Curr. Opin. Genet. Dev. 2005;15:507–514. doi: 10.1016/j.gde.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 70.Johnson RC, Johnson LM, Schmidt JW, Gardner JF. Major Nucleoid Proteins in the Structure and Function of the Escherichia coli Chromosome. In: Patrick Higgins N, editor. The Bacterial Chromosome. Washington, DC: ACM Press; 2005. pp. 65–131. [Google Scholar]
- 71.Murphy LD, Zimmerman SB. Stabilization of compact spermidine nucleoids from Escherichia coli under crowded conditions: implications for in vivo nucleoid structure. J. Struct. Biol. 1997;119:336–346. doi: 10.1006/jsbi.1997.3884. [DOI] [PubMed] [Google Scholar]
- 72.Dame RT, Goosen N. HU: promoting or counteracting DNA compaction? FEBS Lett. 2002;529:151–156. doi: 10.1016/s0014-5793(02)03363-x. [DOI] [PubMed] [Google Scholar]
- 73.Skoko D, Wong B, Johnson RC, Marko JF. Micromechanical analysis of the binding of DNA-bending proteins HMGB1, NHP6A, and HU reveals their ability to form highly stable DNA-protein complexes. Biochemistry. 2004;43:13867–13874. doi: 10.1021/bi048428o. [DOI] [PubMed] [Google Scholar]
- 74.Zimmerman SB. Cooperative transitions of isolated Escherichia coli nucleoids: implications for the nucleoid as a cellular phase. J. Struct. Biol. 2006;153:160–175. doi: 10.1016/j.jsb.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 75.Azam TA, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wery M, Woldringh CL, Rouviere-Yaniv J. HU-GFP and DAPI co-localize on the Escherichia coli nucleoid. Biochimie. 2001;83:193–200. doi: 10.1016/s0300-9084(01)01254-8. [DOI] [PubMed] [Google Scholar]
- 77.Rouviere-Yaniv J, Yaniv M, Germond JE. Escherichia Coli DNA-binding protein HU Forms nucleosome-like structure with circular double-stranded DNA. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 78.Zimmerman SB. Shape and compaction of Escherichia coli nucleoids. J. Struct. Biol. 2006;156:255–261. doi: 10.1016/j.jsb.2006.03.022. [DOI] [PubMed] [Google Scholar]