Abstract
SET domain-containing proteins of the SU(VAR)3-9 class are major regulators of heterochromatin in several eukaryotes, including mammals, insects, plants and fungi. The function of these polypeptides is mediated, at least in part, by their ability to methylate histone H3 on lysine 9 (H3K9). Indeed, mutants defective in SU(VAR)3-9 proteins have implicated di- and/or trimethyl H3K9 in the formation and/or maintenance of heterochromatin across the eukaryotic spectrum. Yet, the biological significance of monomethyl H3K9 has remained unclear because of the lack of mutants exclusively defective in this modification. Interestingly, a SU(VAR)3-9 homolog in the unicellular green alga Chlamydomonas reinhardtii, SET3p, functions in vitro as a specific H3K9 monomethyltransferase. RNAi-mediated suppression of SET3 reactivated the expression of repetitive transgenic arrays and reduced global monomethyl H3K9 levels. Moreover, chromatin immunoprecipitation (ChIP) assays demonstrated that transgene reactivation correlated with the partial loss of monomethyl H3K9 from their chromatin. In contrast, the levels of trimethyl H3K9 or the repression of euchromatic sequences were not affected by SET3 downregulation; whereas dimethyl H3K9 was undetectable in Chlamydomonas. Thus, our observations are consistent with a role for monomethyl H3K9 as an epigenetic mark of repressed chromatin and raise questions as to the functional distinctiveness of different H3K9 methylation states.
INTRODUCTION
Transcriptional gene expression in eukaryotes is regulated at two major levels: the operation of the transcription machinery and the modulation of chromatin structure (1). Indeed, the regulation of chromatin packaging has recently emerged as an important mechanism for maintaining gene expression patterns. Eukaryotic genomes are commonly organized into two main types of chromatin: euchromatin, consisting of transcriptionally permissive or active domains, and heterochromatin, characterized by densely packed silent regions (2). These functionally and structurally distinct chromatin states are marked by distinctive covalent modifications on the DNA and on the nucleosomal histones. For instance, active euchromatin is usually characterized by the presence of histone H3 methylated on lysines 4 and 36; whereas silenced heterochromatin is often distinguished by the presence of histone H3 methylated on lysines 9 and 27, and histone H4 methylated on lysine 20 (3,4). This organization of chromosomes into domains with distinct histone methylation patterns appears to be common across the eukaryotic spectrum (5–12) and the histone modifications are proposed to influence directly chromatin structure and/or bring about the recruitment of chromatin modulators (3,4). However, it is becoming apparent that the specific modifications associated with functionally equivalent regions (such as pericentric heterochromatin) can be quite variable among different species.
Histone H3 on lysine 9 (H3K9) methylation is carried out by the SET domain of the SU(VAR)3–9-related proteins, which belong to the histone methyltransferase (HMTase) superfamily (13,14). The first SU(VAR)3-9-encoding gene was identified in a genetic screen for suppressors of position effect variegation in Drosophila melanogaster (15), providing clues as to its function as a key regulator of repressive heterochromatic organization. Indeed, Drosophila SU(VAR)3-9 and its mammalian homologs, SUV39H1 and SUV39H2, were latter found to be enriched in interphase heterochromatin and to accumulate transiently at centromeric loci during mitosis (16–18). SU(VAR)3-9-related proteins are now known to be widely distributed among eukaryotes. For instance, the model plant Arabidopsis thaliana contains 29 genes encoding SET domain proteins (19). Ten of these polypeptides have been classified as SU(VAR)3-9 homologs (SUVHs), based on the degree of similarity between their SET domains and those of animal SU(VAR)3-9 proteins. SUVH genes have also been identified in a number of other plant species such as tobacco (20), maize (21) and rice (http://www.chromdb.org) as well as in several fungi (22,23). However, they have not been described as yet in algal systems.
Seven plant SUVH genes have been characterized in some detail: tobacco NtSET1 (20,24,25), and Arabidopsis SUVH1 (8), SUVH2 (8), SUVH3 (26), SUVH4 (KRYPTONITE) (27,28), SUVH5 (29) and SUVH6 (30,31). Several of the corresponding proteins have been shown to possess HMTase activity targeting H3K9 (and H4K20 or H2A in the cases of SUVH2 and SUVH5, respectively) and to associate preferentially with putative heterochromatic regions (8,24,29). Indeed, SUVH2 appears to play a major role in heterochromatin formation and/or maintenance in Arabidopsis since loss-of-function mutants display defects in gene silencing and a reduction in multiple heterochromatin-specific histone methylation marks (8). In contrast, SUVH4, SUVH5 and SUVH6 seem to have partly overlapping functions and control H3K9 methylation and gene silencing at specific loci (29,31).
Methylatable lysine residues can exist in monomethylated, dimethylated or trimethylated states, increasing the coding potential of modified histone lysines as epigenetic marks (4). Interestingly, in mammals, pericentric heterochromatin is enriched in trimethyl H3K9 (H3K9me3), a modification carried out predominantly by the SUV39H1 and SUV39H2 HMTases (4–6,9). Conversely, mono- and dimethyl H3K9 (H3K9me1 and H3K9me2) are enriched in certain euchromatic domains, which have been postulated to be transcriptionally silent (4–6,9,32). The latter modifications are mediated in part by the G9a HMTase, which plays an essential role in developmental regulation of gene expression (5,6,9,33). In Arabidopsis, in contrast to mammals, heterochromatic chromocenters are marked by H3K9me1 and H3K9me2, whereas H3K9me3 is predominantly found in euchromatic regions (7,8,27). Yet in another plant species, maize, only H3K9me1 was observed in heterochromatic domains (as well as in some euchromatic regions) while H3K9me2 and H3K9me3 were restricted to euchromatin and the centromeres (11).
These observations suggest that H3K9 mono-, di- and trimethylation states may serve distinct functions since they reside at least partly in separate chromosomal domains. Moreover, in mammals and Arabidopsis, different HMTases are responsible for specific degrees of H3K9 methylation at certain locations (5,6,9,29,34) and different HMTase mutants have distinct phenotypes (5,33), implying that the HMTases (and the histone modifications that they catalyze) are differentially targeted to diverse genomic regions for regulatory purposes. Yet, seemingly equivalent domains (such as pericentric heterochromatin) are characterized by species-specific H3K9 methylation states, raising questions as to the functional significance of different degrees of H3K9 methylation. In addition, genetic and biochemical analyses in a variety of eukaryotes have established a role for H3K9me2 and H3K9me3 in the formation and/or maintenance of transcriptionally repressive chromatin (3,4,8), whereas H3K9me1 lacks a defined functional assignment at present, partly because a HMTase exclusively responsible for this modification has not been identified thus far.
To gain further insight into the role of H3K9 methylation in chromatin organization, we undertook a reverse genetics approach to the study of SUVHs in the green alga Chlamydomonas reinhardtii. BLAST searches of the almost fully sequenced genome (http://genome.jgi-psf.org/Chlre3/Chlre3.home.html) revealed the existence of two SU(VAR)3-9-related proteins in Chlamydomonas. We report here on the characterization of one of these homologs, named SET3p. This polypeptide showed the same domain organization as plant SUVH proteins and its suppression by RNA interference (RNAi) released the transcriptional silencing of tandem transgenes. In contrast, repressed single-copy euchromatic sequences and dispersed transposons were not reactivated. Recombinant SET3p behaved, in vitro, as an exclusive monomethyl H3K9 HMTase. Moreover, chromatin immunoprecipitation (ChIP) assays demonstrated that, in vivo, H3K9me1 was dependent on the SET3p activity and was associated with silent multiple-copy transgenes. Conversely, H3K9me3 was mainly detected in a euchromatic gene and its intensity was not altered by SET3 suppression. Thus, our results provide direct evidence for a functional role of monomethyl H3K9 in the maintenance of repressed chromatin and add to the growing body of evidence suggesting that seemingly equivalent chromatin states may be characterized by species-specific combinations of histone modifications.
MATERIALS AND METHODS
Chlamydomonas reinhardtii strains and culture conditions
Chlamydomonas cells were routinely grown in Tris-acetate-phosphate (TAP) medium under moderate light conditions (35,36). The 1-P[300] strain, containing over 100 integrated copies of the RbcS2:aadA:RbcS2 transgene, has been previously described (35). The Set3-IR and Maa7-IR strains are derivatives of 1-P[300] in which the expression of SET3 or MAA7 (encoding tryptophan synthase β subunit) has been suppressed by RNAi, by using the approach described by Rohr et al. (37). Briefly, 1-P[300] was transformed by the glass bead method (35) with transgenes that produce double-stranded RNA (dsRNA) capable of inducing the degradation of homologous transcripts. For SET3, a 630-bp fragment corresponding to part of the coding sequence and the 3′ UTR of the transcript was amplified by PCR with primers Set3-F1 (5′-GCGACGGCAACCTGACCATCC-3′) and Set3-R1 (5′−CTGACCCCACACCCACGCTCTGAC-3′). This segment was then inserted in sense and antisense orientations, flanking a spacer sequence, in vector Maa7/X IR (37). The Maa7-IR3 construct, utilized as a negative control, has already been described (37). Reactivation of the RbcS2:aadA:RbcS2 transgenes was tested by spotting serial dilutions of cells on TAP-agar plates with or without 50 mg/l of spectinomycin.
Reverse transcriptase-polymerase chain reaction (RT-PCR) analyses
Total RNA was isolated using the TRIZOL reagent, according to the manufacturer's instructions (Molecular Research Center, Cincinnati, OH, USA), and contaminant DNA was removed by DNase-I treatment (Ambion, Austin, TX, USA). First-strand cDNA synthesis and PCR reactions were performed as previously described by Rohr et al. (37). PCR products were resolved on 1.5% agarose gels and visualized by ethidium bromide staining. The numbers of cycles showing a linear relationship between input RNA and the final product were determined in preliminary experiments. Controls included the use as template of reactions without RT and verification of PCR products by hybridization with specific probes (data not shown). The primer sequences were as follows: for SET3, Set3-RTF6 (5′-GGTGTGCAAGTTCCTGATGCAC-3′) and Set3-RTR7 (5′-TGAACTGCAGCATCTCCTCGTC-3′); for aadA, aadA-CodL, (5′-TCTGGCTATCTTGCTGACAAAA-3′); and aadA-CodR, (5′-TAGTGATCTCGCCTTTCACGTA-3′); and for MUT9, Mut9-5 (5′-GCTGTACATCTCGTGCGTGT-3′) and Mut9-2 (5′-ATGGCGGTCACGTAGAAGC-3′).
Phylogenetic analysis
Individual domains present in SET3p (AAV84356) and other SU(VAR)3-9-related proteins were identified using the SMART database (38). The SET domain amino acid sequences were aligned using CLUSTAL X version 1.81 (39) and manually corrected with the GENEDOC program (http://www.psc.edu/biomed/genedoc). Phylogenetic relationships between these sequences were inferred by the neighbor-joining (NJ) method (40). The MEGA program version 3.1 (41) was used to obtain the NJ trees, using Poisson-corrected amino acid distances, and the bootstrap support values for 1000 pseudoreplicates.
Partial protein purification and immunoblotting
Chromatin-associated proteins were partially purified by differential centrifugation prior to immunoblotting. Approximately 5 × 108 TAP-grown cells were resuspended in nuclear isolation buffer (20 mM PIPES, pH 7.0, 0.25 M sucrose, 10 mM MgCl2, 2.0 mM spermidine, 100 mM sodium butyrate, 0.1% Triton X-100, 5 mM β-mercaptoethanol, 2 mM benzamidine and 0.1 mM PMSF) and broken by two passages through a French press at 5000 psi. Lysed cells were then centrifuged for 10 min at 30 000 g. The supernatant was discarded and proteins in the pellet (including nuclear chromatin) were solubilized in high salt buffer (20 mM HEPES, pH 7.5, 2 M NaCl, 1 mM EDTA, 1 mM DTT, 2 mM benzamidine and 2 µl/ml of plant protease inhibitor cocktail [Sigma-Aldrich, Saint Louis. MO, USA]). Soluble proteins were then separated from insoluble debris by centrifugation for 10 min at 10 000 g, resolved by SDS-PAGE and transferred to nitrocellulose membranes. Specific methylated states of histone H3 lysine 9 were detected with antibodies against monomethyl H3K9 (Upstate, 07-395; or Abcam, ab9045), dimethyl H3K9 (Upstate, 07-212; or Abcam, ab7312) or trimethyl H3K9 (Abcam, ab1186 or ab8898). A modification-insensitive anti-H3 antibody (Abcam, ab1791) was used to adjust the amount of histone H3 loaded in each lane.
Histone methyltransferase assays
The histone methylating activity of recombinant SET3p was assayed as described earlier (42). Briefly, 10 µg of core histones or purified H3 from calf thymus and 1 µg of recombinant SET3p protein or a protein extract from an empty vector control were incubated with 250 nCi of S-adenosyl-l-(methyl-14C)methionine (14C-SAM) (GE Healthcare, Piscataway, NJ, USA) in a final volume of 40 µl for 2 h at 30°C in MAB buffer. Samples were then resolved by SDS-PAGE on 15% polyacrylamide gels, stained with Coomassie Brilliant Blue and dried onto filter paper. The incorporated radioactivity was detected with a phosphor imager (Amersham). The methylation activity of SET3p on wild-type (H3N) and lysine mutated (K4R, K9R, K27R or 3K-R) histone H3 tails fused to GST was assayed in a similar way. For time course experiments, 8 µg of recombinant SET3p protein, 40 µg of recombinant human histone H3 (43) and 20 µM of unlabeled SAM were incubated at 30°C in MAB buffer in a final volume of 80 µl. Ten microliters of aliquots were taken at different times (Figure 3D) and the reactions stopped by the addition of 10 µl of 2× SDS-PAGE loading buffer. Samples were then examined by western blotting with antibodies against specific H3K9 methylation states.
Figure 3.
In vitro histone methyltransferase activity of recombinant SET3p. (A) His-tagged SET3p exclusively methylates histone H3. The recombinant protein was used in HMTase assays with calf thymus core histones or purified histone H3 as substrates. Samples were resolved by SDS-PAGE and stained with Coomassie blue (Bottom panel). Incorporation of the radiolabeled methyl group was detected by phosphor imaging (Top panel). pET30a indicates an empty vector protein extract used as a negative control. (B) HMTase assays of SET3p using as substrates wild-type and lysine mutants of the histone H3 N-tail fused to GST. Radiolabeled proteins were detected as in (A). Bands observed below the full-length substrates are likely degradation products. Bottom panel, Coomassie blue staining; Top panel, phosphor imager scan. (C) Alignment of the region corresponding to the pseudoknot in the SET domain of several SU(VAR)3-9 proteins. The position of the first amino acid in the alignment is indicated on the left. The Phe/Tyr (F/Y) switch, determining the degree of methylation by HMTases (51), is also shown. (D) SET3p specifically monomethylates H3K9 in vitro. Purified SET3p was incubated with recombinant human histone H3 in the presence of cold SAM for different amounts of time. Samples were resolved by SDS-PAGE, transferred to nitrocellulose and probed with the indicated antibodies. Equivalent loading of the lanes was checked by Ponceau S staining. C, purified calf thymus (non-recombinant) H3; M, molecular weight markers.
Chromatin immunoprecipitation (ChIP) assays
To examine the methylation status of histone H3 at specific chromosomal loci, 5 × 107 TAP-grown cells were cross-linked for 10 min with 1% formaldehyde and then quenched by adding glycine to 0.1 M. Cells were then pelleted, washed with TBS and frozen in liquid nitrogen (42). The cell pellet was resuspended in 6 ml of ChIP lysis buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 2 µl/ml of plant protease inhibitor cocktail [Sigma-Aldrich]), and cells were lysed by two passages through a French press at 5000 psi. Chromatin was sheared by sonication to an average size of 500 bp and then immunoprecipitated using a ChIP assay kit (Upstate) and a modification-insensitive anti-H3 antibody (15 µl, Abcam ab1791). The precipitated chromatin was eluted by incubation for 10 min at 68°C with 100 µl of ChIP elution buffer (50 mM Tris-HCl, pH 7.5, 10 mM EDTA and 1% SDS) (44). This procedure was repeated once and 10% of the combined eluate was removed to be used as the H3 secondary input (see reference 44 for a justification). The remaining eluate was diluted to 2 ml with ChIP lysis buffer containing 25 µg/ml of sheared salmon testes DNA (Sigma-Aldrich), and 5 mg/ml BSA (Fraction V, Sigma-Aldrich); and then the chromatin was immunoprecipitated with a second antibody against monomethyl H3K9 (15 µl, Upstate, 07-395), trimethyl H3K9 (3 µg, Abcam ab1186) or rabbit IgG (5 µl at 2 µg/µl Sigma-Aldrich, I5006). After cross-link reversal and DNA purification by phenol/chloroform extractions, the immunoprecipitated DNA was quantified by real-time PCR on a Bio-Rad iCycler iQ using SYBR Green. After each run, a melting curve was performed to ensure that no primer dimers interfered with the quantification. Serial dilutions of the primary input (DNA prior to anti-H3 immunoprecipitation) were used to generate a calibration curve with which the amounts of target DNA in the secondary inputs (DNA co-immunoprecipitated with anti-H3, anti-H3K9me1, anti-H3K9me3 or IgG antibodies) were calculated. The levels of the H3K9 modifications or of the IgG negative control were then normalized relative to the anti-H3 controls for each examined sequence. Primers used for amplification of the target loci were: for RbcS2:aadA:RbcS2, aadA-CodL (5′-TCTGGCTATCTTGCTGACAAAA-3′) and aadA-CodR (5′-TAGTGATCTCGCCTTTCACGTA-3′); and for RPS3, C_20102proL1 (5′-AAGGGCGCTGCTAGTATAACCA-3′) and C_20102proR1 (5′-CCTTTGTTCCCGAGAGAGAGAA-3′).
DNA methylation analysis
Genomic DNA from Chlamydomonas was isolated, digested, resolved in agarose gels, blotted and hybridized following standard procedures (35,45). The aadA and psbA probes used for Southern hybridization correspond to the coding sequence of the RbcS2:aadA:RbcS2 transgene and the 3′ end of the chloroplast psbA gene, respectively (35).
RESULTS
Chlamydomonas reinhardtii SET3p is a homolog of plant SUVH proteins
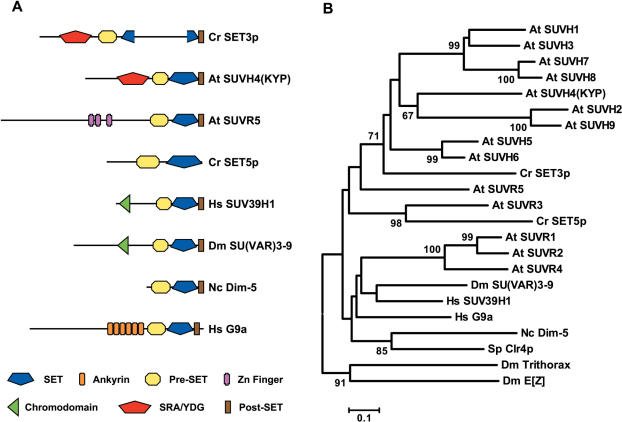
SET3 encodes a SET domain-containing protein with a predicted size of 957 amino acids. The carboxyl terminal region of this polypeptide is well conserved relative to that of SU(VAR)3-9 proteins and consists of the SET domain flanked by two cysteine-rich motifs, the pre-SET and the post-SET domains (Figure 1A). However, SET3p also includes a unique insertion, of approximately 300 amino acids, within the SET domain that separates the conserved sub-motifs II and III (46) (Figure 1A). At its N-terminal end, SET3p displays the configuration characteristic of plant SUVH proteins (Figure 1A). These polypeptides contain, instead of the characteristic chromodomain of the metazoan enzymes, a conserved motif designated YDG or SRA (SET and RING finger associated) (19,47).
Figure 1.
SU(VAR)3-9 homologs in Chlamydomonas and several eukaryotes. (A) Domain organization of SU(VAR)3-9-related proteins. (B) Phylogenetic relationship between Chlamydomonas SET3p and SET5p and histone methyltransferases from several species. Numbers indicate bootstrap values, as percentage, based on 1000 pseudoreplicates. At SUVH1 to At SUVH9, Arabidopsis thaliana SU(VAR)3-9 homologs 1 to 9 (AAK28966 to AAK28974, respectively); At SUVR1 to At SUVR5, A. thaliana SU(VAR)3-9-related 1 to 5 (AAK77165, AAK92218, NP_974212, AAL01113 and NP_179954, respectively); Cr SET3p, Chlamydomonas reinhardtii SET3p protein (AAV84356); Cr SET5p, C. reinhardtii SET5p protein (145254 at http://genome.jgi-psf.org/Chlre3/Chlre3.home.html); Dm E[Z], Drosophila melanogaster Enhancer of zeste protein (P42124); Dm SU(VAR)3-9, Drosophila Suppressor of variegation 3-9 protein (P45975); Dm Trithorax, Drosophila Trithorax protein (P20659); Hs G9a, human SET domain protein G9a (NP_006700); Hs SUV39H1, human SU(VAR)3-9 homolog 1 (O43463); Nc Dim-5, Neurospora crassa Dim-5 (Q8X225); Sp Clr4, Schizosaccharomyces pombe Cryptic loci regulator 4 protein (AAC18302).
A phylogenetic tree constructed using sequences corresponding to the SET domain of multiple HMTases shows that SET3p clusters with the plant SUVH proteins (Figure 1B). These polypeptides have been reported to fall into four distinct subgroups, suggesting a possible functional differentiation that might have preceded the angiosperm/gymnosperm divergence (8,19,21). However, SET3p behaves as an outgroup to all of the Arabidopsis SUVH proteins, implying that most of the functional diversification of SUVH polypeptides likely occurred within the plant lineage after the evolutionary separation of green algae and land plants. In addition, higher plants also contain SU(VAR)3-9-RELATED (SUVR) proteins, which lack the YDG/SRA domain but are grouped with the SUVHs into the Class V of plant SET domain polypeptides (19,21). The Chlamydomonas genome also encodes a SU(VAR)3-9-related protein, named SET5p (145254 at http://genome.jgi-psf.org/Chlre3/Chlre3.home.html), that lacks the YDG/SRA domain (Figure 1A). Interestingly, whereas Chlamydomonas SET3p is closely related to plant SUVHs, SET5p associates with plant SUVRs in phylogenetic analyses (Figure 1B), suggesting that differentiation of the SUVH and SUVR genes likely occurred prior to the divergence of the green algae and plant lineages.
RNAi-mediated suppression of SET3 results in defective silencing of multiple-copy transgenes
Plant SUVH genes have been implicated in gene silencing, heterochromatin formation and the partial control of non-CpG DNA methylation. For instance, SUVH4 mutants derepress the silenced SUPERMAN and PAI genes (27,28) and loss-of-function of SUVH2 reactivates a LUCIFERASE transgene (8). To examine whether Chlamydomonas SET3 has a similar role in gene silencing, we tested the effect of downregulating its expression on the transcriptional activity of repressed transgenes. Chlamydomonas strain 1-P[300] contains over 100 silenced copies of the RbcS2:aadA:RbcS2 transgene (conferring resistance to spectinomycin), integrated as head-to-head, tail-to-tail and head-to-tail concatamers at two chromosomal loci (35). Based on the repeated conformation of the transgenic arrays, their transcriptionally silenced state (35) and the presence of DNA methylation (see below), it is possible that the chromatin associated with these loci is (hetero)chromatic in nature, as reported for equivalent concatamers in other eukaryotes (8,48–50).
To suppress SET3 expression, the 1-P[300] strain was transformed with an inverted repeat construct that generates dsRNA homologous to the 3′ end of the SET3 mRNA. We recovered several independent transformants showing reduced SET3 transcript levels and two strains, Set3-IR1 and Set3-IR2, were examined in detail for reactivation of the RbcS2:aadA:RbcS2 transgenes as well as for their ability to survive on medium containing spectinomycin (Figure 2 and data not shown). RT-PCR analyses indicated that RNAi-mediated SET3 suppression resulted in increased RbcS2:aadA:RbcS2 mRNA levels (Figure 2A). In addition, the Set3-IR strains were able to grow on medium containing spectinomycin whereas the non-transgenic wild-type (CC-124) and the silenced parental strain (1-P[300]) were not (Figure 2B). These effects were specific for the Set3-IR transformants since RNAi-mediated downregulation of an unrelated gene, MAA7 (encoding tryptophan synthase β subunit), did not reactivate RbcS2:aadA:RbcS2 expression in the 1-P[300] strain nor allow survival in the presence of spectinomycin (Figure 2, Maa7-IR3 strain). Yet, SET3 suppression did not release the silencing of an unmethylated, single-copy transgene (in strain 11-P[300]) or of dispersed transposable elements, such as the TOC1 retrotransposon (data not shown). We have reported earlier that the 11-P[300] transgene and the TOC1 loci appear to be repressed by euchromatin-specific epigenetic marks (42). Thus, our results suggest that downregulation of SET3 expression leads to defects in the transcriptional silencing of multiple-copy cis-tandem transgenes, presumably associated with a (hetero)chromatic conformation (see discussion).
Figure 2.
Suppression of SET3 expression and reactivation of RbcS2:aadA:RbcS2 transgenes in Set3-IR strains. (A) RT-PCR analysis of SET3 and RbcS2:aadA:RbcS2 (aadA) expression in the indicated strains. Amplification of MUT9 (encoding a Ser/Thr protein kinase) transcripts was used as a control for equivalent RNA input and efficiency of the RT-PCR reactions. (B) Growth and survival of the indicated strains on medium with (TAP + Spec) or without (TAP) spectinomycin. Strains: CC-124, wild-type strain; 1-P[300], silenced parental strain; Maa7-IR3, 1-P[300] transformant expressing dsRNA complementary to the MAA7 gene; Set3-IR1 and Set3-IR2, 1-P[300] transformants expressing dsRNA complementary to SET3.
SET3p possesses H3K9 monomethyltransferase activity in vitro
In order to investigate whether SET3p might be a determinant of repressed chromatin structure, we first tested its ability to methylate core histones in vitro. Recombinant, His-tagged SET3p was purified from Escherichia coli and used in a methylation assay with a mix of calf thymus histones or purified histone H3 as the substrates. When SET3p was incubated with histones in the presence of 14C-SAM, as the methyl donor, a 17-kDa band corresponding to methylated histone H3 was readily detected by phosphor imager scanning (Figure 3A). On the contrary, no signal was observed when core histones were incubated with an empty vector protein extract.
We next examined the histone H3 lysine residue that was targeted by the SET3p activity. We carried out in vitro HMTase assays using as substrates the histone H3 N-terminal tail (amino acids 1–52) with the wild-type sequence (H3N) or with mutations to arginine in specific lysines (K4R, K9R or K27R) or in the three lysine residues (3K-R = K4R, K9R and K27R). These polypeptides were expressed as GST fusions in E. coli and purified as recombinant proteins. When incubated in the presence of SET3p and 14C-SAM, H3N was readily methylated, whereas recombinant GST alone did not serve as a substrate (Figure 3B, lanes 1 and 2). The K4R and K27R fusions, carrying mutations in H3 lysine 4 or 27, were methylated as well as H3N (Figure 3B, lanes 3 and 5). Conversely, mutation of H3 lysine 9 (K9R) or of the three lysine residues (3K-R) completely eliminated methylation by SET3p (Figure 3B, lanes 4 and 6). Thus, SET3p appears to have HMTase activity specific for lysine 9 of histone H3.
Several residues within the SET domain have been shown to be important for the degree of methylation carried out by an HMTase, with the Phe/Tyr switch being the most extensively characterized (51). The SET domains of proteins with di- and/or trimethylating activity generally posses a Phe residue, corresponding to positions 281 in Neurospora crassa Dim-5 and 943 in human G9a (Figure 3C). In contrast, a Tyr residue at this position inhibits trimethylation and Phe-to-Tyr mutants of both Dim-5 and G9a show altered specificity behaving as mono- or dimethyltransferases (51,52). SET3p contains a Tyr in the position corresponding to the Phe/Tyr switch (Figure 3C), suggesting that it might be restricted in its capacity to carry out trimethylation. To address this question, we incubated SET3p with recombinant (unmethylated) histone H3 in the presence of unlabeled SAM and proceeded to detect the degree of SET3p-catalyzed H3K9 methylation by immunoblotting with antibodies raised specifically against mono-, di- or trimethyl H3K9. Interestingly, SET3p carried out monomethylation of H3K9, which reached a plateau after 2–3 h of incubation (Figure 3D). In contrast, neither di- nor trimethylation of H3K9 could be detected, even after 21 h of incubation. These observations, taken together, indicated that SET3p behaves in vitro as an exclusive H3K9 monomethyltransferase.
SET3p is responsible for H3K9 monomethylation, but not H3K9 trimethylation, in vivo
Given its in vitro role as a H3K9 HMTase, we next examined whether SET3p affected this modification in vivo. Immunoblots of extracts enriched in chromatin proteins, probed with antibodies raised against the different forms of methylated H3K9, revealed the presence of H3K9me1 and H3K9me3 in Chlamydomonas (Figure 4A). In contrast, as reported earlier (42), dimethyl H3K9 was undetectable even though the anti-H3K9me2 antibody was capable of recognizing this modification in calf thymus histone H3 (Figure 3D, control). Interestingly, significantly lower levels of monomethyl H3K9 were observed in the strains undergoing suppression of SET3 expression, whereas trimethyl H3K9 remained unchanged, in comparison with the parental 1-P[300] strain (Figure 4A). Global H3K9me1 and H3K9me3 levels in the control Maa7-IR3 strain were similar to those in the 1-P[300] strain (Figure 4A). Hence, in agreement with the in vitro data, Chlamydomonas SET3p seems to be at least partly responsible for global H3K9 monomethylation in vivo, whereas H3K9me3 depends on another yet unidentified HMTase.
Figure 4.
Monomethyl H3K9 is dependent on SET3p and is associated with the transcriptionally silenced RbcS2:aadA:RbcS2 transgenes. (A) Immunoblot analysis of in vivo H3K9 methylation states. Partially purified chromatin proteins from the indicated strains were separated by SDS-PAGE, transferred to nitrocellulose and probed with antibodies raised against mono-, di- or trimethyl H3K9. Sample loading was calibrated based on immunoblots with an anti-H3, modification-insensitive antibody. Numbers below the panels indicate relative levels of a specific histone modification normalized to the histone H3 amount. (B) Association of mono- and trimethyl H3K9 with the silent RbcS2:aadA:RbcS2 transgenes and the transcriptionally active RPS3 gene. Sequential ChIP assays were performed on TAP-grown cells of the indicated strains using an antibody against histone H3, and then antibodies against monomethyl H3K9, trimethyl H3K9 or rabbit IgG (negative control). Immunoprecipitated DNA was examined by real-time PCR. For illustration purposes, enrichment was calculated relative to the anti-H3 co-immunoprecipitated DNA and then normalized to the level of monomethyl H3K9 associated with the RbcS2:aadA:RbcS2 transgenes in the 1-P[300] strain. Results represent the mean ± SD of three independent experiments.
Monomethyl H3K9 is associated with the chromatin of silenced, multiple-copy transgenes
To determine if the loss of H3K9me1 in the Set3-IR strains was directly associated with reactivation of the silenced RbcS2:aadA:RbcS2 transgenes, we analyzed their chromatin environment by sequential ChIP assays. By using primers specific for the 5′ end of the coding sequence of the RbcS2:aadA:RbcS2 transgene and real-time PCR, we found that monomethyl H3K9 was highly enriched in the silenced, multiple-copy transgenes (Figure 4B). In contrast, H3K9me1 was virtually absent from an equivalent region in the constitutively expressed RPS3 gene (encoding ribosomal protein S3) (Figure 4B). ChIP analysis also revealed a substantial decrease in monomethyl H3K9 associated with the RbcS2:aadA:RbcS2 transgenes in the Set3-IR2 strain, as compared with the parental 1-P[300] strain (Figure 4B). Unexpectedly, trimethyl H3K9 was almost undetectable in the RbcS2:aadA:RbcS2 transgenes (independently of their transcriptional state), whereas low levels of H3K9me3 were observed in the transcriptionally active RPS3 gene. Furthermore, no differences in H3K9me3 signals were discernable between the Set3-IR2 and the 1-P[300] strains at the tested loci. Thus, in Chlamydomonas, monomethyl H3K9 may function as an epigenetic mark for the silenced chromatin, presumably heterochromatic-like, typical of tandem transgenes. Conversely, trimethyl H3K9 was not associated with the repressed RbcS2:aadA:RbcS2 transgenes and the low levels detected on the active RPS3 gene suggest that this modification may be present in euchromatic domains.
CpG DNA methylation of the RbcS2:aadA:RbcS2 transgenes is increased in strains experiencing RNAi-mediated suppression of SET3
Since in some species cytosine methylation appears to be dependent on H3K9 methylation (4,23,27,29) and direct interaction between H3K9 HMTases and DNA methyltransferases has been demonstrated in mammals (53), we also studied the effect of SET3 downregulation on the DNA methylation of the RbcS2:aadA:RbcS2 transgenes. The methylation-sensitive isoschizomers HpaII and MspI recognize the same DNA sequence (5′-CCGG-3′), but HpaII is inhibited by methylation of either cytosine, whereas MspI is only sensitive to methylation of the outer cytosine residue. Thus, digestion of an unmethylated RbcS2:aadA:RbcS2 transgene with these enzymes and HindIII generates a fragment of 530 bp and three fragments smaller than 160 bp that can be detected by hybridization with a probe encompassing the aadA coding sequence (Figure 5A). In contrast, if some of the HpaII/MspI sites become methylated, the inability of the enzymes to cleave will result in the appearance of DNA fragments of higher molecular weight. By using this approach, we observed that the multiple copies of the RbcS2:aadA:RbcS2 transgene were nearly fully digested with MspI but only partly cleaved with HpaII in the 1-P[300] parental strain (Figure 5B), suggesting the presence of CpG DNA methylation in the aadA coding sequence and, possibly, in the upstream RbcS2 promoter. The almost complete digestion with MspI indicated that CpNpG methylation (at least in a CCG context) is virtually absent from the RbcS2:aadA:RbcS2 transgenes. Interestingly, in the Set3-IR strains we detected a noticeable decrease in the cleaving capability of HpaII, indicative of a higher degree of CpG DNA methylation in comparison to the 1-P[300] strain, whereas the patterns of MspI digestion remained unchanged (Figure 5B). As a control, Southern hybridization analysis of the same blots with a psbA (encoding a Photosystem II component) probe showed that the chloroplast DNA was equally digested in all lanes, substantiating that differences in aadA restriction patterns were not due to defective enzymatic activity in some samples. Similar results were observed with other methylation sensitive restriction enzymes, such as HhaI and ScrFI (data not shown). Thus, RNAi-mediated downregulation of SET3 appears to result in increased levels of CpG DNA methylation associated with the RbcS2:aadA:RbcS2 transgenes.
Figure 5.
Effect of SET3 suppression on the DNA methylation of the RbcS2:aadA:RbcS2 transgenes. (A) Schematic representation of the plasmid containing the RbcS2:aadA:RbcS2 transgene. The SacI restriction site, used to linearize the vector prior to transformation, and the probe used for Southern blot analyses are shown. H, HpaII/MspI restriction sites. (B) Total DNA of the indicated strains was digested with HindIII/MspI or HindIII/HpaII, separated by agarose gel electrophoresis, transferred to a nylon membrane and hybridized sequentially with probes corresponding to the aadA coding sequence (Top panel) or the 3′ end of the chloroplast psbA gene (Bottom panel). DNA size markers in kilobases are indicated on the left.
DISCUSSION
SUVHs are conserved proteins directly implicated in chromatin-mediated silencing in a variety of eukaryotes. Mammalian SUV39H1 and G9a act as transcriptional repressors in reporter-based transient expression assays (54,55) and fission yeast Clr4 is a key component in the silencing of transgenes integrated into heterochromatic regions (22). Arabidopsis mutants defective in SUVH4 were repeatedly isolated as suppressors of silencing of certain alleles of the SUPERMAN and PAI2 genes (27,28); and SUVH2 was shown to affect the transcriptional repression of LUCIFERASE transgenic repeats in a dosage-dependent manner (8). Consistent with its relatedness to plant SUVH proteins, Chlamydomonas SET3p is also involved in the transcriptional silencing of tandemly repeated transgenes. However, the expression of single-copy transgenes and dispersed endogenous transposons, previously shown to be repressed by euchromatic histone marks (42), was not affected by RNAi-mediated suppression of SET3. Although, we cannot rule out that partial downregulation of SET3 may have not reduced the corresponding protein to a level low enough to compromise the silencing of euchromatic sequences. Nonetheless, in a number of species, transgenic repeat arrays appear to become silenced by condensation into heterochromatin (8,48,49,50), an intrinsic property of tandem arrays not attributable to position effects of nearby sequences (56,57). Indeed, it has been proposed that this form of repression may reflect a genomic defense response against invading foreign sequences such as transposable elements (56). Likewise, we speculate that concatameric transgenes in Chlamydomonas may be characterized by a (hetero)chromatic configuration that is at least partly dependent on SET3p activity. Although, the current lack of sequence data on heterochromatic Chlamydomonas repeats prevented us from testing more directly whether SET3p is required for the maintenance of natural heterochromatin.
The role of SU(VAR)3-9 proteins in the organization of transcriptionally repressive chromatin is mediated, at least in part, by their specific H3K9 methyltransferase activity (8,10,58). However, the ε-amino group of H3K9 can be mono-, di- or trimethylated, thereby adding to the coding complexity of this particular histone modification. In mammals, as already mentioned, pericentric heterochromatin is characterized by H3K9me3 and this modification is almost entirely eliminated in suv39h1/suv39h2 double mutants, where H3K9me1 becomes predominant (4–6,9). Based on these and other observations it has been proposed that SUV39H-mediated methylation of heterochromatin is primed by an unidentified H3K9 monomethylating HMTase (59). In wild-type mammalian cells, mono- and dimethyl H3K9 are mostly found in euchromatin (4–6,9,12,32). Monomethylated forms of H3K9 and H4K20 partition together into discrete nuclear compartments that have been proposed to represent silent chromatin states, functionally different from those characterized by the di- and/or trimethylated forms (32). This organization of chromosomes into domains with distinct H3K9 methylation states has also been observed in plants (7,8,11,27). Moreover, isotope-labeling of mammalian cells to detect half lives and histone exchange rates revealed differential stability of the H3K9 methylation states: fast turnover for H3K9me1, intermediate turnover for H3K9me2 and very slow turnover for H3K9me3 (60). Thus, different degrees of H3K9 methylation may be functionally relevant. Yet, the biological significance of monomethyl H3K9 has remained unexplained because of the lack of mutants exclusively defective in this modification.
Interestingly, Chlamydomonas SET3p behaved as an exclusive H3K9 monomethyltranferase in vitro and RNAi-mediated downregulation of SET3 significantly reduced global H3K9me1 levels in vivo. ChIP assays demonstrated that H3K9me1 was specifically associated with silent, multiple-copy transgenes. Moreover, the reactivation of these transgenes in the SET3 RNAi strains correlated with the partial loss of monomethyl H3K9 from their chromatin. Somewhat surprisingly, we were unable to detect dimethyl H3K9, an abundant H3K9 modification in Arabidopsis (61), in whole-cell extracts or partially purified histone extracts from Chlamydomonas, indicating that this mark is either absent or present at very low levels in this alga. This is in agreement with an earlier report that examined bulk histone H3 modifications in Chlamydomonas by protein sequencing (62). On the other hand, trimethyl H3K9 was readily observable but independent of the SET3p activity. In addition, H3K9me3 was virtually absent from the silenced multiple-copy transgenes, although it was associated at low levels with the transcriptionally active RPS3 gene. In higher plants, reports on the presence and distribution of trimethyl H3K9 have been somewhat contradictory (8,30,61) but recent observations suggest that it may be enriched in euchromatin (8,11). Conversely, monomethyl H3K9 is found preferentially in heterochromatin both in Arabidopsis and maize (7,8,11). Our findings in Chlamydomonas are also consistent with a role for H3K9me1 in indexing silent, presumably (hetero)chromatic domains, which may be typical of tandemly repeated sequences (56,57). Thus, our results, taken together, implicate SET3p as the first exclusive H3K9 monomethyltransferase and provide direct evidence that H3K9me1 may function as an epigenetic mark for repressive chromatin.
In several organisms, heterochromatin is also characterized by high levels of methylation of the underlying DNA (64,65). In plants, cytosine methylation is found predominantly at symmetric CpG and CpNpG sequences and at a lower frequency at asymmetric CpNpN sites (where N = A, T or C) (66). In Arabidopsis, CpG methylation appears to direct H3K9me2 to silent chromosomal regions (64,67). Conversely, several SUVH proteins seem to control the deposition of non-CpG methylation, mediated by the CMT3 chromomethylase, at distinct loci (27–29,31). Likewise, in Neurospora, the H3K9 HMTase Dim-5 is required for DNA methylation (23). In mammals, H3K9 methylation and CpG methylation show a complex interplay, perhaps as part of a self-reinforcing loop for repressive chromatin (53,68). The suv39h1/suv39h2 double mutant cells display an altered DNA methylation profile at pericentric satellite repeats, but not at other repetitive sequences (68); whereas mouse embryonic stem cells deficient in maintenance or de novo DNA methyltransferases show no alteration in H3K9 methylation at several repeats (69). In green algae, cytosine methylation appears to be restricted to the CpG sequence (35,70) and a possible relationship between DNA methylation and H3K9 methylation had not been previously examined. To our surprise, we detected an increase in DNA methylation of the reactivated RbcS2:aadA:RbcS2 transgenes in the SET3 RNAi strains. This suggests that CpG methylation in Chlamydomonas is not dependent on the H3K9me1 mark. Indeed, we speculate that a reduction in H3K9 monomethylation may result in an opening of the chromatin structure and increased accessibility of the DNA to CpG methyltransferases and/or to other factors that control DNA methylation. Transgene reactivation in this context is likely the net result of a reduction in (hetero)chromatic structure, due to suppression of SET3p activity, partly counteracted by an increase in DNA-methylation-dependent silencing.
With the present results all H3K9 methylation states have now been implicated in the establishment and/or maintenance of repressive chromatin, yet their precise roles remain unclear since their distribution in nuclear domains is species specific. Moreover, H3K9me3 has also been found associated with active genes in some species (this work and 63). The different H3K9 methylation states are in all likelihood bound by specific chromatin proteins that determine the higher order levels of chromatin organization. For instance, HETEROCHROMATIN PROTEIN1 (HP1), a key factor for heterochromatin formation in Drosophila, Neurospora and mammals, is partly localized to specific nuclear domains by preferential binding to H3K9me3 (and with lower affinity to H3K9me2); however it will not recognize chromatin characterized by H3K9me1 (65,71,72). Interestingly, in Arabidopsis, characterized by H3K9me1 and H3K9me2 in its heterochromatin, the putative HP1 homolog (LIKE HETEROCHROMATIN PROTEIN1) appears to be dispensable for heterochromatic silencing (28,73). Thus, there must be differences in the molecular mechanisms of recognition and interpretation of epigenetic histone modifications that ultimately produce the same functional outcome (for instance a silent heterochromatic domain) from varying methylation states in different species. Within this context, how H3K9me1 is recognized and transduced into a repressive chromatin domain, or whether it has other roles, remains to be explored. Furthermore, the cumulative evidence from multiple eukaryotes (10,59) strongly supports the notion that a combination of several histone lysine methylation marks (and probably other histone modifications as well as DNA methylation), rather than a single epigenetic mark, is required to discriminate active from inactive chromatin.
ACKNOWLEDGEMENTS
We are grateful to D. Rokhsar and Joint Genome Institute scientists for allowing access to the Chlamydomonas genome sequence prior to publication and to members of the Cerutti's lab for helpful discussions. This work was supported in part by a grant from the National Science Foundation (MCB-0544448). Funding to pay the Open Access publication charge was provided by the National Science Foundation.
Conflict of interest statement. None declared.
References
- 1.Kornberg RD, Lorch Y. Chromatin and transcription: where do we go from here. Curr. Opin. Genet. Dev. 2002;12:249–251. doi: 10.1016/s0959-437x(02)00293-9. [DOI] [PubMed] [Google Scholar]
- 2.Farkas G, Leibovitch BA, Elgin SC. Chromatin organization and transcriptional control of gene expression in Drosophila. Gene. 2000;253:117–136. doi: 10.1016/s0378-1119(00)00240-7. [DOI] [PubMed] [Google Scholar]
- 3.Sims RJ, III, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Lachner M, Sengupta R, Schotta G, Jenuwein T. Trilogies of histone lysine methylation as epigenetic landmarks of the eukaryotic genome. Cold Spring Harb. Symp. Quant. Biol. 2004;69:209–218. doi: 10.1101/sqb.2004.69.209. [DOI] [PubMed] [Google Scholar]
- 5.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 6.Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 7.Mathieu O, Probst AV, Paszkowski J. Distinct regulation of histone H3 methylation at lysines 27 and 9 by CpG methylation in Arabidopsis. EMBO J. 2005;24:2783–2791. doi: 10.1038/sj.emboj.7600743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naumann K, Fischer A, Hofmann I, Krauss V, Phalke S, Irmler K, Hause G, Aurich AC, Dorn R, et al. Pivotal role of AtSUVH2 in heterochromatic histone methylation and gene silencing in Arabidopsis. EMBO J. 2005;24:1418–1429. doi: 10.1038/sj.emboj.7600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu R, Terry AV, Singh PB, Gilbert DM. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol. Biol. Cell. 2005;16:2872–2881. doi: 10.1091/mbc.E04-11-0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebert A, Lein S, Schotta G, Reuter G. Histone modification and the control of heterochromatic gene silencing in Drosophila. Chromosome Res. 2006;14:377–392. doi: 10.1007/s10577-006-1066-1. [DOI] [PubMed] [Google Scholar]
- 11.Shi J, Dawe RK. Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics. 2006;173:1571–1583. doi: 10.1534/genetics.106.056853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zinner R, Albiez H, Walter J, Peters AH, Cremer T, Cremer M. Histone lysine methylation patterns in human cell types are arranged in distinct three-dimensional nuclear zones. Histochem. Cell Biol. 2006;125:3–19. doi: 10.1007/s00418-005-0049-1. [DOI] [PubMed] [Google Scholar]
- 13.Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, et al. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- 15.Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3–9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, et al. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3–9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aagaard L, Schmid M, Warburton P, Jenuwein T. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J. Cell Sci. 2000;113:817–829. doi: 10.1242/jcs.113.5.817. [DOI] [PubMed] [Google Scholar]
- 18.Schotta G, Reuter G. Controlled expression of tagged proteins in Drosophila using a new modular P-element vector system. Mol. Gen. Genet. 2000;262:916–920. doi: 10.1007/pl00008659. [DOI] [PubMed] [Google Scholar]
- 19.Baumbusch LO, Thorstensen T, Krauss V, Fischer A, Naumann K, Assalkhou R, Schulz I, Reuter G, Aalen RB. The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 2001;29:4319–4333. doi: 10.1093/nar/29.21.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen WH. NtSET1, a member of a newly identified subgroup of plant SET-domain-containing proteins, is chromatin-associated and its ectopic overexpression inhibits tobacco plant growth. Plant J. 2001;28:371–383. doi: 10.1046/j.1365-313x.2001.01135.x. [DOI] [PubMed] [Google Scholar]
- 21.Springer NM, Napoli CA, Selinger DA, Pandey R, Cone KC, Chandler VL, Kaeppler HF, Kaeppler SM. Comparative analysis of SET domain proteins in maize and Arabidopsis reveals multiple duplications preceding the divergence of monocots and dicots. Plant Physiol. 2003;132:907–925. doi: 10.1104/pp.102.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivanova AV, Bonaduce MJ, Ivanov SV, Klar AJ. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat. Genet. 1998;19:192–195. doi: 10.1038/566. [DOI] [PubMed] [Google Scholar]
- 23.Tamaru H, Selker EU. A histone H3 methyltransferase controls DNA methylation in Neurospora. crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- 24.Yu Y, Dong A, Shen WH. Molecular characterization of the tobacco SET domain protein NtSET1 unravels its role in histone methylation, chromatin binding, and segregation. Plant J. 2004;40:699–711. doi: 10.1111/j.1365-313X.2004.02240.x. [DOI] [PubMed] [Google Scholar]
- 25.Shen WH, Meyer D. Ectopic expression of the NtSET1 histone methyltransferase inhibits cell expansion, and affects cell division and differentiation in tobacco plants. Plant Cell Physiol. 2004;45:1715–1719. doi: 10.1093/pcp/pch184. [DOI] [PubMed] [Google Scholar]
- 26.Casas-Mollano JA, Lao NT, Kavanagh TA. Intron-regulated expression of SUVH3, an Arabidopsis Su(var)3–9 homologue. J. Exp. Bot. 2006;57:3301–3311. doi: 10.1093/jxb/erl093. [DOI] [PubMed] [Google Scholar]
- 27.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 28.Malagnac F, Bartee L, Bender J. An Arabidopsis SET domain protein required for maintenance but not establishment of DNA methylation. EMBO J. 2002;21:6842–6852. doi: 10.1093/emboj/cdf687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ebbs ML, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell. 2006;18:1166–1176. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jackson JP, Johnson L, Jasencakova Z, Zhang X, Perez-Burgos L, Singh PB, Cheng X, Schubert I, Jenuwein T, et al. Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma. 2004;112:308–315. doi: 10.1007/s00412-004-0275-7. [DOI] [PubMed] [Google Scholar]
- 31.Ebbs ML, Bartee L, Bender J. H3 lysine 9 methylation is maintained on a transcribed inverted repeat by combined action of SUVH6 and SUVH4 methyltransferases. Mol. Cell Biol. 2005;25:10507–10515. doi: 10.1128/MCB.25.23.10507-10515.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sims JK, Houston SI, Magazinnik T, Rice JC. A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J. Biol. Chem. 2006;281:12760–12766. doi: 10.1074/jbc.M513462200. [DOI] [PubMed] [Google Scholar]
- 33.Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorstensen T, Fischer A, Sandvik SV, Johnsen SS, Grini PE, Reuter G, Aalen RB. The Arabidopsis SUVR4 protein is a nucleolar histone methyltransferase with preference for monomethylated H3K9. Nucleic Acids Res. 2006;34:5461–5470. doi: 10.1093/nar/gkl687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cerutti H, Johnson AM, Gillham NW, Boynton JE. A eubacterial gene conferring spectinomycin resistance on Chlamydomonas reinhardtii: integration into the nuclear genome and gene expression. Genetics. 1997;145:97–110. doi: 10.1093/genetics/145.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jeong B-R, Wu-Scharf D, Zhang C, Cerutti H. Suppressors of transcriptional transgenic silencing in Chlamydomonas are sensitive to DNA-damaging agents and reactivate transposable elements. Proc. Natl. Acad. Sci. U.S.A. 2002;99:1076–1081. doi: 10.1073/pnas.022392999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohr J, Sarkar N, Balenger S, Jeong B-R, Cerutti H. Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 2004;40:611–621. doi: 10.1111/j.1365-313X.2004.02227.x. [DOI] [PubMed] [Google Scholar]
- 38.Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 41.Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;2:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 42.van Dijk K, Marley KE, Jeong B-R, Xu J, Hesson J, Cerny RL, Waterborg JH, Cerutti H. Monomethyl histone H3 lysine 4 as an epigenetic mark for silenced euchromatin in Chlamydomonas. Plant Cell. 2005;17:2439–2453. doi: 10.1105/tpc.105.034165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Griffin K, Mondal N, Parvin JD. Phosphorylation of histone H2A inhibits transcription on chromatin templates. J. Biol. Chem. 2004;279:21866–21872. doi: 10.1074/jbc.M400099200. [DOI] [PubMed] [Google Scholar]
- 44.Geisberg JV, Struhl K. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res. 2004;32:e151. doi: 10.1093/nar/gnh148. 10.1093/nar/gnh148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook J, Russell DW. Molecular Cloning – A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 46.Cheng X, Collins RE, Zhang X. Structural and sequence motifs of protein (histone) methylation enzymes. Annu. Rev. Biophys. Biomol. Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alvarez-Venegas R, Avramova Z. SET-domain proteins of the Su(var)3–9, E(z) and trithorax families. Gene. 2002;285:25–37. doi: 10.1016/s0378-1119(02)00401-8. [DOI] [PubMed] [Google Scholar]
- 48.Ye F, Signer ER. RIGS (repeat-induced gene silencing) in Arabidopsis is transcriptional and alters chromatin configuration. Proc. Natl. Acad. Sci. U.S.A. 1996;93:10881–10886. doi: 10.1073/pnas.93.20.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Probst AV, Fransz PF, Paszkowski J, Mittelsten Scheid O. Two means of transcriptional reactivation within heterochromatin. Plant J. 2003;33:743–749. doi: 10.1046/j.1365-313x.2003.01667.x. [DOI] [PubMed] [Google Scholar]
- 50.Wang F, Koyama N, Nishida H, Haraguchi T, Reith W, Tsukamoto T. The assembly and maintenance of heterochromatin initiated by transgene repeats are independent of the RNA interference pathway in mammalian cells. Mol. Cell Biol. 2006;26:4028–4040. doi: 10.1128/MCB.02189-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins RE, Tachibana M, Tamaru H, Smith KM, Jia D, Zhang X, Selker EU, Shinkai Y, Cheng X. In vitro and in vivo analyses of a Phe/Tyr switch controlling product specificity of histone lysine methyltransferases. J. Biol. Chem. 2005;280:5563–5570. doi: 10.1074/jbc.M410483200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X, Yang Z, Khan SI, Horton JR, Tamaru H, Selker EU, Cheng X. Structural basis for the product specificity of histone lysine methyltransferases. Mol. Cell. 2003;12:177–185. doi: 10.1016/s1097-2765(03)00224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fuks F, Hurd PJ, Deplus R, Kouzarides T. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31:2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Firestein R, Cui X, Huie P, Cleary ML. Set domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3–9. Mol. Cell Biol. 2000;20:4900–4909. doi: 10.1128/mcb.20.13.4900-4909.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snowden AW, Gregory PD, Case CC, Pabo CO. Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Curr. Biol. 2002;12:2159–2166. doi: 10.1016/s0960-9822(02)01391-x. [DOI] [PubMed] [Google Scholar]
- 56.Henikoff S. Conspiracy of silence among repeated transgenes. Bioessays. 1998;20:532–535. doi: 10.1002/(SICI)1521-1878(199807)20:7<532::AID-BIES3>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 57.Martienssen RA. Maintenance of heterochromatin by RNA interference of tandem repeats. Nat. Genet. 2003;35:213–214. doi: 10.1038/ng1252. [DOI] [PubMed] [Google Scholar]
- 58.Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jenuwein T. The epigenetic magic of histone lysine methylation. FEBS J. 2006;273:3121–3135. doi: 10.1111/j.1742-4658.2006.05343.x. [DOI] [PubMed] [Google Scholar]
- 60.Fodor BD, Kubicek S, Yonezawa M, O'Sullivan RJ, Sengupta R, Perez-Burgos L, Opravil S, Mechtler K, Schotta G, et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson L, Mollah S, Garcia BA, Muratore TL, Shabanowitz J, Hunt DF, Jacobsen SE. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 2004;32:6511–6518. doi: 10.1093/nar/gkh992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waterborg JH, Robertson AJ, Tatar DL, Borza CM, Davie JR. Histones of Chlamydomonas reinhardtii: synthesis, acetylation, and methylation. Plant Physiol. 1995;109:393–407. doi: 10.1104/pp.109.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 64.Soppe WJ, Jasencakova Z, Houben A, Kakutani T, Meister A, Huang MS, Jacobsen SE, Schubert I, Fransz PF. DNA methylation controls histone H3 lysine 9 methylation and heterochromatin assembly in Arabidopsis. EMBO J. 2002;21:6549–6559. doi: 10.1093/emboj/cdf657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 2004;5:296–304. doi: 10.1038/nrm1355. [DOI] [PubMed] [Google Scholar]
- 66.Gruenbaum Y, Naveh-Many T, Cedar H, Razin A. Sequence specificity of methylation in higher plant DNA. Nature. 1981;292:860–862. doi: 10.1038/292860a0. [DOI] [PubMed] [Google Scholar]
- 67.Tariq M, Saze H, Probst AV, Lichota J, Habu Y, Paszkowski J. Erasure of CpG methylation in Arabidopsis alters patterns of histone H3 methylation in heterochromatin. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8823–8827. doi: 10.1073/pnas.1432939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen T, Li E, Jenuwein T, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 2003;13:1192–200. doi: 10.1016/s0960-9822(03)00432-9. [DOI] [PubMed] [Google Scholar]
- 69.Martens JH, O'Sullivan RJ, Braunschweig U, Opravil S, Radolf M, Steinlein P, Jenuwein T. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 2005;24:800–812. doi: 10.1038/sj.emboj.7600545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Babinger P, Kobl I, Mages W, Schmitt R. A link between DNA methylation and epigenetic silencing in transgenic Volvox carteri. Nucleic Acids Res. 2001;29:1261–1271. doi: 10.1093/nar/29.6.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Freitag M, Hickey PC, Khlafallah TK, Read ND, Selker EU. HP1 is essential for DNA methylation in Neurospora. Mol. Cell. 2004;13:427–434. doi: 10.1016/s1097-2765(04)00024-3. [DOI] [PubMed] [Google Scholar]
- 73.Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, et al. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4286–4296. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]