Abstract
DNA mapping is an important analytical tool in genomic sequencing, medical diagnostics and pathogen identification. Here we report an optical DNA mapping strategy based on direct imaging of individual DNA molecules and localization of multiple sequence motifs on the molecules. Individual genomic DNA molecules were labeled with fluorescent dyes at specific sequence motifs by the action of nicking endonuclease followed by the incorporation of dye terminators with DNA polymerase. The labeled DNA molecules were then stretched into linear form on a modified glass surface and imaged using total internal reflection fluorescence (TIRF) microscopy. By determining the positions of the fluorescent labels with respect to the DNA backbone, the distribution of the sequence motif recognized by the nicking endonuclease can be established with good accuracy, in a manner similar to reading a barcode. With this approach, we constructed a specific sequence motif map of lambda-DNA. We further demonstrated the capability of this approach to rapidly type a human adenovirus and several strains of human rhinovirus.
INTRODUCTION
DNA mapping is an important analytical tool in genomic sequence assembly, medical diagnostics and pathogen identification (1–3). The current strategy for DNA mapping is based on sizing DNA fragments generated by enzymatic digestion of genomic DNA with restriction endonucleases. Many techniques have been developed for DNA fragment sizing, including pulsed field gel electrophoresis (4), capillary and microchannel electrophoresis (5,6) and flow cytometry (7). Gel electrophoresis-based restriction mapping is the most widely used DNA mapping technique and has played an important role in the human genome project by fingerprinting BAC clones to build BAC contiqs (8). More recently, a high-throughput DNA fingerprinting system based on the use of fluorescently labeled terminators had been developed for a number of new applications (9).
The DNA molecular combing and optical mapping techniques, which interrogate multiple sequence sites on single DNA molecules deposited on a glass surface (10,11), not only provide the location of restriction sites and the length of the restriction fragments, but also preserve the order of the restriction sites within the DNA molecule. The DNA molecular combing technique has been used to map the human genome and to detect disease-related mutations (10). In addition, optical restriction maps for several microbial genomes have been constructed successfully (12). More recently, a DNA direct linear analysis (DLA) technique has been developed, in which a long double-stranded DNA (dsDNA) molecule was tagged at specific sequence sites with fluorescent dyes and stretched into linear form as it flowed through a microfluidic channel. In the DLA approach, the sequence motif map is obtained by recording the fluorescent tags while the DNA molecules pass through the detectors placed above the microchannel (13,14). These technologies demonstrate the power of single molecule mapping for genomic applications.
In this report, we describe a DNA mapping method based on direct imaging of individual DNA molecules and localization of multiple sequence motifs on these molecules. Our method starts with introducing nicks in dsDNA at specific sequence motifs recognized by nicking endonucleases, which cleave only one strand of a dsDNA substrate (15). DNA polymerase then incorporates fluorescent dye-terminators at these sites. The labeled DNA molecules are stretched into linear form on a modified glass surface and individually imaged using multicolor total internal reflection fluorescence (TIRF) microscopy, a technique capable of localizing single fluorescent dye molecules with nanometer-scale accuracy (16). By determining the positions of the fluorescent labels along the DNA backbone, the distribution of the sequence motifs recognized by the nicking endonuclease can be established with great accuracy, in a manner similar to reading a barcode (17) (Figure 1). With this approach, we constructed sequence motif maps of lambda-phage, a strain of human adenovirus and several strains of human rhinoviruses. Because of the simplicity of this mapping strategy (single DNA molecule analysis, high accuracy and potential of high-throughput), it will likely find applications in DNA mapping, medical diagnostics and especially in rapid identification of microbial pathogens.
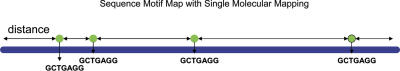
Figure 1.
Sequence motif map based on nick-labeling of Nb.BbvC I sites (GCTGAGG) together with measurement of distances between the sites. The nicking sites are labeled with a green fluorescent dye while the DNA backbone is labeled with a blue dye for TIRF microscopic imaging.
MATERIALS AND METHODS
DNA sample preparation
Lambda-DNA was purchased from New England Biolabs (Ipswich, MA). Adenovirus type 2 DNA was obtained from Invitrogen (Carlsbad, CA). Micrococcal nuclease-treated viral culture supernatants of the human rhinovirus (HRV) strains 14, 15, 28, 36, 73 and 89 were obtained from the DeRisi group (University of California - San Francisco, San Francisco, CA). Total RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA) and was then prepared with a standard ethanol precipitation protocol. The total RNA was resuspended in 50 μl nuclease-free water to a concentration of ∼30 ng/μl and 300 ng of total RNA was used in the first-strand cDNA synthesis step. Following the vendor's recommended protocol, 1 μl of the oligo(dT)20 primer was used with the SuperScript III RT mix in a 20 μl reaction: 1 μl of 10 mM dNTP mix, 4 μl of 5× first-strand buffer, 1 μl of 0.1 M DTT, 1 μl of RNaseOUT and 2 μl (400 U) of the SuperScript III RT (all from Invitrogen, Carlsbad, CA). After incubating at 50°C for 1 h and 70°C for 15 min, 2 U of RNase H (Promega, Madison, WI) was added and incubated at 37°C for 20 min. The reaction mix served as the template in the long-range PCR without purification.
The Eppendorf TripleMaster PCR system was used to generate the double-stranded HRV DNA (Eppendorf, Westbury, NY). A 10 μl aliquot of the above cDNA mix was added to the reaction mix according to the manufacturer's protocol. The nucleotide sequences of the forward and reverse primers for strain 14 are 5′-GGATCTCACGAAAATCAAAACA-3′ and 5′-TTTTCTCCTGAGTGCCATGC-3′, respectively; and for strain 89 are 5′-TGGGACACACTCCACACAAA-3′ and 5′-CATGTCTACCATTATGCCACA-3′. Steps for the thermalcycler are as follows: 93°C for 5 min, 93°C for 15 s, 60°C (63°C for strain 89) for 30 s, 68°C for 10 min and repeat steps 2–4 for 29 cycles.
The size of the PCR products was checked with 0.5% agarose gel electrophoresis.
Tagging sequence motif with fluorescent dye molecules
Both the lambda-DNA and nicking enzyme NBbvc1 were obtained from New England Biolabs (Ipswich, MA). The template-directed incorporation (TDI) reaction was performed utilizing cy3-acyclo-T dye, 10× Acyclo buffer and AcycloPol enzyme from Perkin Elmer (Boston, MA).
Lambda-or adenovirus-DNA was diluted to 50 ng/μl for use in the nicking reaction. 170 μl of lambda-DNA (50 ng/μl) were added to a 0.2 ml PCR centrifuge tube followed by 20 μl of 10× NE buffer no. 2 and 10 μl of Nb.BbvC1 enzyme. The mixture was incubated at 37°C for 1 h. After the nicking reaction is completed, the dye mix (consisted of 15 μl of nicking product and 5 μl of dye mix containing 2 μl of 10× Acyclo buffer, 0.5 μl of AcycloPol enzyme, 1 μl cy3-acyclo-T dye and 1.5 μl of water) was added to the mixture. The TDI reaction mixture was incubated at 55°C for 30 min. The labeled DNA molecules of lambda and adenovirus were used directly without purification. 8 μl HRV PCR products were used in the nicking reaction and labeled products were imaged without purification.
Preparation of the glass coverslips and DNA mounting
This protocol was adapted from Braslavsky et al. and Kartalov et al., with some modifications (18,19). Briefly, Fisher premium coverslips (22 × 30 mm2) were sonicated in 2% MICRO-90 soap (Cole-Parmer, Vernon Hills, IL) for 20 min and then cleaned by boiling in RCA solution (6:4:1 high-purity H2O/30% NH4OH/30% H2O2) for 1 hour. Poly(allylamine) (PAll) and Poly(acrylic acid) (PAcr) (Sigma-Aldrich, St Louis, MO) were dissolved at 2 mg/ml in high-purity water. The solutions were adjusted to pH 8.0 using HCl and NaOH. The polyelectrolyte solutions were then passed through a 0.22 μm filter. The RCA-cleaned coverslips were immersed in the positive (PAll) and the negative (PAcr) polyelectrolytes according to the scheme +/wash/−/wash/+/wash. Each polyelectrolyte incubation was 30 min on a shaker at 150 r.p.m. and 35°C, and each wash step involved rinsing with high-purity water three times. The polyelectrolyte-coated coverslips were stored in high-purity water at room temperature.
DNA mounting for lambda- and adenovirus-DNA was performed by a procedure similar to the one published by the Schwartz group (20). DNA was diluted to ∼100 pM in imaging buffer (10 mM TRIS, 1 mM EDTA, 2 μM YOYO-1 and 20% 2-mercaptoethanol, pH 7.5). Glass slides were passed through a propane torch flame to remove impurities and moisture. A coated coverslip was placed on the glass slide, and 7 μl of diluted DNA was pipetted onto the edge. The solution was drawn under the coverslip by capillary action, resulting in a strong flow, which caused the DNA fragments to be stretched and aligned on the coverslip surface. The coverslip was sealed with clear nail polish (Revlon Extra Life Top Coat 950). The imaging buffer consisted of 300 pM YOYO-1 iodide (Molecular Probes, Eugene, OR) and 20% 2-mercaptoethanol in sterile TE buffer. YOYO-1 iodide is an intercalating dye that stains the DNA backbone and makes it possible to visualize the DNA. 2-Mercaptoethanol is a strong reducing agent that retards photobleaching of the YOYO-1 and cyanine dyes by scavenging oxygen from the solution.
Human rhinovirus DNA was stretched with a slightly modified protocol, which is more suitable for stretching shorter DNA fragments. A coated cover slip was placed over the glass slide with one edge touching the slide at an angle of 15°, and 7 μl of diluted DNA was pipetted onto the interface between slide and the cover slip. The solution was pushed between the cover slip and slide by dropping the cover slip onto the slide, resulting in a strong flow, which caused the DNA fragments to be stretched and aligned on the cover slip surface.
Total internal reflection microscopy
The total internal reflection fluorescence microscope was based on an Olympus IX-71 microscope (Olympus America Inc) with a custom-modified Olympus TIRFM Fiber Illuminator, and a 100× SAPO objective (Olympus SAPO 100×/1.40 oil) as shown in Figure S4 of the supplemental material. YOYO-1 was excited using 488 nm wide-field excitation from a mercury lamp mounted on the rear port of the TIRFM illuminator. The Cy3 labels were excited with 543-nm laser (JDS Uniphase, San Jose, CA), which were combined by a dichroic mirror and expanded to a 7-mm diameter beam. The 543-nm excitation was focused via objective-type TIR through a translation stage. The emitted photons were collected through two separate filter cubes which contained either emission filter HQ510LP for blue dye YOYO-1 or a dual-band pass emission filter (Z543-633M) for Cy3. The HQ510LP and Z543-633M emission filters were wedge matched to reduce pixel shift during filter cube switching. A polychroic mirror (Z543-633 RPC) was used in conjunction with the Z543-633M emission filter. All filters were obtained from Chroma Technology (Rockingham, VT). The image was magnified by a 1.6× tube lens and recorded by a back-illuminated, TE-cooled, frame-transfer charge-coupled device (CCD) detector BV887 (Andor, Ireland). Both the DNA backbone and the cy3 label images were integrated for 1 s. Each dye was illuminated separately in sequence, and the series of two images formed a single image set.
DATA ANALYSIS
Custom written software in IDL (Research Systems Inc., Boulder, CO) was used for data analysis. First, raw images were combined into a false color two-channel composite image, and DNA fragments with adequate stretching and labeling were selected and extracted for analysis. After a DNA fragment was extracted, the contour of the backbone was computed. The fragment boundaries were determined by threshold segmentation. Depending on the orientation of the molecule, the image was then split into either horizontal or vertical ‘strips’ of pixels. Each strip was fit to a Gaussian distribution plus an offset, and the backbone contour was determined by the line joining the centroids of each distribution.
The dye-terminator labels in the green channels were then individually localized either by fitting to Gaussian point-spread functions (PSFs), or in cases where the signal-to-noise ratio was too low, the centroid was determined by finding the center of mass of the intensity distribution. The label position was then mapped to the closest point on the DNA backbone contour. The label position, in terms of base pairs, was then computed as the distance ratio of full-length along the contour from top right end of the DNA molecules.
Once the label positions have been determined, it is necessary to determine the orientation of the DNA molecules to construct the map. Each individual label was matched to the closest known nicking site maintaining the observed linear order of the labels (lambda-DNA with at least three labels, and adenovirus type with at least four labels) with two orientations separately. The total standard N deviation was calculated S = ∑ d2i, where di is the distance between label i and i = 1 their closest matched known nicking sites, and N is the number of labels. The orientation with the smaller S was assigned as the DNA orientation. All the DNA molecules were then forced to align in one orientation to construct the sequence motif map.
RESULTS
Sequence specific labeling of dsDNA with fluorescent dyes
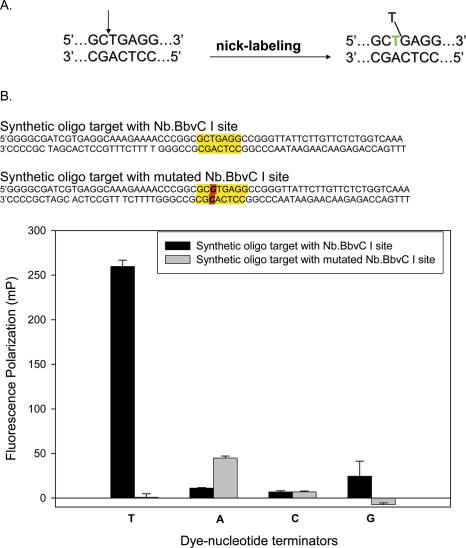
In the first set of experiments, we established the sequence specificity of the nicking endonuclease. Two synthetic DNA targets were prepared: one containing the recognition site (GCTGAGG) for the nicking endonuclease Nb.BbvCI and one with a mutated recognition site (GCGTGAGG). Figure 2A shows the recognition sequence motif and the nicking site of Nb.BbvCI, together with the dye-terminator incorporation scheme.
Figure 2.
Nick-labeling scheme. (A) Recognition sequence of nicking endonuclease Nb.BbvCI and the labeling scheme. After nicking (arrow), a fluorescent nucleotide terminator is incorporated at the nicking site (green T) as the native T is displaced. (B) The specificity of the nick-labeling scheme was demonstrated with synthetic oligos containing the Nb.BbvC I recognition sequence. In eight parallel experiments containing all possible combinations of synthetic oligos with or without the Nb.BbvC I site and the four dye-terminators, only the synthetic oligo with the Nb.BbvC I site was nick-labeled with the T-terminator, as monitored by a steep rise in fluorescence polarization value.
Dye-terminator incorporation was monitored by fluorescence polarization (FP). Because the FP value of a large fluorescent molecule is high while that of a small fluorescent molecule is low, the reaction mixture containing mainly free dye-terminators has a low FP value and that containing mainly incorporated dye-terminators has a high FP value. Figure 2B shows that only fluorescent Tamra-ddUTP-terminator was incorporated for the synthetic double-stranded oligos containing the GCTGAGG recognition sequence, with a substantial increase in FP to >200 mP, compared with FP value of 20 mP for reaction mixtures containing the other terminators. If an extra base G is inserted into the recognition sequence for the target oligos, none of the terminators were incorporated.
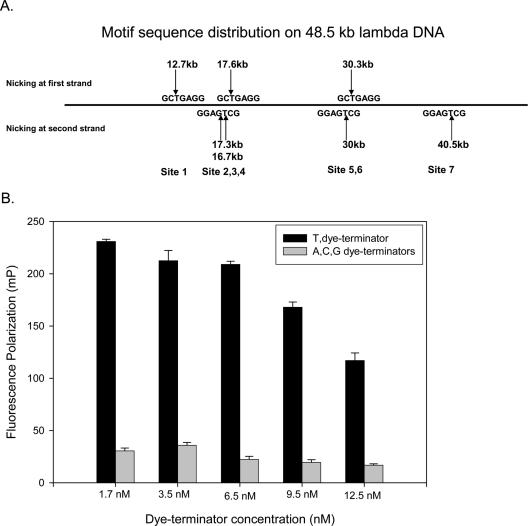
The labeling specificity and efficiency were further verified by titration of the nick-labeling of Tamra-ddUTP on a lambda-DNA substrate, which contains seven nicking sites as shown in Figure 3A. Different amounts of Tamra-ddUTP were added to a nick-labeling reaction containing 1 nM of lambda-DNA. The incorporation of Tamra-ddUTP was saturated at terminator concentrations of ≥6.5 nM (Figure 3B). As with the synthetic targets, none of the other fluorescent ddNTPs was incorporated, regardless of their concentration. The same results were obtained when the experiments were repeated with a different nicking endonuclease Nb.BsmI (data not shown). These results provide strong evidence that the nick-labeling scheme is very specific and efficient.
Figure 3.
The sequence motif distribution on lambda-DNA and nick-labeling. (A) The sequence motif distribution on lambda-DNA. There are seven Nb.BbvC I sites along the 48.5 kb lambda-DNA. (B) The results of titration of dye-terminator incorporation for 1 nM lambda-DNA after nicking. Only the T-dye-terminator was incorporated at the entire range of terminator concentrations. As monitored by fluorescence polarization, where high FP values correspond to high proportion of incorporated dye-terminator in the reaction, dye-terminator incorporation was complete at ≤6.5 nM of dye-terminators, showing that labeling occurred at all seven Nb.BbvC I sites.
Mapping of lambda-DNA
To show that single molecule detection of nick-labeled DNA can be used for DNA mapping, we used lambda-DNA as a model system to construct a sequence motif map. The distribution of the seven nick endonuclease Nb.BbvC I recognition sites of lambda-DNA are shown in Figure 4A. The solid black line represents the backbone of the lambda-DNA and the black arrow indicates the positions of the predicted Nb.BbvC I sites. The nick-labeling was done with Tamra-ddUTP (green) and the DNA backbone was stained with YOYO (blue). The labeled lambda-DNA was then stretched on a glass cover slip and imaged with TIRF microscopy (see materials and methods). Two images (with the green and blue channels) were taken and superimposed to produce a composite picture of the DNA molecules.
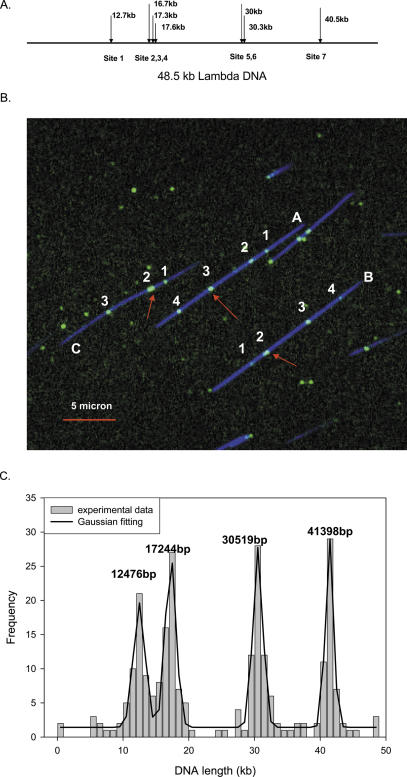
Figure 4.
Sequence motif map of lambda-DNA. (A) The predicted Nb.BbvC I map of lambda-DNA. Positions of the nicking sites are indicated by arrows. Nicking sites 2–4 and 5–6 are closely clustered and are not resolvable due to the limits of optical diffraction. (B) In the intensity scaled composite image of linear lambda-DNA, the Nb.BbvC I sites (labeled with Tamra-ddUTP) are shown as green spots and the DNA backbone (labeled with YOYO) is shown as blue lines. Due to the diffraction limits of the microscope, only four labels can be fully resolved. In this field, two DNA fragments (A and B) are fully labeled while one fragment (C) has three labels. Red arrows point to clustered sites, some of them are brighter than other because of the presence of multiple labels. (C) The sequence motif map in the bottom graph was obtained by analyzing 61 single molecule fluorescence images. The solid line is the Gaussian curve fitting and the peaks correspond well to the predicted locations of the sequence motif. The inset shows the labeling efficiency, 81 DNA molecules out of a total 112 DNA molecules have more than three labels.
Figure 4B is a false color two-channel composite image showing the stretched DNA contours (YOYO in blue) and labeled sites (Tamra-ddUTP in green). Three DNA molecules are nearly fully stretched (A, B and C) with contour lengths of 19.8, 19.5 and 16.9 μ, respectively. Although the data suggest that DNA molecules A and B are overstretched at 0.41 nm/bp, compared with the solution conformation of 0.34 nm/bp, this may be due to the effect of YOYO staining (21). The rest of the DNA molecules are either broken or folded back onto themselves, giving lengths much shorter than that predicted. There are also occasional Tamra dye signals (green) not associated with the DNA backbone. These are most likely the result of either fluorescent impurities on the coverslip or free Tamra-ddUTP. DNA fragments A and B in Figure 4B have four Tamra labels (green) along the DNA backbone, and DNA fragment C has three green labels. The signal for two of the green labels (red arrows) is much stronger and occupies more pixels than that corresponding to a single fluorescent dye, indicating that several green labels have clustered together and cannot be resolved due to light diffraction limits of the instrument, for example, one of the pixels in label 2 of DNA C shows maximum counts of 374, while the maximum count on label 1 is only 171. The two clusters most likely correspond to the predicted sites 2, 3 and 4 and sites 5 and 6, as they are separated by no more than 1000 bp. Accordingly, the seven Nb.BbvC I sites of lambda-DNA are collapsed to four resolvable sites, with the middle two signals stronger than the outer two signals. The distances between the labels were calculated with respect to the DNA backbone (see Materials and Methods) starting from the top right end of the DNA backbone. The positions of four green labels on DNA molecule A starting from the right end are 12.6, 17.9, 31.0 and 40.3 kb, respectively, but they are at 6.8, 17.4, 31.0 and 35.8 kb for DNA molecule B. Clearly, the two DNA molecules were in opposite orientation.
To obtain a robust map, DNA molecules that fulfill the following two criteria were selected for analysis. First, the labeled DNA fragments must be nearly fully stretched so that the label positions can be accurately determined. In the case of lambda-DNA, DNA molecules longer than 15 microns were used in the analysis. Second, DNA fragments must have at least three labels so that the relative distances between the labels can be used to establish the orientation of the DNA molecules (see Material and Methods). Figure 4C is the sequence motif map of lambda-DNA based on the analysis of 81 molecules that met the criteria we set. Four peaks were calculated as 12 476, 17 244, 30 519 and 41 398 bp from one end, with good agreement with the predicted distribution of sequence motif. The closest two peaks are ∼5 kb and they are clearly distinguishable. More than 70% of the fully stretched DNA molecules have more than three labels, indicating that the labeling efficiency is relatively high. By randomly selecting DNA molecules to construct the map, we determined that a minimal of 45 DNA molecules is needed to obtain good Gaussian fits and the pattern.
Applications in rapid identification of viral genomes
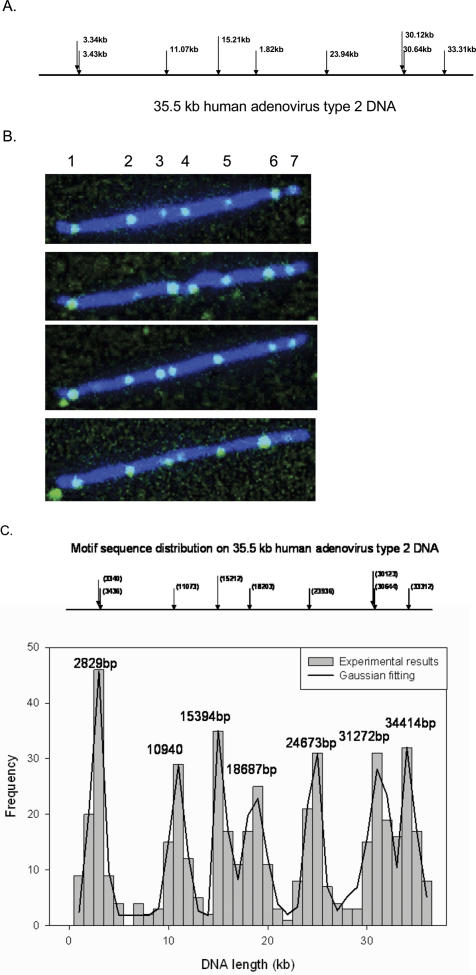
The sequence motif maps of human adenovirus and rhinovirus genomes were constructed to demonstrate the capability of our approach for rapid mapping and identification of pathogen genomes. The genome of human adenovirus type 2 is 35.5 kb in length and the distribution of Nb.BbvC I sites are shown in Figure 5A. The genome contains nine Nb.BbvC I sites, of which seven are resolvable. Figure 5B is a false color two-channel composite image showing several fully stretched DNA molecules (YOYO in blue) with all seven resolvable Nb.BbvC I sites labeled with Tamra-ddUTP (green). A total of 105 DNA molecules were used in constructing the sequence motif map shown in the bottom graph of Figure 5C. The seven peaks were found at 2.8, 10.9, 15.4, 18.7, 24.7, 31.3 and 34.4 kb from one end, compared with the expected position of 3.3, 11.1, 15.2, 18.2, 23.9, 30.1 and 33.3 kb. Based on previously published phylogenetic analyses of the genome sequences of the human adenovirus (22,23), virtual sequence motif maps of Nb.BbvC I sites of several human adenovirus genomes were constructed. Clearly, the Nb.Bbv C I maps of six major type (A–F) of human adenoviruses are quite different. Even the strains of closely related subtype can be quite easily distinguished. Therefore, one can distinguish the different viral strains by just comparing the map obtained experimentally with the known virtual sequence motif maps.
Figure 5.
Sequence motif map of human adenovirus type 2. (A) Predicted Nb.BbvC I map of human adenovirus type 2. Nine sites are found on the 35.5 kb viral DNA with two sets of clustered sites (1–2 and 7–8), leading to seven resolvable labels. (B) In the intensity scaled composite images of 4 fully labeled human adenovirus type 2 DNA, the Nb.Bbvc I sites (labeled with Tamra-ddUTP) are shown as green spots and the DNA backbone (labeled with YOYO) is shown as blue lines. Due to the diffraction limits of the microscope, only seven labels can be fully resolved. Labels 1 and 6 are generally brighter than the other labels due to clustering. (C) The sequence motif map in the graph was obtained by analyzing 63 single molecule fluorescence images. The solid line is the Gaussian curve fitting and the peaks correspond well to the predicted locations of the sequence motif.
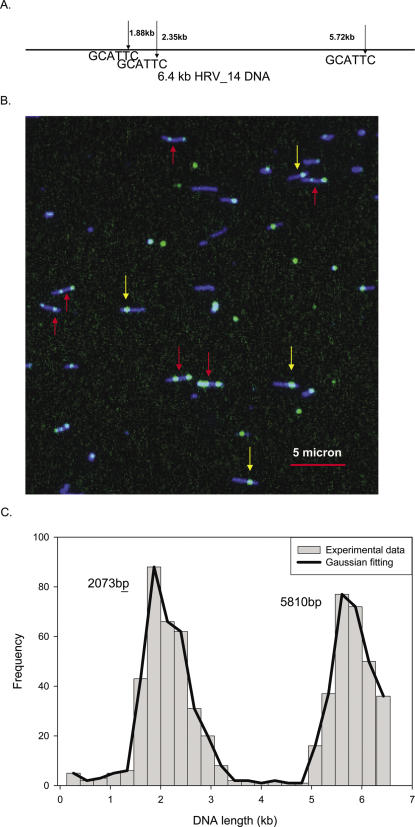
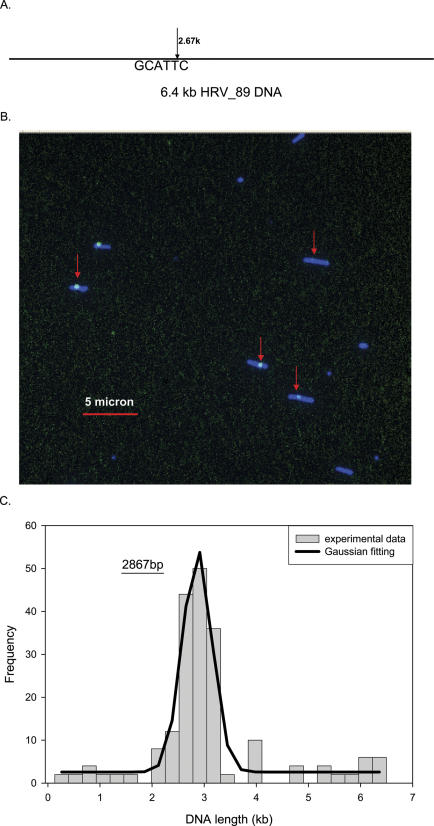
Human rhinovirus is an RNA virus and the length of the genomes of different types of rhinovirus is ∼7.2 kb. Before constructing its Nb.Bsm I map, nearly full-length dsDNAs (6.4 of 7.2 kb) were generated by reverse transcription followed by PCR (see Materials and Methods). Two strains, HRV14 and HRV89, were mapped initially and the predicted Nb.Bsm I (GCATTC) maps are shown in Figures 6A and 7A. In this case Tamra-ddCTP was used for the labeling. Figures 6B and 7B are false color two-channel composite images showing the stretched DNA contours and the Nb.Bsm I sites (Tamra-ddCTP in green and YOYO in blue) for HRV14 and HRV89, respectively. As expected, many of the DNA fragments of HRV14 in Figure 6B clearly contain two individual internal green labels (red arrows), while some fragments have only one green label (yellow arrows). On the other hand, almost all the DNA fragments of HRV89 in Figure 7B show only one internal green label, indicating that only one Nb.Bsm I site is in the HRV89 genome. A total of 356 and 187 DNA molecules, respectively, were used in constructing the Nb.Bsm I maps for HRV14 and HRV89 (Figures 6C and 7C). Two peaks, at 2.1 and 5.8 kb, are observed for HRV14, which agree well with predicted Nb.Bsm I sites at 1.9 and 5.7 kb, since nicking sites at 1.88 and 2.35 kb are not resolvable and merge as one peak. Only one peak was observed for HRV89, at 2.9 kb, compared with the expected position at 2.7 kb. The Nb.Bsm I maps can be used to distinguish and identify these two strains of the HRV virus.
Figure 6.
Sequence motif map of human rhinovirus 14 DNA. (A) The predicted Nb.Bsm I (GCATTC) map of human rhinovirus 14 DNA. Only two sites are present in this small RNA virus. (B) In the large field of the intensity scaled composite image, numerous molecules are found. Some molecules have two green labels (red arrows) and some have only one green label (yellow arrows). (C) The sequence motif map in the graph was obtained by analyzing 56 single molecule fluorescence images. The solid line is the Gaussian curve fitting and the peaks correspond well to the predicted locations of the sequence motif.
Figure 7.
Sequence motif map of human rhinovirus 89. (A) The predicted Nb.Bsm I (GCATTC) map of human rhinovirus 89 DNA. Only one site is present in this small RNA virus. (B) In the large field of the intensity scaled composite image, a number of singly labeled molecules are observed (red arrows). (C) The sequence motif map in the graph was obtained by analyzing 50 single molecule fluorescence images. The solid line is the Gaussian curve fitting and the peaks correspond well to the predicted locations of the sequence motif.
To see if one could use this approach to identify viral isolates, we conducted a set of studies in which the identities of the viral strains were unknown to those performing the experiments and data analysis. Four anonymous strains of human rhinoviruses were obtained from our collaborators. Full-length dsDNA was generated by reverse transcription and long-range PCR as before, using conserved sequences as PCR primers. After DNA nicking with Nb.Bsm I (GCATTC) and labeling with Tamra-ddCTP, the optical maps of the four viral strains were obtained and analyzed. Based on the maps we constructed (see Supplementary data), the strains were identified as HRV15, HRV28, HRV36 and HRV73, which were indeed the viral strains sent by our collaborators
DISCUSSION
Our approach of single molecule DNA mapping combines several different technologies: covalently tagging the sequence motif sites on long genomic dsDNA; stretching DNA molecules into linear form on a solid glass surface; and efficient detection and accurate localization of single fluorescent dye molecules on the dsDNA backbone. Its usefulness in DNA analysis will depend on its overall robustness, which depends in part on the efficiency of each of the individual steps and the degree of automation one can achieve.
A number of methods are available for labeling specific DNA sequence on dsDNA duplex, such as hybridization of short oligos to form Watson–Crick duplexes, using of triplex formation-oligonucleotides, and PNAs to form triplex (24,25); RecA protein-coated complementary sDNA (26); binding of polyamides to specific sequences (27) and some DNA binding proteins (e.g. zinc finger protein). Chan et al. have used bis-PNAs to map lambda-DNA (13). All of the above labeling strategies are based on non-covalent labeling and the sequence recognition depends on the relative binding affinity of the probe. At the single molecule level, the dissociation of the labeled probe is significant if the unbound probes have to be removed, thus significantly reducing the labeling efficiency.
In our nick-labeling scheme, the specificity is determined by both the enzymatic nicking reaction and the fluorescent nucleotide incorporation reaction. Furthermore, the single fluorescent dye molecule is covalently bound to the dsDNA, and is therefore not subjected to the variation of binding constants. In addition, our data show that the approach has high labeling efficiency and specificity, for example, >70% of the lambda-DNA molecules nicked by Nb.BbvCI have more than three labels. However, the labeling efficiency with nicking endonuclease Nb.Bsm I is somewhat lower, possibly due to compatibility issues of the buffer system since the PCR products were directly used in the nicking reaction.
One limitation of our approach is that some DNA molecules may contain natural nicks and give false signals if the labeled nucleotides are incorporated at these sites. Although we were able to obtain high quality lambda-DNA with almost no natural nicks, and the long-range PCR products were similarly free of non-specific nicks, natural nicks could be a real problem when mapping larger genomes. However, if necessary, the natural nicks can be terminated with non-fluorescent ddNTP, which could then be removed by Alkaline Phosphatase before performing the nicking reaction. Another limitation of our approach is that there are only a handful of nicking endonucleases available commercially. Xu et al. (28) however, reported a new way of generating the nicking endonuclease by swapping the putative dimerization domain with a non-functional dimerization domain, which could greatly increase the number of nicking endonucleases available that recognize many more sequence motifs.
The degree of DNA stretching also directly affects our mapping results, as the spatial localization of fluorescent tags with respect to the DNA backbone is more accurate with fully stretched DNA fragments. There are a number of ways of mounting and stretching dsDNA molecules on a glass surface, including the methods used in karyotyping, fluorescent in situ hybridization (FISH), optical mapping and nanowire (10,29,30). Here we combine a glass polymer coating system (18,19) with a DNA mounting strategy to allow us to observe single fluorescent dye molecules attached to fully stretched DNA molecules. The percentage of fully stretched DNA molecules depends on their size. The longer the DNA fragment, the better it stretches; for example, ∼70% DNA molecules (longer than 18 kb) were fully stretched. For shorter DNA molecules, such as those derived from HRV (7 kb), only ∼25% of them can be fully stretched (31). Even at 25%, there are plenty of fully stretched DNA molecules to analyze and construct the sequence motif maps. Furthermore, because data analysis is based on the relative ratio of labeling positions versus total length of DNA molecules, the DNA sizing error has limited impact on the accuracy of the motif map.
Fluorescence detection with TIRF microscopy is very efficient. The use of a high NA objective and a cooled, back-thinned CCD allows highly efficient photon collection and excellent signal-to-noise ratio (16). In addition, the use of oxygen scavengers such as 2-mercaptoethanol significantly reduced the rate of dye photobleaching. Over 90% of the dye molecules could be detected in the illuminated area. The combination of TIRF and the use of a polymer coated surface reject most free dye molecules, resulting in very low fluorescent background even without a clean-up step to remove the free labels (18,19). The spatial localization of individual dye molecules at the polymorphic sites is based on centroid analysis (2D Gaussian fitting) (32), a highly accurate way to localize a fluorescent signal. However, the resolution is limited by the light diffraction limit, which is on the order of 250 nm or ∼800 bp of a dsDNA.
The two-color system employed in our studies includes a blue channel (YOYO-1) for the DNA backbone and a different color channel for detecting the labels, which is required to construct sequence motif maps. If a map based on multiple sequence motifs is needed, more colors can be added to the system, for example, the HRV15 and HRV36 strains contain one additional Nb.BbvC I site besides two Nb.Bsm I sites. Two nucleotide terminators bearing different fluorescent labels, such as Cy3-ddUTP and Cy5-ddCTP, can be used to label the two sites generated by the nicking endonucleases. A three-color system can therefore provide more flexible and accurate mapping of genomes.
Each of the steps mentioned above, such as DNA labeling, DNA stretching and fluorescence imaging, can be automated to improve the throughput. In addition, the throughput will become even better with higher labeling and stretching efficiency, because when most of the DNA molecules are fully labeled and fully stretched, only a handful of fluorescent images are needed to construct the map.
Schwartz and his colleagues have developed an elegant optical mapping technique (20), in which a long double-stranded genomic DNA is stretched into linear form on a modified glass surface, cut with a restriction endonuclease, stained and imaged with fluorescence microscopy. The gaps at the cutting sites along the DNA backbone indicate the order of the DNA map. Crucial to the success of their method is the map-assembly algorithm developed by Anantharaman et al. (33). Using this optical mapping technique, they have been able to map several microbial genomes (12,34–38). In this proof-of-principle report, we have shown that our method improves the chemistry, DNA stretching and optical imaging aspects of the optical mapping approach. With a simple analysis algorithm, we can map small genomes (<40 kb), such as the viral genomes, quite easily. Unlike optical mapping, where immobilized DNA molecules are cut with restriction endonucleases in an inefficient reaction, the nick-labeling experiment is a one tube reaction, where the labeling efficiency is high.
The current throughput of our approach is comparable to those of gel electrophoresis-based restriction mapping restriction, with the nick-labeling reaction of many samples done in parallel in 90 min. DNA stretching and image capture can be done in ∼15 min/sample. With manual operation, the most time consuming step at present is the selection of fully stretched DNA molecules for measurements. With automation currently under development in our group, this bottleneck will be removed in the near future.
In conclusion, the nick-labeling approach reported here improves on the chemistry of optical mapping and can be used in many applications in genomics, especially in the identification of small microbes by mapping their genomes. This technology provides the linear ordered map of sequence motifs, which cannot be directly obtained with gel based restriction mapping. The single fluorescent molecule localization analysis can generate a map with high resolution within one single molecule. While the length measurements of gel based restriction mapping are averaged over many fragments and the accumulation of measurements error could result in incorrect map. As <100 molecules are needed to construct the sequence motif map in our approach, the drastic reduction of DNA material used in labeling step could be possible, once the labeling procedures can be miniaturized. By comparing the sequence motif map of a microbial isolate against the predicted (virtual) map in the database, one can determine the identity of a microbe accurately and efficiently. If one can address the problem of natural nicks in the DNA and utilize the powerful optical mapping algorithms developed by others, this approach can also be used to build sequence motif maps of much larger genomes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by National Institutes of Health (NIH).
Conflict of interest statement. None declared.
REFERENCES
- 1.Ried T., Liyanage M., du Manoir S., Heselmeyer K., Auer G., Macville M., Schrock E. Tumor cytogenetics revisited: comparative genomic hybridization and spectral karyotyping. J. Mol. Med. 1997;75:801–814. doi: 10.1007/s001090050169. [DOI] [PubMed] [Google Scholar]
- 2.van Belkum A. DNA fingerprinting of medically important microorganisms by use of PCR. Clin. Microbiol. Rev. 1994;7:174–184. doi: 10.1128/cmr.7.2.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wong G.K., Yu J., Thayer E.C., Olson M.V. Multiple-complete-digest restriction fragment mapping: generating sequence-ready maps for large-scale DNA sequencing. Proc. Natl Acad. Sci. USA. 1997;94:5225–5230. doi: 10.1073/pnas.94.10.5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birren B.W., Lai E., Hood L., Simon M.I. Pulsed field gel electrophoresis techniques for separating 1- to 50-kilobase DNA fragments. Anal. Biochem. 1989;177:282–286. doi: 10.1016/0003-2697(89)90052-3. [DOI] [PubMed] [Google Scholar]
- 5.Foquet M., Korlach J., Zipfel W., Webb W.W., Craighead H.G. DNA fragment sizing by single molecule detection in submicrometer-sized closed fluidic channels. Anal. Chem. 2002;74:1415–1422. doi: 10.1021/ac011076w. [DOI] [PubMed] [Google Scholar]
- 6.Lindberg P., Righetti P.G., Gelfi C., Roeraade J. Electrophoresis of DNA sequencing fragments at elevated temperature in capillaries filled with poly(N-acryloylaminopropanol) gels. Electrophoresis. 1997;18:2909–2914. doi: 10.1002/elps.1150181531. [DOI] [PubMed] [Google Scholar]
- 7.Goodwin P.M., Johnson M.E., Martin J.C., Ambrose W.P., Marrone B.L., Jett J.H., Keller R.A. Rapid sizing of individual fluorescently stained DNA fragments by flow cytometry. Nucleic Acids Res. 1993;21:803–806. doi: 10.1093/nar/21.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soderlund C., Humphray S., Dunham A., French L. Contigs built with fingerprints, markers, and FPC V4.7. Genome Res. 2000;10:1772–1787. doi: 10.1101/gr.gr-1375r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson W.M., Bharti A.K., Butler E., Wei F., Fuks G., Kim H., Wing R.A., Messing J., Soderlund C. Whole-genome validation of high-information-content fingerprinting. Plant Physiol. 2005;139:27–38. doi: 10.1104/pp.105.061978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrick J., Bensimon A. Imaging of single DNA molecule: applications to high-resolution genomic studies. Chromosome Res. 1999;7:409–423. doi: 10.1023/a:1009276210892. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz D.C., Li X., Hernandez L.I., Ramnarain S.P., Huff E.J., Wang Y.K. Ordered restriction maps of Saccharomyces cerevisiae chromosomes constructed by optical mapping. Science. 1993;262:110–114. doi: 10.1126/science.8211116. [DOI] [PubMed] [Google Scholar]
- 12.Reslewic S., Zhou S., Place M., Zhang Y., Briska A., Goldstein S., Churas C., Runnheim R., Forrest D., Lim A., et al. Whole-genome shotgun optical mapping of Rhodospirillum rubrum. Appl. Environ. Microbiol. 2005;71:5511–5522. doi: 10.1128/AEM.71.9.5511-5522.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan E.Y., Goncalves N.M., Haeusler R.A., Hatch A.J., Larson J.W., Maletta A.M., Yantz G.R., Carstea E.D., Fuchs M., Wong G.G., et al. DNA mapping using microfluidic stretching and single-molecule detection of fluorescent site-specific tags. Genome Res. 2004;14:1137–1146. doi: 10.1101/gr.1635204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phillips K.M., Larson J.W., Yantz G.R., D'Antoni C.M., Gallo M.V., Gillis K.A., Goncalves N.M., Neely L.A., Gullans S.R., Gilmanshin R. Application of single molecule technology to rapidly map long DNA and study the conformation of stretched DNA. Nucleic Acids Res. 2005;33:5829–5837. doi: 10.1093/nar/gki895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morgan R.D., Calvet C., Demeter M., Agra R., Kong H. Characterization of the specific DNA nicking activity of restriction endonuclease N.BstNBI. Biol Chem. 2000;381:1123–1125. doi: 10.1515/BC.2000.137. [DOI] [PubMed] [Google Scholar]
- 16.Yildiz A., Forkey J.N., McKinney S.A., Ha T., Goldman Y.E., Selvin P.R. Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 17.Kwok P.Y., Xiao M. Single-molecule analysis for molecular haplotyping. Hum. Mutat. 2004;23:442–446. doi: 10.1002/humu.20020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braslavsky I., Hebert B., Kartalov E., Quake S.R. Sequence information can be obtained from single DNA molecules. Proc. Natl Acad. Sci. USA. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kartalov E.P., Unger M.A., Quake S.R. Polyelectrolyte surface interface for single-molecule fluorescence studies of DNA polymerase. Biotechniques. 2003;34:505–510. doi: 10.2144/03343st02. [DOI] [PubMed] [Google Scholar]
- 20.Aston C., Hiort C., Schwartz D.C. Optical mapping: an approach for fine mapping. Methods Enzymol. 1999;303:55–73. doi: 10.1016/s0076-6879(99)03006-2. [DOI] [PubMed] [Google Scholar]
- 21.Sischka A., Toensing K., Eckel R., Wilking S.D., Sewald N., Ros R., Anselmetti D. Molecular mechanisms and kinetics between DNA and DNA binding ligands. Biophys. J. 2005;88:404–411. doi: 10.1529/biophysj.103.036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allard A., Albinsson B., Wadell G. Rapid typing of human adenoviruses by a general PCR combined with restriction endonuclease analysis. J. Clin. Microbiol. 2001;39:498–505. doi: 10.1128/JCM.39.2.498-505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casas I., Avellon A., Mosquera M., Jabado O., Echevarria J.E., Campos R.H., Rewers M., Perez-Brena P., Lipkin W.I., Palacios G. Molecular identification of adenoviruses in clinical samples by analyzing a partial hexon genomic region. J. Clin. Microbiol. 2005;43:6176–6182. doi: 10.1128/JCM.43.12.6176-6182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Felsenfeld G., Rich A. Studies on the formation of two- and three-stranded polyribonucleotides. Biochim. Biophys. Acta. 1957;26:457–468. doi: 10.1016/0006-3002(57)90091-4. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen P.E., Egholm M. An introduction to peptide nucleic acid. Curr. Issues Mol. Biol. 1999;1:89–104. [PubMed] [Google Scholar]
- 26.Seong G.H., Niimi T., Yanagida Y., Kobatake E., Aizawa M. Single-molecular AFM probing of specific DNA sequencing using RecA-promoted homologous pairing and strand exchange. Anal. Chem. 2000;72:1288–1293. doi: 10.1021/ac990893h. [DOI] [PubMed] [Google Scholar]
- 27.Dervan P.B., Burli R.W. Sequence-specific DNA recognition by polyamides. Curr. Opin. Chem. Biol. 1999;3:688–693. doi: 10.1016/s1367-5931(99)00027-7. [DOI] [PubMed] [Google Scholar]
- 28.Xu Y., Lunnen K.D., Kong H. Engineering a nicking endonuclease N.AlwI by domain swapping. Proc. Natl Acad. Sci. USA. 2001;98:12990–12995. doi: 10.1073/pnas.241215698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai W., Jing J., Irvin B., Ohler L., Rose E., Shizuya H., Kim U.J., Simon M., Anantharaman T., Mishra B., et al. High-resolution restriction maps of bacterial artificial chromosomes constructed by optical mapping. Proc. Natl Acad. Sci. USA. 1998;95:3390–3395. doi: 10.1073/pnas.95.7.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoltenberg R.M., Woolley A.T. DNA-templated nanowire fabrication. Biomed. Microdevices. 2004;6:105–111. doi: 10.1023/b:bmmd.0000031746.46801.7d. [DOI] [PubMed] [Google Scholar]
- 31.Chan T.F., Ha C., Phong A., Cai D., Wan E., Leung L., Kwok P.Y., Xiao M. A simple DNA stretching method for fluorescence imaging of 710 single DNA molecules. Nucleic Acids Res. 2006;34:e113. doi: 10.1093/nar/gkl593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thompson R.E., Larson D.R., Webb W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anantharaman T.S., Mishra B., Schwartz D.C. Genomics via optical mapping. II: ordered restriction maps. J. Comput. Biol. 1997;4:91–118. doi: 10.1089/cmb.1997.4.91. [DOI] [PubMed] [Google Scholar]
- 34.Lai Z., Jing J., Aston C., Clarke V., Apodaca J., Dimalanta E.T., Carucci D.J., Gardner M.J., Mishra B., Anantharaman T.S., et al. A shotgun optical map of the entire Plasmodium falciparum genome. Nature Genet. 1999;23:309–313. doi: 10.1038/15484. [DOI] [PubMed] [Google Scholar]
- 35.Lim A., Dimalanta E.T., Potamousis K.D., Yen G., Apodoca J., Tao C., Lin J., Qi R., Skiadas J., Ramanathan A., et al. Shotgun Optical Maps of the Whole Escherichia coli O157:H7 Genome. Genome Res. 2001;11:1584–1593. doi: 10.1101/gr.172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin J., Qi R., Aston C., Jing J., Anantharaman T.S., Mishra B., White O., Daly M.J., Minton K.W., Venter J.C., et al. Whole-genome shotgun optical mapping of Deinococcus radiodurans. Science. 1999;285:1558–1562. doi: 10.1126/science.285.5433.1558. [DOI] [PubMed] [Google Scholar]
- 37.Zhou S., Deng W., Anantharaman T.S., Lim A., Dimalanta E.T., Wang J., Wu T., Chunhong T., Creighton R., Kile A., et al. A whole-genome shotgun optical map of Yersinia pestis strain KIM. Appl. Environ. Microbiol. 2002;68:6321–6331. doi: 10.1128/AEM.68.12.6321-6331.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou S., Kile A., Kvikstad E., Bechner M., Severin J., Forrest D., Runnheim R., Churas C., Anantharaman T.S., Myler P., et al. Shotgun optical mapping of the entire Leishmania major Friedlin genome. Mol. Biochem. Parasitol. 2004;138:97–106. doi: 10.1016/j.molbiopara.2004.08.002. [DOI] [PubMed] [Google Scholar]