Abstract
The mini-chromosome maintenance (MCM) complex is the principal candidate for the replicative helicase of archaea and eukaryotes. Here, we describe a functional dissection of the roles of the three principal structural modules of the homomultimeric MCM of the hyperthermophilic archaeon Sulfolobus solfataricus. Our results include the first analysis of the central AAA+ domain in isolation. This domain possesses ATPase and helicase activity, defining this as the minimal helicase domain. Reconstitution experiments show that the helicase activity of the AAA+ domain can be stimulated by addition of the isolated N-terminal half in trans. Addition of the N-terminus influences both the processivity of the helicase and the choice of substrate that can be melted by the ATPase domain. The degenerate helix-turn-helix domain at the C-terminus of MCM exerts a negative effect on the helicase activity of the complex. These results provide the first evidence for extensive regulatory inter-domain communication within the MCM complex.
INTRODUCTION
The core replication machineries of Eukarya and Archaea are fundamentally related, indicative of their descent from a common ancestor. Given the relative simplicity of the archaeal machinery, it serves as a valuable model system for dissection of the shared archaeo-eukaryotic replication machinery (1,2). The archaeal MCM is a homo-multimeric protein, while the MCM complex found in all eukarya is a heterohexamer composed of one copy of each of MCM 2, 3, 4, 5, 6 and 7. MCM 2-7 display considerable sequence homology to each other, and are presumably derived by gene duplication events from a single, archaeal-like, ancestral MCM (3). In addition to MCM 2-7, recent work has described and characterised MCM 8, a homo-multimeric complex found exclusively in higher eukaryotes (4,5). MCM 8 plays an important role in the elongation phase of replication, while MCM 2-7 is required for both initiation and elongation phases (5,6). It is believed that MCM 2-7 serves as the replicative helicase and, in support of this, a sub-complex of MCM 4,6 and 7, but not the entire MCM 2-7 complex, possesses 3′ to 5′ helicase activity in vitro (7). Similarly, a number of archaeal MCMs have been demonstrated to have processive 3′-5′ helicase activity (8–11).
Electron microscopy has revealed that the MCM from the archaeon M. thermautotrophicum (Mth) can adopt a number of configurations, with hexamer, double hexamer, heptamer and filamentous structures observed (8,12–14). In solution, Mth MCM appears to exist primarily as a double hexamer (8–10). In contrast, studies to date indicate that the MCMs from Archaeoglobus fulgidus and Sulfolobus solfataricus (Sso) appear to be hexameric in solution (11,15). Further, a hexameric form of Sso MCM has been shown to bind model DNA substrates (15). The double hexameric nature of Mth MCM in solution has been supported by the X-ray crystal structure of the N-terminal 286 amino acids of the protein (16). The structure revealed a sixfold symmetric arrangement, with hexameric rings forming double hexamers via a head-to-head interaction. A zinc-binding motif in the N-terminus of each subunit was positioned at the site of interaction between hexamers, suggesting an important role for this motif in mediating double hexamerisation. Intriguingly, however, mutational analysis of this motif did not reveal any alteration to multimeric status, but did show reduced DNA binding and helicase activity (17). The structure of Mth MCM (2-286) revealed three domains, A, B and C (16). Only domain C is essential for activity of Mth MCM and forms the core fold mediating multimerisation of the N-terminal portion of Mth MCM (18).
Here, we present an analysis of the Sso MCM and reveal that the C-terminal AAA+ domain, although dimeric in solution, retains helicase activity. The isolated AAA+ domain has promiscuous helicase activity, being able to catalyse melting of a broader range of model substrates than the full-length enzyme, including pure duplex and 5′ tailed molecules. This promiscuity can be alleviated by addition of isolated N-terminal domains in trans. In addition to modulating substrate specificity, the N-terminal domains, when added in trans, enhance the processivity of the enzyme.
MATERIALS AND METHODS
Cloning and mutagenesis
The amino acids to which the different constructs correspond are listed in Figure 1C. Oligonucleotide primers were purchased from Sigma Genosys, Cambridge, UK; sequences are available on request. The full length, ▵A, ▵HTH, ▵A/HTH, and N-half constructs were cloned into the pET30a vector as described in related literature(15). The C-half, AAA+ core and HTH constructs were cloned into a modified pRSET vector (Invitrogen) using the BamHI and EcoRI sites resulting in a hexahistidine tag and thrombin recognition site N-terminal to the protein sequence. The Walker A mutant K346A (with reference to the position in the full-length protein) was introduced into the AAA+ core construct using the Stratagene Quick Change protocol.
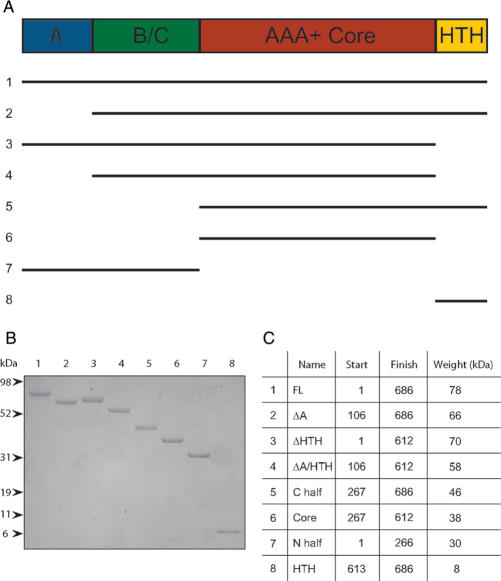
Figure 1.
(A) Demonstrates the domains contained within each of the eight constructs. (B) 2 µg of each protein were run on a 4–20% Tris-Glycine gel in MES buffer, before staining with Coomassie blue. (C) Shows the boundaries of each construct relative to the full-length protein and their molecular weights.
Glutaraldehyde crosslinking
Glutaraldehyde was added to a final concentration of 0.01% to 25 μM of the indicated proteins in 1xPBS, total reaction volume 30 μl. The reactions were incubated for 15 min at room temperature before being quenched by addition of 3 μl IM Tris (pH 8) 1 M glycine. After incubation at room temperature for a further 10 min, 30 μl loading buffer (1.05% (w/v) SDS, 62.5 mM Tris-HCl (pH 6.8), 10% (v/v) glycerol, 0.35 M β-mercaptoethanol, 0.0125% (w/v) bromophenol blue) was added to each tube. Reactions were boiled for 5 min before loading 20 μl on a NuPage 4–12% Bis-Tris gel (Invitrogen). After electrophoresis, proteins were visualised by staining with Coomassie brilliant blue.
Expression and purification
All constructs were expressed as described in related literature(15). Purification of the full-length, ▵A, ▵HTH, ▵A/HTH and N-half fragments was also performed in the same manner as in this study. Extracts containing the C-half, AAA+ core and HTH constructs were treated at 75°C for 30 min and centrifuged [17,000 rpm, 30 min SS34 rotor (Beckman)] before being purified over an Ni-NTA agarose (Qiagen) column followed by cleavage with thrombin (Sigma) and gel-filtration over a Superdex 75 column (GE Healthcare). This resulted in the presence of additional glycine-serine two amino acid extension at the N-termini of these proteins.
Gel filtration
50 µl of protein at the indicated concentration was applied to either a Superose 6 or Superdex 200 gel filtration column (PC3.2/30; Amersham Biosciences, Inc.) equilibrated with 20 mM Tris (pH 8), 150 mM sodium chloride and 1 mM dithiothreitol. This was then eluted at 40 µl/min for one column volume (2.4 ml). Absorbance at 280 nm and 215 nm was monitored and 80 µl fractions collected. The amplitude of the resulting peaks was normalized, using the Unicorn program, to allow comparison.
Pull down interaction assays
75 pmoles amount C-terminally his-tagged AAA+ core domain was pre-incubated with 50% Ni-NTA (Qiagen) slurry in TBST in a volume of 30 μl. BSA was added, to a final concentration of 0.1 mg/ml. After pre-incubation at room temperature for 10 min, 5.2 nmoles untagged N-terminal domain was added, and the reaction was incubated for 1 h at room temperature. After washing beads in TBST, protein was removed by boiling in the presence of SDS-PAGE loading buffer (1% (w/v) SDS, 62.5 mM Tris-HCl (pH 6.8) 10%, 0.35 M β-mercaptoethanol, (v/v) glycerol, 0.0125% (w/v) bromophenol blue) + 0.25 M imidazole for 10 min. 20 μl was resolved by SDS-Polyacrylamide gel electrophoresis (SDS-PAGE).
ATPase assay
Reactions were performed in a 20 µl reaction volume containing 30 mM Tris acetate (pH 8), 75 mM NaCl, 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM dithiothreitol, 100 µM ATP (Sigma), 10 nM [γ-32P]ATP (3000 Ci/mmol) and 1 µM of the indicated protein. Reactions were incubated at 65°C for 10 min before 1 µl aliquots of each sample were analyzed using thin-layer chromatography on PEI cellulose TLC plates (Machery Nagel) developed with 0.8 M lithium chloride 1 M formic acid buffer. Analysis was performed by phosphorimaging and quantification using ImageGauge.
EMSA and helicase assays
These were performed using a Y-shaped construct as previously described (15). The sequences for the 3′ tailed, 5′ tailed and flush duplex substrate oligonucleotides are available on request. Assays were performed at least three times, the results quantified by phosphorimager and averaged. The error bars on the graphs are the standard error of the mean.
Processivity assays
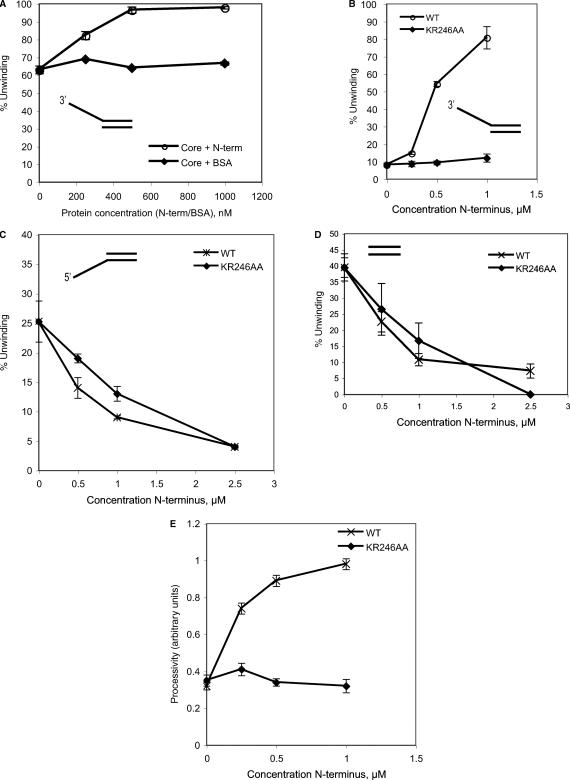
A range of lengths of radiolabelled DNA fragments annealed to single-stranded M13 DNA were produced using a Sequenase version 2 DNA sequencing kit (USB) according to manufacturer's instructions. The indicated proteins were then incubated in the presence of 1 μl of this substrate in a 20 μl reaction volume containing 30 mM Tris acetate (pH 8), 75 mM NaCl, 50 mM potassium acetate, 10 mM magnesium acetate, 1 mM dithiothreitol and 10 mM ATP for 1 h at 50°C. Products were resolved on a 6% gel in TBE with 0.2% SDS before drying and phosphorimaging. Band intensities of molecules in the size range 75 to 150 nt (short) and 600 to 800 (long) were calculated using Image Gauge and expressed as a ratio of long:short for plotting in Figure 7.
Figure 7.
(A) 600 nM core domain MCM was incubated with 3′-tailed substrate in the presence of the indicated concentrations of BSA or N-half. (B) 300 nM core domain was incubated with 3′-tailed substrate in the presence of the indicated concentrations of wild-type N-half (WT) or N-half with a mutation in the DNA-binding β-hairpin motif (KR246AA). (C) 300 nM core domain was incubated with 5′-tailed substrate in the presence of the indicated concentrations of wild-type N-half (WT) or N-half with a mutation in the DNA-binding β-hairpin motif (KR246AA). (D) 300 nM core domain was incubated with a double-stranded DNA substrate in the presence of the indicated concentrations of wild-type N-half (WT) or N-half with a mutation in the DNA binding β-hairpin motif (KR246AA). (E) The core domain was incubated with various length substrates in the presence of the indicated concentrations of wild-type N-half (WT) or N-half with a mutation in the DNA binding β-hairpin motif (KR246AA), and the ratio of long:short fragments unwound was calculated.
RESULTS
Generation of deletion constructs
Using homology modelling, we have identified the domain boundaries in the N-terminal region of Sso MCM and designed expression constructs for production of recombinant protein in E. coli (Figure 1A and C). For the C-terminal half of the protein, we made a series of constructs based on fragments from limited proteolysis of the isolated C-terminal domain in the presence of nucleotide (data not shown). Recombinant proteins were expressed in E. coli and purified as described in the section titled Materials and Methods, and the resultant proteins are shown in Figure 1B. In addition to the constructs we describe, we also attempted to make deletion constructs in which either C1 or C2 sub-domains of the AAA+ module were deleted, however, the resultant proteins were highly insoluble (data not shown) and so were not pursued further.
Multimeric status of Sso MCM derivatives
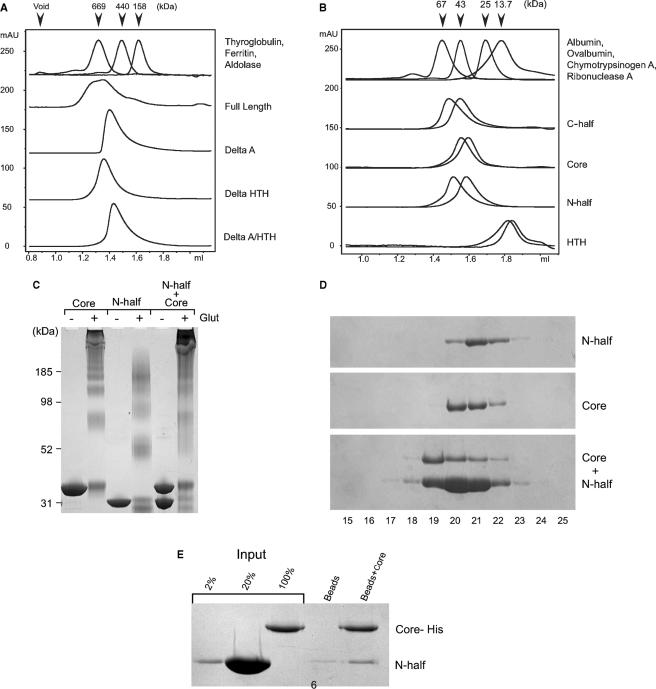
We first tested the multimeric status of the various purified proteins by subjecting them to hydrodynamic analyses in gel filtration chromatography (Figure 2). Our results reveal that when 25 µM full-length, wild-type MCM was assayed, we could detect a complex and broad elution profile, compatible with the presence of a mixed population with species of up to double hexamer in size (Figure 2A). This is the first time that greater-than-hexameric forms of the Sso MCM have been detected; lower concentrations of this construct give rise to primarily hexameric species (data not shown). However, previous reports have used substantially lower concentrations of protein; indeed, at lower protein concentrations, we see an elution profile compatible with generation of single hexamers (15). In contrast to the above, analyses of 25 µM of mutant Sso MCMs lacking the A domain, the HTH or both showed more homogeneous profiles, compatible with the formation of single hexamers of these species, suggesting that the N- and C-terminal domains of the MCM are important for formation of higher-order structures.
Figure 2.
(A) Figure 2A shows the UV 280 nm trace from gel filtration over a Superose 6 column using 25 µM of each of the indicated proteins. (B) Figure 2B shows the UV 280 nm trace from gel filtration over a Superdex 200 column using two concentrations for each protein. N-half: 200 µM, 25 µM. C-half 25 µM, 2.5 µM. AAA+ Core 25 µM, 2.5 µM. HTH 60 µM, 1 µM. The absorbance of each trace was normalised using the UNICORN program. (C) 25 µM Core domain, N-half or a mixture of both were incubated in the presence (+) or absence (−) of glutaraldehyde prior to SDS-PAGE on a 4–12% polyacrylamide gradient gel. (D) 25 µM AAA+ Core were run alone or mixed with 200 µM N-half on a Superdex 200 column. 80 µl samples were collected and electrophoresed on a 10% polyacrylamide gel. (E) His-tagged AAA+ domain was used in pull-down experiments with untagged N-terminal domain. 2–1% N-terminal domain input, 20–10% N-terminal domain input, Bead—Ni-NTA beads incubated with N-terminus, Beads + Core – Ni-NTA beads bound to AAA+ domain incubated with N-terminus.
Next, we assayed the multimeric status of the isolated C-terminal half, core AAA+ domain, N-terminal half and HTH domain of the Sso MCM. The profiles shown for each mutant in Figure 2B show superimposed elution profiles for two concentrations of protein. The profiles obtained indicate that, at the lower concentrations, these proteins have an elution profile consistent with monomeric species; however, at higher concentrations, we observe a reduced retention time on the gel filtration column, suggestive of either conformational alteration or formation of multimeric forms of the protein. In support of the latter, when we perform glutaraldehyde-mediated crosslinking of the isolated N-half and core domains of MCM, we observe the generation of higher-order multimers (Figure 2C). When the same concentrations of BSA were treated identically, the majority of the BSA stayed monomeric, with only trace amounts of dimer and trimer species detectable (data not shown). Gel filtration analyses of the N-terminal half of Mth MCM revealed that it forms an extremely stable double hexamer over a broad range of concentrations, our data indicate that this property is not shared by the N-half of SsoMCM. We next tested whether mixing the isolated N and C-terminal halves of the protein could result in the generation of higher-order forms. As can be seen in Figure 2D, addition of the AAA+ domain to an excess of the N-terminal fragment results in a shift in the elution profile of the AAA+ domain to a higher-molecular-weight form, indicative of interaction between these species. We were additionally able to detect weak interactions in affinity chromatography approaches, employing a C-terminally hexahistidine-tagged version of the C-terminal half of the protein and a non-tagged version of the N-terminal half of the protein (Figure 2E). Finally, glutaraldehyde-mediated crosslinking of a sample containing both N-half and core domain proteins resulted in an increased yield of very-low-mobility complexes that barely entered the resolving gel during SDS-PAGE (Figure 2C).
DNA binding of Sso MCM derivatives
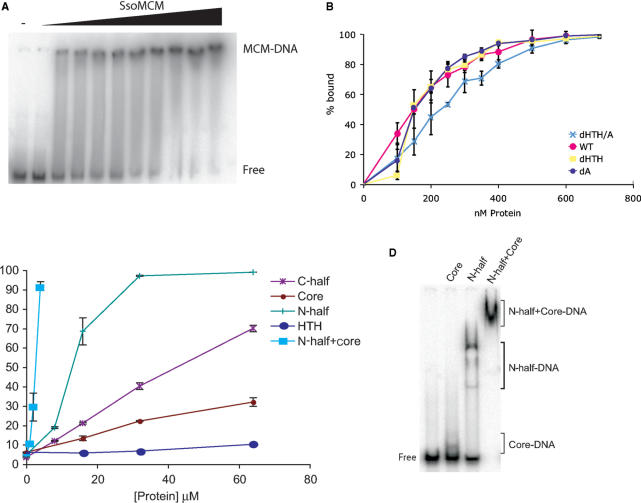
Next, we assayed the ability of the various mutant Sso MCMs to bind DNA with the electrophoretic mobility shift assay (EMSA), using a radiolabelled fork substrate (15), Figure 3A and B. The results indicate that the wild-type protein and constructs lacking A domain, HTH or both all have very similar DNA binding activities, with the construct lacking both the A-domain and HTH, having a slightly lower affinity than that of the other constructs (Figure 3B). Importantly, however, removal of the HTH—a candidate DNA binding domain—had no effect on the DNA binding affinity of the protein.
Figure 3.
EMSA assays were performed as described (15), (A) Representative EMSA assay with 0, 50, 100, 150, 200, 250, 300, 350, 400, 500 and 600 nM wild-type MCM (B) Figure 3B demonstrates the percentage of DNA bound at the indicated concentration for constructs: full length, ▵A, ▵HTH and ▵A/HTH. (C) Figure 3C demonstrates the results for N-half, C-half, AAA+ core, HTH and an equimolar mix of N-half and AAA+ core. (D) EMSA assays employing 6 µM Core and/or N-half protein. The positions of free DNA and the relevant complexes are indicated.
Similar assays were performed with the C-terminal half of the protein, AAA+ core domain, N-terminal half and isolated HTH domain. As can be seen in Figure 3C, the isolated N-half of the protein has detectable DNA binding activity; however, the affinity is at least 50-fold lower than the full-length, wild-type protein. The two proteins containing the AAA+ domain had even lower affinity, whereas the isolated HTH domain had no detectable DNA binding activity at all.
In light of the gel filtration and affinity chromatography data suggesting that the N- and C-terminal halves of the protein could interact (Figure 2), we tested the effect of mixing the N-half and AAA+ core domain constructs of the protein in EMSA. This gave a highly synergistic effect with substantially enhanced DNA binding compared to either domain in isolation, Figures 3C and D, suggestive of a functional interaction on DNA between these two regions of Sso MCM.
ATPase activity of the Sso MCM derivatives
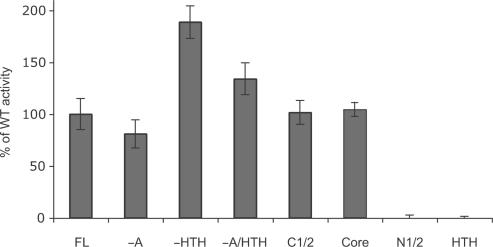
As Sso MCM has an intrinsic ATPase activity, we tested the effect of the truncation mutations on this activity of the protein (Figure 4). The N-half and HTH constructs, lacking the AAA+ domain had no detectable activity. Deletion of the N-terminal A domain resulted in a modest reduction of ATPase activity. However, deletion of the HTH domain resulted in an almost twofold stimulation of the ATPase activity; as seen with wild-type (15), this activity could be further stimulated by the addition of DNA (data not shown).
Figure 4.
ATPase activity of the different constructs is shown relative to the activity of the full-length construct (wild-type activity of 1 µM MCM is 35 nM s−1).
Surprisingly, the C-terminal half of the protein, containing the AAA+ and HTH domains, has essentially wild-type activity, even though this protein, as a dimer, has only a single functional ATPase site. Finally, the AAA+ core domain has an ATPase activity slightly greater than wild-type. Thus, it appears that the HTH domain plays complex context-dependent roles; in the presence of the N-terminal B and C domains, removal of the HTH results in a stimulation of ATPase activity; this, however, is not seen when these domains are present.
Helicase activity of the Sso MCM mutants
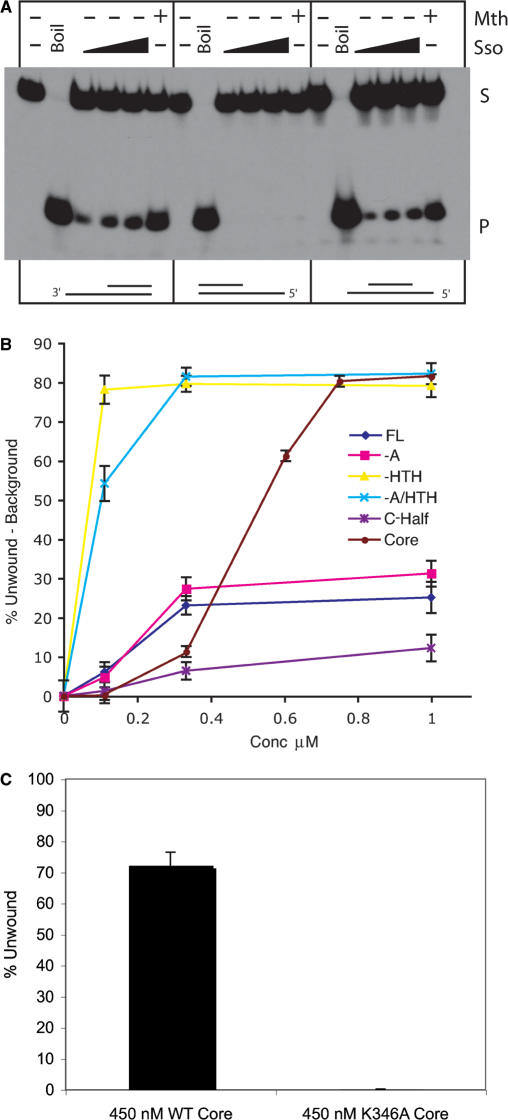
Wild-type Sso MCM displays 3′ to 5′ helicase activity (Figure 5A). We tested the ability of the various truncated Sso MCM constructs to mediate melting of a flayed duplex DNA substrate. As can be seen in Figure 5B, over the range of concentrations tested (0.1–1 µM), wild-type MCM effects melting of a maximum of 30% of substrate. Deletion of the HTH domain causes a large stimulation of the helicase activity of MCM, as compared to wild-type. This stimulation is particularly apparent at 111 nM protein, where the helicase activity of the mutant lacking the HTH is 15-fold higher than the wild-type protein. Removal of the A domain does not significantly alter helicase activity, and if the HTH domain is additionally removed then a significant stimulation is observed, in agreement with the results for ATPase activity described above.
Figure 5.
(A) Determination of the polarity of Sso MCM helicase activity. 25, 50 or 100 ng of Sso MCM or 100 ng of Mth MCM were incubated with substrates designed to have 3′, 5 or 3′ and 5′ single–stranded extensions. The positions of substrate (S) and product (P) are indicated. (B) Figure 5B shows the helicase activity of the different deletion constructs at various concentrations, N-half and HTH are omitted as no activity was detected. (C) Figure 5C compares the activity of the AAA+ core with an equivalent construct possessing a K > A mutation in the Walker A motif.
We also tested the more extensive deletion constructs for helicase activity. Neither the N-terminal half nor the HTH construct showed detectable activity (data not shown). However, we could detect significant helicase activity mediated by the C half and AAA+ core domain proteins (Figure 5B). This activity was abrogated when assays were performed with an identically purified construct, in which we introduced a point mutation into the Walker A box of the AAA+ domain (Figure 5C). Thus, it appears that the isolated AAA+ domain of Sso MCM has intrinsic DNA helicase activity.
The AAA+ core domain can utilise a broad spectrum of substrates
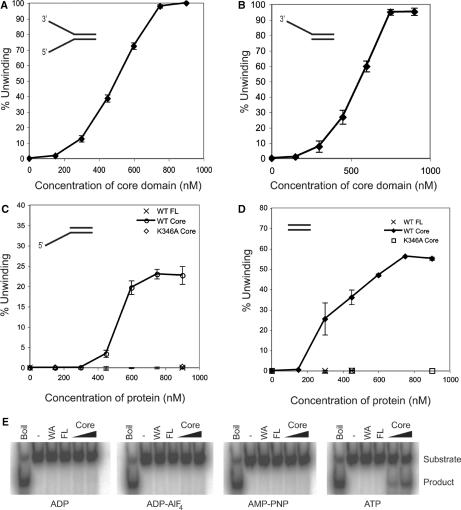
Next, we sought to determine the range of substrates that could be melted by the AAA+ core domain of Sso MCM. As with full-length Sso MCM, the AAA+ core domain was capable of melting both Y-shaped and 3′ tailed substrates (Figure 6A and B). However, and in marked contrast to the behaviour of the full-length MCM, the isolated AAA+ core domain construct was able to catalyse melting of a 5′ tailed construct (Figure 6C). We note that the absolute level of product detected with the 5′ tailed construct plateaus at approximately 25% of the level detected with 3′ tailed and Y-shaped substrates. This activity is abrogated by the introduction of a Walker A (K-A) mutation in the core domain (Figure 6C), allowing us to rule out trivial explanations of contaminating helicases in our MCM core domain preparation.
Figure 6.
Indicated concentrations of the core AAA+ domain were incubated with substrates with (A) both 3′ and 5′ single-stranded extensions (B) 3′ single-stranded extensions. (C and D) In Figures 5C and D, indicated concentrations of wild-type full-length MCM (WT FL), wild-type core AAA+ domain (WT Core) or core AAA+ domain with a mutation in the Walker A motif (K346A Core) were incubated in the presence of substrates containing (C) a 5′ single-stranded extension, (D) no single-stranded extensions. (E) 2.5 μM wild-type full-length MCM (FL), 2.5 μM core AAA+ domain with a mutation in the Walker A motif (WA), or 0.25 μM and 0.5 μM wild-type core AAA+ domain were incubated with a double-stranded DNA substrate in the presence of 10 mM ADP, ADP-AlFl4, AMP-PNP or ATP as indicated.
This unprecedented ability of the AAA+ core of the protein to utilise a 5′ tailed substrate could arise in two ways. First, deletion of the N-terminal domains and the HTH could uncover a cryptic 5′ to 3′ helicase activity of the MCM. A second possibility is that the truncated construct is still acting as a 3′ to 5′ helicase, but is able to engage with the 3′ ends of the substrate either in the context of a double-strand end or 3′ recessed end. To test this latter possibility, we examined whether the Sso MCM or AAA+ domain could mediate melting of a duplex substrate lacking single-stranded tails. As can be seen in Figure 6D, while the full-length protein cannot melt this substrate, the AAA+ core domain can do so efficiently; as with the 5′ tailed substrate, this activity is abrogated by the introduction of Walker A (K-A) mutation in the AAA+ core construct.
Work on Mth MCM has indicated that deletion of parts of that protein could result in filamentation of the truncated protein. It was, therefore, formally possible that the apparent helicase activity we observe could in fact be due to nucleation of filamentous polymerisation of the MCM AAA+ core domain along a single strand of DNA, thereby displacing the other strand. We believe that this is unlikely because the core domain construct is dimeric in solution and the Walker A mutant is devoid of DNA melting activity. Nevertheless, we tested whether ATP hydrolysis was required for the observed melting of duplex DNA. To do this, we incubated the core domain with either ATP, a non-hydrolysable analog, AMP-PNP, ADP or the hydrolysis transition state analog, ADP-AlF4. As can be seen in Figure 6E, only ATP is able to facilitate unwinding of duplex DNA by the AAA+ core domain.
Although we cannot rule out the formal possibility that the AAA+ core domain possesses a novel 5′ to 3′ activity in addition to 3′ to 5′ helicase activity, we believe that the above data indicate that the ability of the AAA+ core domain construct to unwind a 5′ tailed substrate most likely arises from the protein's ability to engage with a 3′ end in the context of a double-stranded end, then proceed in an ATP-dependent manner in the conventional 3′–5′ polarity. It may be significant that, as Sso MCM is from a hyperthermophilic organism, we perform the helicase assays at 50°C. This may lead to transient melting of base pairs at the DNA double-strand end, facilitating engagement by the core AAA+ domains.
The N-terminal processivity clamp
Finally, because our gel filtration, affinity pull down and EMSA data indicate that there is functional interaction between the N- and C-terminal domains, we tested the effect of adding back the N-half construct in trans on the helicase activity of the core AAA+ domain. As can be seen in Figure 7A, the N-half protein stimulates the helicase activity of the AAA+ domain on a 3′ tailed substrate. Similar results were obtained with a Y-shaped substrate (data not shown). This stimulation is specific to the N-half protein, as addition of either BSA or isolated HTH had no significant effect (Figure 7A and data not shown). It should be noted that the experiments in Figure 5B indicate that when the N-half and core domain are present in their natural cis configuration (-HTH construct), considerably higher activity is detectable, presumably reflective of the relatively weak interaction between these domains in trans (Figure 2). No helicase activity was detected upon addition of N-half to the Walker A mutant AAA+ domain (data not shown). We and others have shown previously that the N-half contains a DNA binding site in the form of a β-hairpin (15,16). We made point mutations in the isolated N-terminal domain construct, changing the conserved lysine and arginine residues at the tip of the hairpin to alanine (15). EMSA assays confirmed that this mutant derivative had lost the ability to bind DNA (data not shown). Addition of the resultant protein showed no stimulation of helicase activity, indicating the importance of the N-terminal DNA binding site in stimulating the activity of the core domain (Figure 7B). We next tested the effect of adding back the N-half protein to the range of substrates that could be unwound by the core domain. Significantly, addition of the N-half protein resulted in clear inhibition of the ability of the AAA+ core domains to unwind both non-tailed duplex and 5′ tailed substrates. We then tested the effect of the addition of DNA-binding-defective N-half on the ability of the core domain to melt 5′ tailed and flush duplex substrates (Figure 7C and D). In contrast to the result obtained with 3′ tailed or Y-shaped substrates (Figure 7B), we saw essentially identical inhibition of core-domain-mediated helicase activity with both wild-type and DNA binding defective N-half. This observation allows us to rule out the trivial explanation that the N-half is simply blocking access to the DNA substrate by the core domain by virtue of its higher DNA binding affinity. Thus, we can discriminate two mechanistically distinct consequences of interaction of the N-half with the core domain; first, a DNA-binding-independent inhibition of the ability to use 5′ tailed and flush duplex substrates and, second, a DNA-binding-dependent stimulation of 3′ tail dependent helicase activity.
In light of our data showing cooperative DNA binding of N- and C-terminal halves of MCM and the dependence of the stimulatory effect of the N-half on its ability to bind DNA, we speculated, therefore, that the addition of the N-half may facilitate productive interaction of the complex with DNA. We, therefore, wished to test the processivity of the complex. To this end, we performed helicase assays with an M13 template that had a variety of lengths of duplex DNA annealed. To quantify the ability of the MCM to remain template committed, we measured the relative ratio of long versus short strands displaced from the substrate. As can be seen in Figure 7D, addition of the N-half significantly enhances the relative yield of longer products, a result compatible with the N-half assisting the template commitment of the core domain. Addition of N-half protein containing point mutations in the DNA-binding β-hairpin did not result in a similar stimulation of processivity.
DISCUSSION
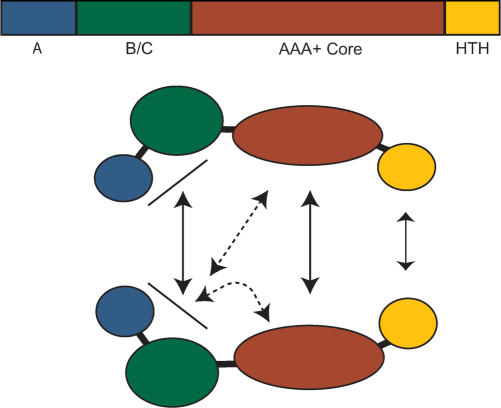
Our data reveal the functional modules that make up the Sso MCM and the complex net of interactions linking them (Figure 8). We have defined the AAA+ module as the minimal domain required for both helicase and ATPase activities. Previous studies using the Mth MCM were unable to test this directly due to an inability to express this domain in isolation. Our gel filtration and glutaraldehyde crosslinking data indicate that the AAA+ domain is capable of forming a range of multimeric forms. The helicase activity of the isolated AAA+ domain suggests that it is capable of forming the active hexameric complex in the presence of substrate. This may be similar to the situation found in other AAA+ family members that only achieve their full oligomeric status in the presence of their target.
Figure 8.
Summary of interactions detected within the MCM complex. The dotted lines indicate that the N-half-core interactions could be occurring in cis and/or trans.
The presence of the N-half, although not essential, is clearly stimulatory to the 3′ end dependent helicase activity of the AAA+ domain. This is evident either when the domains are covalently joined or mixed together as isolated fragments. Previously, the highly stable double hexameric nature of the N-terminal half of Mth MCM had implicated it as the organising centre of the complex. However, we find that the exactly analogous region of Sso MCM forms a range of lower-order multimeric forms in solution, suggesting that the stable formation of a double hexamer is not key to the role of the N-half in SsoMCM. Overall, we find evidence for four interfaces within the Sso complex: N-half to N-half, AAA+ to AAA+, HTH to HTH and N-half to AAA+, suggesting a more distributed pattern of complex formation than that seen in Mth MCM.
The isolated AAA+ domain has a Kd greater than 65 µM by EMSA compared to a Kd of around 12 µM for the N-half. However, a synergistic effect was seen in the presence of both domains. Relative to the isolated N-half, a fivefold increase was seen for a mix of the separate fragments and a 65-fold increase was seen for the covalently linked domains. This indicates that both sets of DNA binding sites are able to bind simultaneously, presenting an extended interface for interaction and reducing the probability of dissociation. This, combined with the ring structure of the hexamer topologically linking MCM to its track, would result in a highly stable complex. However, an independent DNA binding site from that necessary for helicase progression could conceivably have a detrimental effect, perhaps acting like a brake. The interaction between the motor domain and the N-half opens the possibility of dynamic communication of these two domains, potentially influencing the relative positioning of their DNA binding domains. In this light, it may be significant that we have recently described mutations in the N-terminal DNA-binding β-hairpin that prevent DNA-mediated stimulation of MCM ATPase activity (15). This proposed interplay between N-terminal domains and the ATPase domain may be reflected in our observation of N-half-mediated repression of helicase activity on 5′-tailed and flush duplex substrates.
The formation of a double hexamer has been suggested to be critical to the function of Mth MCM at the origin, analogous to the situation seen with the SV40 LTag (8,16). Our data indicate that Sso MCM can also form higher-order complexes, presumably including this species. Its existence seems dependant on the presence of the most peripheral domains, the A domain of the N-half and the C-terminal HTH domain, neither of which are at the hexamer–hexamer interface seen in the crystal structure of Mth MCM. The yeast bob1 mutation, which deregulates initiation, results in a change of the orientation of the A domain (16). An EM comparison of the N-half and full-length Mth MCM also shows a large realignment of the A domain (12). Other structural studies suggest that the A and HTH domains are close to each other in space, possibly allowing a physical interaction between them (14). This suggests a model in which formation of the double hexamer, and thus initiation, is controlled by the position of the A domain relative to the rest of the N-half, which in turn is regulated by the HTH domain. Further evidence of the regulatory properties of the HTH domain comes from the enhancement of helicase and ATPase activity upon its deletion. Interestingly, a recent study of Mth MCM has also revealed that removal of the HTH domain stimulates the DNA-dependent ATPase and helicase activities of the Mth protein, suggesting that the repressive nature of the HTH may be a general feature of MCMs (19).
These results support the idea of the AAA+ domain as a coherent module that supplies the driving force for helicase action, with other domains recruited to optimise or regulate it. This observation is presumably reflective of the evolutionary derivation of the MCM helicase. The progenitor form of the protein may have been the AAA+ domain in isolation that then associated with additional partners to enhance its intrinsic activity. During the course of evolution, gene fusion events may have occurred that “locked” these domains together. In this regard, we have recently described a bacteriophage-encoded protein that has a eukaryal primase domain fused to an MCM ATPase domain (20). Our finding in the current work that the N-terminal domains augment the processivity of the AAA+ helicase activity is reminiscent of the function of molecules such as the bacterial β-clamp and archaeo-eukaryal PCNA. Indeed, this analogy has already been proposed, based on gross structural similarities between the ring shapes of sliding clamps and the Mth MCM N-terminal domains (16). Sliding clamps enhance the processivity of DNA polymerases and additionally serve as docking sites for further accessory factors (21). We believe that this analogy is further strengthened by our observation that the Sulfolobus GINS complex binds to the N-terminal domains of Sso MCM (22).
ACKNOWLEDGEMENTS
We are grateful to James Chong for communicating work prior to publication and for informative discussions. Work in SDB's laboratory is funded by the Medical Research Council. Funding to the pay the Open Access publication charge was provided by the Medical Research Council.
REFERENCES
- 1.Kelman LM, Kelman Z. Archaea: an archetype for replication initiation studies? Mol Micro. 2003;48:605–616. doi: 10.1046/j.1365-2958.2003.03369.x. [DOI] [PubMed] [Google Scholar]
- 2.Duggin IG, Bell SD. The chromosome replication machinery of the Archaeon Sulfolobus solfataricus. J. Biol. Chem. 2006;281:15029–15032. doi: 10.1074/jbc.R500029200. [DOI] [PubMed] [Google Scholar]
- 3.Tye BK. MCM proteins in DNA replication. Ann. Rev. Bioch. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- 4.Johnson EM, Kinoshita Y, Daniel DC. A new member of the MCM protein family encoded by the human MCM8 gene, located contrapodal to GCD10 at chromosome band 20p12.3-13. Nucleic Acids Res. 2003;31:2915–2925. doi: 10.1093/nar/gkg395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malorano D, Cuvier O, Danis E, Mechali M. MCM8 is an MCM2-7-related protein that functions as a DNA helicase during replication elongation and not initiation. Cell. 2005;120:315–328. doi: 10.1016/j.cell.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 6.Labib K, Diffley JF. Is the MCM2-7 complex the eukaryotic DNA replication fork helicase? Curr. Op. Gen. Dev. 2001;11:64–70. doi: 10.1016/s0959-437x(00)00158-1. [DOI] [PubMed] [Google Scholar]
- 7.Ishimi Y. A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J. Biol. Chem. 1997;272:24508–24513. doi: 10.1074/jbc.272.39.24508. [DOI] [PubMed] [Google Scholar]
- 8.Chong JPJ, Hayashi MK, Simon MN, Xu RM, Stillman B. A double-hexamer archaeal minichromosome maintenance protein is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. U.S.A. 2000;97:1530–1535. doi: 10.1073/pnas.030539597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelman Z, Lee JK, Hurwitz J. The single minichromosome maintenance protein of Methanobacterium thermoautotrophicum Delta H contains DNA helicase activity. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14783–14788. doi: 10.1073/pnas.96.26.14783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shechter DF, Ying CY, Gautier J. The intrinsic DNA helicase activity of Methanobacterium thermoautotrophicum Delta H minichromosome maintenance protein. J. Biol. Chem. 2000;275:15049–15059. doi: 10.1074/jbc.M000398200. [DOI] [PubMed] [Google Scholar]
- 11.Grainge I, Scaife S, Wigley D. Biochemical analysis of components of the pre-replication complex of Archaeoglobus fulgidus. Nucleic Acids. Res. 2003;31:4888–4898. doi: 10.1093/nar/gkg662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen YJ, Yu XO, Kasiviswanathan R, Shin JH, Kelman Z, Egelman EH. Structural polymorphism of Methanothermobacter thermautotrophicus MCM. J. Mol. Biol. 2005;346:389–394. doi: 10.1016/j.jmb.2004.11.076. [DOI] [PubMed] [Google Scholar]
- 13.Yu X, VanLoock MS, Poplawski A, Kelman Z, Xiang T, Tye BK, Egelman EA. The Methanobacterium thermoautotrophicum MCM protein can form heptameric rings. EMBO Rep. 2002;3:792–797. doi: 10.1093/embo-reports/kvf160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pape T, Meka H, Chen SX, Vicentini G, van Heel M, Onesti S. Hexameric ring structure of the full-length archaeal MCM protein complex. EMBO Rep. 2003;4:1079–1083. doi: 10.1038/sj.embor.7400010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGeoch AT, Trakselis MA, Laskey RA, Bell SD. Organization of the archaeal MCM complex on DNA and implications for helicase mechanism. Nat. Struct. Mol. Biol. 2005;12:756–762. doi: 10.1038/nsmb974. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher RJ, Bishop BE, Leon RP, Sclafani RA, Ogata CM, Chen XS. The structure and function of MCM from archaeal M. thermoautotrophicum. Nat. Struct. Biol. 2003;10:160–167. doi: 10.1038/nsb893. [DOI] [PubMed] [Google Scholar]
- 17.Poplawski A, Grabowski B, Long SF, Kelman Z. The zinc finger domain of the archaeal minichromosome maintenance protein is required for helicase activity. J. Biol. Chem. 2001;276:49371–49377. doi: 10.1074/jbc.M108519200. [DOI] [PubMed] [Google Scholar]
- 18.Kasiviswanathan R, Shin JH, Melamud E, Kelman Z. Biochemical characterization of the Methanothermobacter thermautotrophicus minichromosome maintenance (MCM) helicase N- terminal domains. J. Biol. Chem. 2004;279:28358–28366. doi: 10.1074/jbc.M403202200. [DOI] [PubMed] [Google Scholar]
- 19.Jenkinson ER, Chong JP. Minichromosome maintenance helicase activity is controlled by N- and C-terminal motifs and requires the ATPase domain helix-2 insert. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7613–7618. doi: 10.1073/pnas.0509297103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGeoch AT, Bell SD. Eukaryotic/Archaeal primase and MCM proteins encoded in a bacteriophage genome. Cell. 2005;120:167–168. doi: 10.1016/j.cell.2004.11.031. [DOI] [PubMed] [Google Scholar]
- 21.Vivona JB, Kelman Z. The diverse spectrum of sliding clamp interacting proteins. FEBS Letters. 2003;546:167–172. doi: 10.1016/s0014-5793(03)00622-7. [DOI] [PubMed] [Google Scholar]
- 22.Marinsek N, Barry ER, Makarova KS, Dionne I, Koonin EV, Bell SD. GINS, a central nexus in the archaeal DNA replication fork. EMBO Rep. 2006;7:539–545. doi: 10.1038/sj.embor.7400649. [DOI] [PMC free article] [PubMed] [Google Scholar]