Abstract
To identify noncoding RNAs (ncRNAs) in the pathogenic bacterium Listeria monocytogenes, we analyzed the intergenic regions (IGRs) of strain EGD-e by in silico-based approaches. Among the twelve ncRNAs found, nine are novel and specific to the Listeria genus, and two of these ncRNAs are expressed in a growth-dependent manner. Three of the ncRNAs are transcribed in opposite direction to overlapping open reading frames (ORFs), suggesting that they act as antisense on the corresponding mRNAs. The other ncRNA genes appear as single transcription units. One of them displays five repeats of 29 nucleotides. Five of these new ncRNAs are absent from the non-pathogenic species L. innocua, raising the possibility that they might be involved in virulence. To predict mRNA targets of the ncRNAs, we developed a computational method based on thermodynamic pairing energies and known ncRNA–mRNA hybrids. Three ncRNAs, including one of the putative antisense ncRNAs, were predicted to have more than one mRNA targets. Several of them were shown to bind efficiently to the ncRNAs suggesting that our in silico approach could be used as a general tool to search for mRNA targets of ncRNAs.
INTRODUCTION
Noncoding RNAs (ncRNAs), other than ribosomal (rRNAs) and transfer RNAs (tRNAs), are recognized as important regulators in eukaryotes and prokaryotes. In bacteria, ncRNAs usually regulate gene expression either by pairing to mRNAs and affecting their stability and/or translation or by binding to proteins and modifying their activity (1). ncRNAs mostly function as coordinators of adaptation processes in response to environmental changes, integrating environmental signals and controlling target gene expression (2,3). Few ncRNAs controlling virulence in bacteria have been identified, the best-documented example being RNAIII of Staphylococcus aureus (4,5).
Listeria monocytogenes, a Gram-positive bacterium, is the etiologic agent of listeriosis, a severe human infection with an overall 30% mortality rate (6). A number of virulence-related genes have been identified (7). They are regulated either by PrfA, a master transcriptional activator whose expression is under the control of an RNA thermosensor, or by the stress sigma factor SigB, or by two-component systems (8–12). A L. monocytogenes strain deficient for Hfq, a protein involved in the pairing of ncRNAs with their mRNA targets in E. coli, is affected in tissue colonization in mouse, suggesting that ncRNAs might be involved in the control of virulence (13,14). Very recently an Hfq-immunoprecipitation method allowed the identification of three ncRNAs in L. monocytogenes (15). This approach being restricted to Hfq-binding ncRNAs, we had decided to undertake a global search for ncRNAs.
The identification of mRNA targets of ncRNAs is essential to understand their functions, although it remains a challenging issue. Known examples of bacterial ncRNA–mRNA hybrids generally feature internal loops and bulges, a major obstacle in predicting ncRNA targets (16,17). A computational method has been developed in E. coli, to search for complementarities between a ncRNA and mRNAs (18). The empirical alignment score employed in (18) equally weights A–U and C–G pairings, despite the stronger pairing energy of the latter. This may not be optimal for genomes with low GC content, such as that of Listeria (19).
Among possible approaches to search for ncRNAs (reviewed in (20)), we employed computer-based methods, followed by northern blots and 5′ end mapping. This allowed us to identify 12 ncRNAs in the L. monocytogenes genome EGD-e (19). We also developed a novel computational method to predict mRNA targets of ncRNAs and were able to predict mRNA targets for three of our novel ncRNAs. These predictions were experimentally validated, strongly suggesting regulatory functions for these ncRNAs in Listeria.
MATERIALS AND METHODS
In silico analysis of IGR candidates for ncRNAs
- Selection of intergenic region candidates for ncRNA genes. Sequences between annotated open reading frames, i.e. intergenic regions (IGRs), in the L. monocytogenes EGD-e genome available at the ListiList web site (http://genolist.pasteur.fr/ListiList/) were considered in this study (19). We restricted our analysis to IGRs with a minimal size of 150 bp. Regions carrying rRNAs, tRNAs and within the A118 cryptic phage were excluded. A total of 694 IGRs were retained and analyzed using the BLAST program for their degree of conservation with sequences of microbial genomic sequences available at the NCBI web site (http://www.ncbi.nih.gov/BLAST/) (21). Three kinds of conservation were observed:
- IGRs displaying portions of relatively long sequences (≥80 bp) highly conserved between EGD-e and other Listeria genomes (magenta or red according to the BLAST color code), while the remaining parts were not similar. In these alignments (∼8% of the total IGRs), conservations within IGRs concerned the L. monocytogenes genomes and L. innocua occasionally, but no other bacterial genome. Since conservation may reveal the presence of a transcribed region, all of the IGRs in this group were kept for further analysis using folding predictions.
- IGRs entirely conserved between Listeria species (∼2% of the total IGRs), with portions of the sequence repeated elsewhere in the Listeria genomes and in other Gram-positive bacteria, mainly Bacillus species. In general, such type of conservation signed riboswitches (see below). However, three of these IGRs were predicted to carry the ncRNA genes rnpB, ssrA and ssrS and were kept as controls.
- Remaining IGRs (∼90% of the total IGRs) were entirely conserved among L. monocytogenes isolates (EGD-e and F6854, serotypes 1/2a; F2365 and H7858, serotypes 4b) and occasionally in the non-pathogenic species L. innocua (isolate CLIP11262). A few of these IGRs were kept for further analysis on the basis of qualitative criteria such as their size (≥350 bp) and the orientation of flanking genes (either both convergent or both divergent open reading frames (ORFs)). A total of 99 IGRs were thus selected for folding prediction analysis (Table S1).
Screening IGRs by RNA folding predictions. Each strand of the DNA sequence from the 99 IGRs previously selected, was assimilated to a RNA molecule and was folded by using the MFOLD program (22). Folding analysis indicated that 29 IGRs displayed on one of the strands, at least two consecutive stem-loops, distant by at least 50 nt from the adjacent ORFs, spaced by a maximum of 50 nt. Within these stem-loops, at least five bases were forming the stem including at least one G:C pairing and a loop smaller than 10 nt (Table S1).
Orphan transcription terminators. We searched for Rho-independent terminators with standard pattern-recognition algorithms using criteria defined for E. coli (23). The pattern was modified to include stems with at least six pairings containing three G:Cs to compensate for the high AT content of Listeria genome (19). Seven IGRs containing a predicted transcription terminator (indicative of a potential transcriptional unit) flanked by two divergent ORFs, were kept for further analysis (Table S1).
ORF predictions. ORFs were predicted within the IGRs using the NEBcutter program (http://tools.neb.com/NEBcutter2/index.php), setting the minimum ORF length to 30 residues and searching for a potential ribosome binding site preceding the initiation codon (AUG, GUG, UUG, CUG or AUU). Once the 5′ ends of ncRNAs were mapped, translatable ORFs were sought on transcribed sequences, with a threshold on their minimal length set to 10 residues.
Identification of riboswitches. We identified riboswitches and ncRNAs, RnpB, SsrA and SsrS in the selected IGRs using the Rfam database (http://www.sanger.ac.uk/Software/Rfam/index.shtml) (24), (Table S1).
Predictions of mRNA targets
ncRNA targets were found by searching the whole genome for strong ncRNA–mRNA duplexes. The strengths of the duplexes were quantified by a pairing score S, constructed as a sum of both positive contributions due to pairing nucleotides and negative contributions due to bulges and internal loops (Table S2). The contribution of A–U, G–C and G–U pairs was taken as the absolute value of thermodynamic binding energies, considering stacking effects (22). The score S will therefore coincide with the absolute value of the free energy for a perfectly pairing duplex (without bulges and internal loops). As for the cost of bulges and internal loops, we empirically gauged it by maximizing the significance of four known hybrid pairings characterized in vivo, RyhB–sodB mRNA, DsrA–rpoS mRNA, Spot42–galEK mRNA in E. coli and RNAIII–spa mRNA in S. aureus, (17, 42–44) as compared to other genes in the respective genomes. ncRNA targets were sought in 5′ regions spanning 140 bases upstream of the translation start codon and 90 bases within all the coding regions, and in 3′ regions spanning 60 bases upstream of the translation stop codon and 90 bases downstream of all the annotated ORFs of L. monocytogenes genome. DNA regions were converted to RNA and their best alignments with ncRNAs searched by standard dynamic programming. Various pairing lengths were considered since our alignment scores do not have zero average over random sequences (25). Statistical significance of the pairings was assessed with respect to an ensemble of random sequences (230 nt and 150 nt long), generated by a Markov chain model gauged on the pentanucleotide statistics of the aforementioned sequences. The threshold of statistical significance was empirically set to the ratio five between the number of genes with a pairing score ≥S and those expected by chance. Predictions for the trans targets of the RliE ncRNA were below the threshold, yet they were kept for further analysis due to the functional relationship among the target genes. The computer program to compute the best RNA alignments is available as a supplementary material file (Document S3).
Strains and plasmids
All strains used were derivatives of the L. monocytogenes strain EGD-e (BUG1600 (19)). Δhfq (BUG2213), ΔprfA (BUG2214) and ΔsigB (BUG2215) mutants were obtained by deletion of the corresponding ORF using the suicide vector pMAD (26). Deletions were generated by PCR-ligation and amplicons were cloned at the SmaI restriction site of the pMAD vector. Overexpression of RliB and RliI were obtained by cloning the genomic loci into the pAT18 vector (27) resulting in strains BUG 2348(EGD-e/pAT18-rliB), BUG2349(EGD-e/pAT18-rliI) and BUG2347 (EGD-e/pAT18). Oligonucleotides flanking the corresponding rli loci were designed so that ∼200 bp of DNA sequence from the most upstream 5′ extremity mapped by RACE were included in the construct. Amplicons were introduced at the SmaI restriction site of the pAT18. All constructs on the plasmid and on the chromosome were verified by sequencing (Genomexpress). Oligonucleotides used in this study are listed Table S4.
Bacterial growth
Bacteria from a single fresh colony on a brain–heart infusion (BHI) agar plate, were grown aerobically overnight and a 1/500 dilution of the preculture was made into 60 ml of prewarmed liquid BHI at 37°C. Growth was then monitored. Total RNA was extracted as previously described (9), from bacteria grown to an OD600 between 0.4 and 0.55 for the exponential growth phase, and between 1.2 and 1.4 for the stationary growth phase. No significant growth defect was observed for any of the EGD-e derivative strains used. When required, erythromycin was used at 5 μg/ml as final concentration.
Northern blots and 5′ end mapping
Northern blots were performed as described earlier (28,29), using oligonucleotides described in Table S4. Briefly, for northern analysis of IGR candidates in low stringency conditions, double-stranded DNA probes were generated by PCR from a colony of L. monocytogenes EGD-e strain. PCR amplification was realized with annealing at 50°C for 30 cycles in 1× PCR buffer (1 mM each dCTP, dGTP, and dTTP; 2.5 µM dATP; 100 µCi [α32P]dATP; 1U taq polymerase) (INVITROGEN). Probes were purified over G-50 microspin columns (Amersham Pharmacia Biotech) prior to usage. Northern membranes were prehybridized in a 1:1 mixture of Hybrisol I and Hybrisol II (Intergen) at 40°C. DNA probes were heated for 3 min at 95°C and directly added to the prehybridization solution; membranes were hybridized overnight at 40°C. Membranes were washed by rinsing twice with 4× SSC/0.1% SDS at room temperature followed by three washes with 2× SSC/0.1% SDS at 40°C. Northern blot analysis in high stringency conditions was realized using an oligonucleotide probe and Ultrahyb solution (AMBION), as described by the manufactured protocol (AMBION). Oligonucleotides were 5′ labeled with [γ32P]ATP using the T4 Polynucleotide Kinase as recommended by the manufactured protocol (New England Biolabs). Northern membranes were prehybridized in Ultrahyb at 40°C, followed by addition of labeled oligonucleotide probe and hybridization overnight at 40°C. Membranes were washed twice with 2× SSC/0.1% SDS at room temperature followed by two washes with 0.1× SSC/0.1%SDS for 15 min each at 40°C. Northern blots shown in the figures are a representative experiment repeated at least twice for each one of three independent RNA preparations. Signals were quantified by using the NIH Image program (http://rsb.info.nih.gov/nih-image/).
The 5′ RACE method allows discriminating a 5′ end generated by transcription initiation, from a 5′ end provided by RNA processing. 5′ RACE mapping was performed as described earlier (30) with slight modifications. Reactions were conducted on RNA extracted in exponential phase. Once the cDNA was amplified by PCR, products were cloned into the pCRIITOPO vector (INVITROGEN). Clones were screened by PCR using oligonucleotides FR.T7 and FR.SP6 (29). Fifteen clones carrying an insert were selected for each cDNA. Sizes of amplicons were compared and three clones of each size were sequenced (Genomexpress).
Gel shift assays
RNA gel shift assays were performed as described earlier (17). Briefly, uniformly 32P- labeled ncRNA and predicted mRNA target fragments were synthesized in vitro using the T7 RNA polymerase and PCR fragments as template (see Table S2). ncRNA fragments synthesized were from the transcription start site (+1) mapped by 5′ RACE: nt 48 to 113 for RliE; nt 113 to 273 for RliB; nt 55 to 120 for RliI. mRNA targets were from the AUG of corresponding ORFs: comEA (nt −60 to +6), comFA (nt −56 to +6) and lmo0945 (nt −44 to +4) for assays with RliE, lmo1035 (nt +1781 to +1850) for assays with RliI, and lmo2104 (nt +17 to +177) for assays with RliB. Complex formation assays were performed at 37°C for 15 min. in a buffer containing Tris-HCl 25 mM pH 7.5, MgCl2 5 mM, KCl 50 mM in the presence of uniformily labeled ncRNA (<1 nM) and increasing concentrations of the cold target mRNA (10 nM to 1 µM). Before mixing, RNAs were renatured separately in the appropriate buffer.
Supporting information
Table S1: IGRs selected for northern blots.
Table S2: Parameters used for target selection.
Document S3: Computer program for mRNA target predictions.
Table S4: Oligonucleotides used in this study.
Document S5: Rli loci in EGD-e.
Document S6: Predicted folding of Rli ncRNAs by MFOLD.
RESULTS
In silico search for ncRNAs
ncRNA genes are located in IGRs of the genomes, i.e. sequences present between annotated ORFs (29–32), we therefore searched for ncRNAs in the IGRs of the L. monocytogenes EGD-e genome (19) aware that this approach excludes detection of ncRNAs located within annotated genes. We first analyzed the IGRs for their conservation across bacterial species by using BLAST alignments. Out of 694 IGRs, 99 were selected based on the following criteria: (i) IGRs carrying a portion of highly conserved sequence (≥80 bp) among Listeria species, since the conservation may sign a transcribed region and therefore a putative noncoding gene. (ii) Long IGRs (≥350 bp) entirely conserved among Listeria species, flanked by two divergent or two convergent ORFs, since the length of the IGR might increase the chances of finding a non-coding-protein gene. Then we performed predictions for RNA secondary structure and ‘orphan’ transcription terminators (see description in Materials and Methods). Thus, 36 IGRs were selected. Among these, six IGRs containing potential ORFs that had not been annotated in the EGD-e genome due to their relatively short length were removed from the data set, yielding 30 IGR candidates. These IGRs were screened by using the Rfam database (24), enabling us to identify one riboswitch and the three IGRs carrying the predicted three ncRNAs conserved in all bacteria that we kept as controls: RnpB, the ribozyme component of RNaseP (33), SsrA, the RNA rescuing stalled ribosomes (34), and SsrS, the RNA polymerase modulator (35,36). Thus 29 IGRs were kept for further investigation (Table S1).
Detection of IGR-encoded transcripts
To detect ncRNAs in the 29 selected IGRs, we performed northern blots on total RNA extracted from L. monocytogenes EGD-e grown in BHI in exponential and stationary phases, using low stringency hybridization conditions with PCR products corresponding to the entire IGRs as probes. Signals were analyzed by taking into account the length of the transcripts compared to the size of the corresponding IGR, the orientation of the flanking genes and the presence of transcription terminators. The three controls were nicely detected. For four IGRs, transcripts could not be detected. Thirteen IGRs displaying long transcripts that could be part of operons, 5′- or 3′UTR mRNAs were not kept for further analysis.
The nine remaining IGRs encoded transcripts that could not be assigned to any of the above categories, indicating that they could carry previously unknown ncRNAs. These transcripts were named Rli for RNA in Listeria (from A to I).
To determine the transcription orientation of the potential ncRNAs, northern blots in high stringency hybridization conditions were performed using oligonucleotide probes chosen on both strands of the IGRs (Figure 1). A high expression was observed for the predicted ubiquitous ncRNAs, RnpB, SsrA and SsrS. RliB, E, F, G, H and I, showed distinct bands ranging from 110 to 430 nt, consistent with the length of the corresponding IGRs (Figure 1). Three candidates, RliA, C and D, could not be detected using oligonucleotide probes but were kept for further analysis, since highly stable RNA stem-loop structures may prevent their detection (29,30,37).
Figure 1.
Detection of ncRNAs by northern blots. The name of the ncRNA is indicated on top of each panel. Total RNA was extracted from L. monocytogenes EGD-e strain grown to exponential (E) or stationary (S) phase in BHI at 37°C. Blots were performed with 12 µg of total RNA per lane and strand-specific probes, except for RliA, RliC and RliD where PCR products corresponding to the IGRs were used. ‘L’ indicates the DNA ladder used to estimate length. One lane of DNA molecular weight markers is shown for estimation of RNA size, since DNA and RNA run slightly differently on gel. Numbers on the right side of each panel indicate the estimated lengths of the transcripts in nucleotides. Exposure times were optimized for each panel and signal intensity does not indicate relative abundance between ncRNAs.
Expression of ncRNAs
Northern blots presented in Figure 1 allowed to compare the levels of the Rli transcripts in exponential and stationary growth phases in BHI. Levels of RliF and RliI were higher in stationary than in exponential phase, reminiscent of ncRNA expression in other bacteria, i.e. varying according to environmental conditions and growth phases (2,3).
In E. coli, Hfq usually affects the stability of ncRNAs that interact with mRNAs (16). We compared levels of Rli in a Δhfq mutant and the isogenic wild-type EGD-e strain using northern blots. No variations were observed, indicating that Hfq does not affect the abundance of our ncRNAs under the conditions used (not shown).
Since PrfA and SigB are two important transcription regulators involved in the virulence of L. monocytogenes, we also tested the prfA- and sigB-dependent expression of rli genes by northern blots, comparing levels of Rli ncRNAs in ΔprfA and ΔsigB mutants to the isogenic wild-type strain. No PrfA- or SigB-dependent expression was observed in exponential and stationary growth phases at 37°C in BHI medium (not shown).
5′ end mapping
5′ ends of Rli ncRNAs were mapped by RACE experiments to discriminate between primary 5′ ends, i.e. transcriptional start sites, and 5′ extremities generated by processing. 5′ ends were mapped for all candidates except RliF. In particular, we identified 5′ ends for RliA, RliC and RliD, which were not detected earlier by northern blots using oligonucleotide probes, therefore demonstrating their existence. Transcriptional start sites were mapped for rliA, B, C, E, G, H and I, enabling us to predict RpoD-dependent promoters for these genes except for rliE. Based on the 5′ ends mapped, transcript length observed on gel and folding predictions (in particular transcription terminators), 3′ ends could be assigned. Rho-independent transcription terminators could be detected for RliF, G, H and I. In the case of RliA, the observed length on northern blots and the proximity of the downstream ORF (lmo0477) with the same orientation (136 nt), did not allow us to rule out the possibility that RliA may be derived from the processing of lmo0477 mRNA. Collectively, these data enabled us to estimate the lengths for all ncRNAs, e.g. RliA (225 nt), B (360 nt), C (360 nt), D (≥380 nt), E (225 nt), F (∼180 nt) G (280 nt), H (430 nt) and I (240 nt), as well as their positions on the EGD-e genome (see Figure 2, Documents S4 and S5). No translatable ORF with a minimal length of 10 residues could be found on Rli transcripts, indicating that most likely these transcripts do not encode small peptides and constitute bona fide noncoding RNAs.
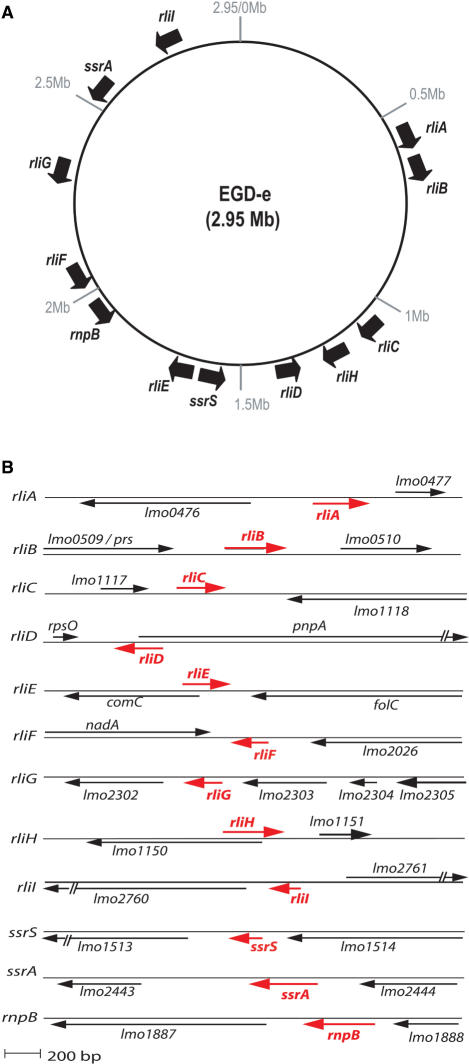
Figure 2.
(A) Distribution of the rli genes along the L. monocytogenes genome of EGD-e strain. Nucleotide position, in Megabase (Mb), is indicated. (B) rli loci in L. monocytogenes. Red arrows represent the rli transcripts and their orientation; black arrows indicate the flanking ORFs. Maps are at scale, based on 5′ RACE mapping data, sizes estimated on gel and transcription terminator predictions. For RliD and RliF, transcription start sites have not be determined; positions and lengths are estimated. Document S4 provides information on sequences and the genomic location of rli genes.
Features of rli genes
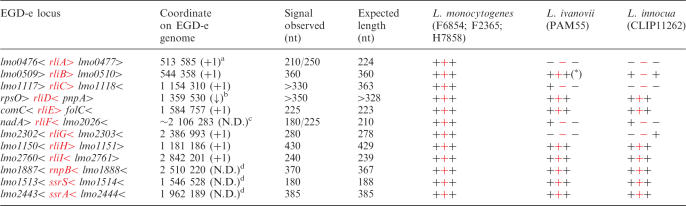
The nine rli sequences were found in the four known L. monocytogenes genomes (strains EGD-e, F6854, F2365 and H7858) (19,38). We also sought for the presence of rli genes in the genome of L. innocua (strain CLIP11262), a non-pathogenic species (19), and L. ivanovii (strain PAM55), an ovine pathogenic species (sequence provided by P. Glaser) (Table 1). Four classes of rli genes could be distinguished according to their conservation across Listeria species, their location on the chromosome, and their structural particularities.
Four rli genes are specific to L. monocytogenes species. rliA, rliC, rliF and rliG were present in L. monocytogenes and absent from L. innocua and L. ivanovii. The absence of rli sequences was associated with the absence of surrounding ORFs, except for the rliF and rliG loci for which upstream genes were present (Table 1).
Three rli genes encode antisense ncRNAs. In contrast to the other rli genes, rliD, rliE and rliH partially overlap with and are transcribed divergently to the ORFs of their flanking genes pnpA, comC and lmo1150, respectively, indicating that these ncRNAs may act as antisense (Figure 2B). pnpA encodes the polynucleotide phosphorylase (PNPase) (39), comC encodes a putative type IV prepilin peptidase analogous to the comC gene of Bacillus subtilis (40), and lmo1150 encodes a putative transcription regulator similar to Salmonella typhimurium PocR (41), (Figure 2B). rliD, E and H are also present in L. innocua and L. ivanovii species (Table 1).
rliB carries five long repeats. In L. monocytogenes, RliB displays five repeats of 29 nt spaced by 35–36 nt (Figure 3). rliB is absent in L. innocua, but two copies were found in L. ivanovii (Table 1): the first one, rliBiv1, is flanked by the same ORFs as in L. monocytogenes and is highly homologous to rliB; the second, rliBiv2 is located elsewhere on the chromosome and is flanked by ORFs absent in L. monocytogenes. rliBiv2 would encode a ncRNA carrying seven repeats of 29 nt.
rliI gene is a single transcription unit within the corresponding IGR (Figure 2) and is conserved in L. innocua and L. ivanovii species (Table 1).
Table 1.
Presence of the rli loci in Listeria species
The first column provides the ncRNA loci in the L. monocytogenes EGD-e genome. ‘<’ and ‘>’ indicate the orientation of the genes marked on the left. ncRNA are noted in red. The second and third columns correspond to the genomic position of the most upstream 5′ end mapped by RACE, and the approximate length estimated on northern blot, respectively. The fourth column indicates the size of the corresponding Rli ncRNA deduced from the 5′ end mapping, the transcription terminator prediction and the length observed on gel. The last three columns correspond to the Listeria species; corresponding isolates are mentioned in parenthesis. ‘+’ and ‘−’ indicate the presence or the absence of the genes.
*rliB is conserved and duplicated in L. ivanovii. The length and the genomic location of Rli transcripts is provided in Document S4.
a5′ end mapped corresponds to a transcription start site;
b5′ end mapped is a processing site; N.D.: not determined;
cnot mapped and estimated from length observed on gel and transcription terminator prediction;
ddeduced from sequence conservation available at Rfam web site.
Figure 3.
Sequence of rliB locus. In red, the rliB-transcribed sequence; arrows show the transcription orientations. The five RliB repeats (guuuuaguuacuuauUGugaaauRuaaau) are underlined and numbered with roman numerals. The transcription start site of rliB (+1) and a processing site (↓) mapped are shown. Deduced –10 and –35 hexamers of a putative RpoD-dependent promoter are boxed.
Prediction of mRNA targets
We developed a computational method to identify mRNA targets of ncRNAs, based on thermodynamic pairing energies and experimentally validated ncRNA–mRNA hybrids (see Materials and Methods). The hybrids used were RyhB–sodB, DsrA–rpoS and Spot42–galEK mRNAs from E. coli and RNAIII–spa mRNA from S. aureus (17,42–44). Our program scans a bacterial genome searching for mRNAs forming stable duplexes with a given ncRNA. Deviations from random expectations indicate putative mRNA targets. This approach biases our predictions to strong duplexes formed between the ncRNA and its targets. We limited our search to genomic regions spanning 140 bases upstream of the translation start codon and 90 bases within the coding region, and to regions spanning 60 bases upstream of the translation stop codon and 90 bases downstream of all the annotated ORFs in L. monocytogenes genome EGD-e.
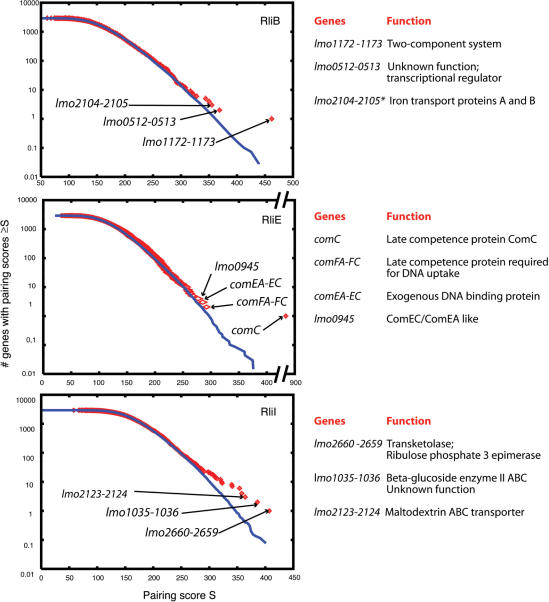
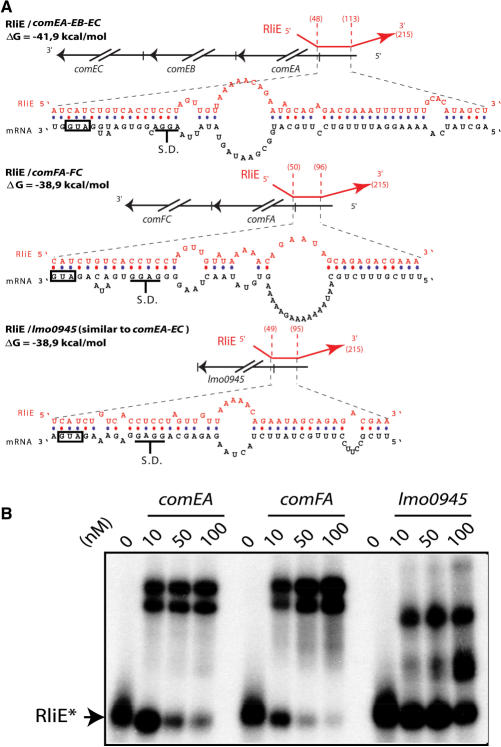
We searched for potential mRNA targets for the nine Rli ncRNAs of L. monocytogenes. Significant predictions were found for RliB, RliE and RliI (Figure 4). For each of these three ncRNAs several targets were identified, suggesting that these ncRNAs could have pleiotropic effects. Besides the antisense pairing with comC, the strongest predicted pairings of RliE were with comEA-EB-EC, comFA-FC and lmo0945 mRNAs. These mRNAs encode proteins highly homologous (>40% identity) to factors of the competence machinery in B. subtilis (40). Remarkably, the same sequence of RliE (nt 49 to 113) would pair with the 5′ leader region of the other mRNA targets, including the translation start codon and the Shine–Dalgarno (S.D.) sequence of the predicted operons (Figure 5A).
Figure 4.
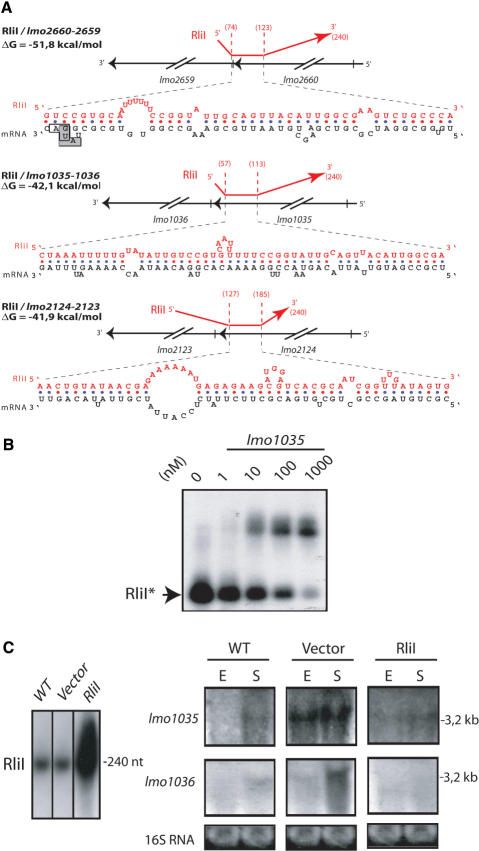
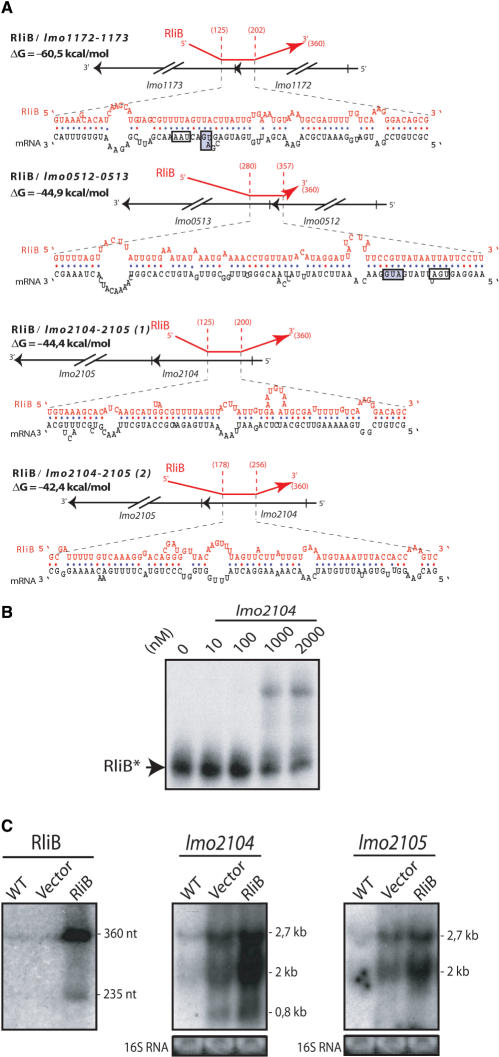
Prediction of mRNA targets for RliE, RliB and RliI. On the left: the pairing scores are computed as described in the text. In blue, the corresponding number of genes expected by chance, according to a null Markov model of length five. In red, the number of genes in EGD-e genome having a pairing strength ≥S, the value on the abscissas. Arrows indicate transcripts showing strongest pairings with the corresponding Rli sequence. On the right: list and function of genes whose mRNA are predicted targets for the corresponding ncRNA. *Two pairing regions are predicted between RliB and lmo2104-2105 mRNA.
Figure 5.
Hybrids between RliE and mRNA targets. (A) For each hybrid, the upper part shows the region of RliE (red) that pairs with the mRNA (black). Numbers in parenthesis indicate the first and the last bases of RliE involved in the pairing. The lower part shows the predicted pairings of RliE with the mRNA at the nucleotide level. Boxes indicate translation start codons, and S.D. the Shine–Dalgarno sequence is shown. (B) In vitro duplex formation between RliE and predicted mRNA targets indicated at the top of each panel. At the top of each lane, numbers indicate the concentration used (nM) of unlabeled RNA with <1 nM of labeled RliE. Unbound radioactive RliE* is indicated.
RliI showed the strongest predicted pairing with the 3′ ends of the first genes of three putative bicistronic transcripts: lmo2660-2659, carrying two overlapping ORFs and encoding a transketolase and a ribulose-phosphate epimerase, lmo1035-1036, encoding a beta-glucoside transporter subunit of a PTS system and a beta-glucosidase, respectively, and lmo2124-2123, encoding components of a maltodextrine ABC transporter system (Figure 4). The location of the predicted pairing on the mRNAs strongly suggests that RliI regulates polarity. Two distinct regions of RliI would be involved in these pairings, one spanning nt 60 to 125 and pairing with the 3′ end of lmo2660 and lmo1035, the other from nt 130 to 185 hybridize with the 3′ end of lmo2124 (Figure 6A). Remarkably, predicted mRNA targets of RliI have biological functions related to sugar metabolism and transport. In relationship with the function of predicted targets of RliI, it is also important to note that rliI is transcribed in the opposite direction to the flanking gene lmo2761 encoding a β-glucosidase located at the end of a putative operon encoding cellobiose and xylose PTS systems, an other set of genes involved in sugar metabolism and transport (19).
Figure 6.
Hybrids between RliI and mRNA targets. (A) For each hybrid, the upper part shows the region of RliI (red) that pairs with the mRNA (black). On the nucleotidic sequence, the translation start codon of lmo2659 is indicated by a grey box, the translation stop codon of lmo2660 by an open box. Data are otherwise presented as in Figure 5A. (B) Duplex formation between RliI and lmo1035 mRNA. Data are presented as in Figure 5B. (C) northern blots on 15 µg of total RNA extracted from L. monocytogenes EGD-e (WT), EGD-e harboring the empty vector pAT18 (vector) and pAT18 carrying the rliI gene (RliI) grown to exponential (E) or stationary (S) phase in BHI at 37°C. The RNA probed is indicated on the left of each panel. RliI is probed with a specific oligonucleotide, lmo1035 and lmo1036 transcripts were probed with PCR fragments. Estimated sizes of the transcripts are indicated on the right side of each panel. As loading control, gels were stained with EtBr before transfer to evaluate 16S rRNA levels (16S RNA).
RliB showed the strongest predicted pairing with the putative bicistronic transcripts lmo1172-1173, encoding a two component system, lmo0512-0513 that encodes a protein of unknown function and a putative transcription regulator, and lmo2104-2105, encoding the ferrous iron transport proteins FeoA and FeoB, respectively (Figure 4). The overlapping regions at the end of lmo1172 and the beginning of lmo1173, and the IGR between lmo0512 and lmo0513 would be engaged in the pairing with RliB, suggesting that the ncRNA might regulate polarity of these mRNAs (Figure 7A). For lmo2104-2105 mRNA, RliB is predicted to pair with two long regions (>70 nt) within the encoding sequence of lmo2104, involving 146 nt out of the 228 nt encoding the short protein FeoA (75 aa), and suggesting a tight regulation of this mRNA by RliB (Figure 7A).
Figure 7.
Hybrids between RliB and mRNA targets. (A) Hybrids between RliB (red) and predicted mRNA targets (black). Data are otherwise presented as in Figure 5A. (B) Duplex formation between RliB and lmo2104 mRNA. Data are presented as in Figure 5B. (C) northern blots on total RNA was extracted from bacteria grown to exponential phase in BHI at 37°C. Data are presented as in Figure 6C.
Gel shift assays of predicted mRNAs with ncRNAs
We then tested in vitro the pairing of RliB, E and I with their predicted mRNA targets by RNA gel shift assays using RNA fragments corresponding to the sequences predicted to interact.
For RliE, in vitro duplex formations were assessed using a 32P-labeled ncRNA fragment and the three unlabeled counterpart comEA-, comFA- and lmo0945 RNAs. Remarkably, RliE was found to bind efficiently the three predicted mRNAs with an apparent dissociation constant ranging between 20 to 100 nM, indicating an interaction as stable as those reported for other validated ncRNA–mRNA hybrids (17,32,45). For the three RliE–mRNA complexes, a double band was visualized that may correspond to different conformers (Figure 5B).
For RliI, among the three predicted mRNA targets, in vitro duplex formation was tested for lmo1035. A complex was observed between a 32P-labeled RliI fragment and an unlabeled RNA fragment containing the predicted RliI targeted region of lmo1035 (Figure 6B). The apparent dissociation constant observed was ∼100 nM, very similar to those observed for RliE and com-like mRNAs.
Since RliB was predicted to pair with two regions within lmo2104 mRNA, we tested the duplex formation with a 173-nt long fragment encompassing the two complementary regions of lmo2104. A complex was observed between a labeled RliB fragment and the unlabeled RNA fragment containing the two predicted RliB-targeted regions (Figure 7B). This result showed the possibility of the interaction between the two RNAs. However, the formation of the complex between RliB and lmo2104 was poorly efficient since the apparent dissociation constant was about 10-fold higher (>1 μM) than that for RliE–com-like mRNAs and RliI–lmo1035-1036 mRNA, indicating that the structures of both RNA are not promoting efficient binding.
In summary, the five predicted mRNA targets tested were able to bind RliE, RliI and RliB in vitro, suggesting that in vivo the ncRNAs may interact and regulate the expression of their mRNA targets.
In vivo effects of the overexpression of ncRNAs
Since in vitro data suggested that hybrids between RliB, E and I and their predicted mRNA targets can form in vivo, we searched for effects of the ncRNAs on their respective targets. Assuming that the action of the ncRNAs could result in changes in the mRNA stability, we analyzed by northern blots levels of the predicted mRNA targets when the ncRNAs were overexpressed. As shown in Figures 6C and 7C, we were able to overexpress RliI and RliB by cloning the corresponding genomic loci in a multicopy plasmid (pAT18) under the control of their endogenous promoters. The expression of these two ncRNAs and their lengths observed by northern blot agreed with the lengths of RNAs expressed from the chromosome, definitively establishing that rliB and rliI genes are contained within the genomic fragment cloned into the vector. For unknown reasons, we were unable to overexpress RliE from its own or from an inducible promoter, suggesting a complex transcriptional control of the rliE gene.
RliI. We first tested the expression of the mRNA targets of RliI in BHI medium at 37°C. northern blots on total RNAs in the wild-type EGD-e strain, showed a single transcript for lmo1035 and lmo1036, migrating around 3.2 kb, indicating a co-transcription of the two genes. An increased level was observed in stationary phase compared to the exponential phase (Figure 6C). The presence of the empty vector (pAT18) increased the basal level of the transcript, but preserved the growth-phase-dependent expression. An effect on the level of lmo1035-1036 mRNA was observed when the vector carrying rliI was used. Overexpression of RliI decreased the level of lmo1035-1036 mRNA, in both exponential (1.5-fold) and stationary growth phases (2.5-fold) as compared to levels obtained with the empty vector (Figure 6C). Thus, this situation is similar to that of many ncRNAs targeting mRNAs that generate ncRNA–mRNA hybrids that are rapidly degraded (16). Together, the effect observed when RliI was overexpressed suggested the possible interaction of the ncRNA with lmo1035-1036 mRNA in vivo, in agreement with the duplex formation observed in vitro (Figure 6B). In the growth conditions that we assayed, we could not detect lmo2124-2123 and lmo2660-2659 mRNAs by northern blots.
RliB. We tested the effect of the overexpression of RliB on its three mRNA targets (Figure 7A). We could not detect any significant effect of the overexpression of RliB on targets lmo0512-0513 and lmo1172-1173 mRNAs (data not shown). Two major bands at 2.7 and 2 kb were detected when probing either for lmo2104 or lmo2105 transcripts in the EGD-e wild-type strain, indicating that the two genes are co-transcribed (Figure 7C). A weaker transcript of 0.8 kb was also observed for lmo2104. As observed earlier for RliI, the vector alone (pAT18) led to an increase of the transcript levels compared to the wild-type strain. However, the presence of rliB gene on that vector, led to a reproducible further increase of the mRNA levels (about 2-fold for each transcript compared to the vector), suggesting that an interaction of RliB with lmo2104-2105 mRNA may occur in vivo (Figure 7C).
DISCUSSION
New noncoding RNAs in Listeria
Here we report the first genomic search for ncRNA in the Gram-positive pathogen L. monocytogenes. We identified and characterized nine novel ncRNAs (from RliA to RliI), whose size ranges from 110 to 430 nt (Figure 2). rli genes appear specific to the Listeria genus since no homologs were detected in other sequenced bacterial genomes. Our rli genes fall into four classes. One class is represented by rliA, C, F and G which are present in L. monocytogenes and absent in L. ivanovii and in the non-pathogenic species L. innocua. rliB is a representative of a second class; it contains five repeats of 29 nt. rliB is duplicated in L. ivanovii, an ovine pathogen, but is absent from L. innocua. It is thus tempting to hypothesize that those two classes of genes, being absent from L. innocua species, might encode ncRNAs involved in adaptation to the host during infection (Table 1). The third class includes antisense ncRNAs (RliD, E and H). The fourth class is represented by RliI which is encoded in the middle of an IGR. These two last classes are present in the three species of Listeria genus, suggesting that these ncRNA genes would control more global adaptation processes (Table 1).
Recently, three ncRNAs (LhrA, LhrB and LhrC) in L. monocytogenes have been described (15). These three ncRNAs did not appear among our ncRNA candidates for the following reasons: LhrA is expressed within an annotated ORF (Lmo2257) and we only analyzed IGRs; LhrB is carried by an IGR entirely conserved between Listeria species, and the five copies of LhrC ncRNAs are located in two IGRs that we did eliminate from our candidates since translatable ORFs can be predicted therein.
mRNA targets
We developed a computational approach to search for mRNA targets of ncRNAs in bacteria. Our method introduces a major improvement over other alternatives based on complementarities between the mRNA target and the ncRNA via BLAST- or FASTA-based programs (e.g. (18)), since the energetic score employed in our alignments permits scanning of genomes with high AT content, e.g. >60% in Listeria.
Using this approach, we have searched for new mRNA targets of already known ncRNAs in bacteria other than Listeria. We discovered new targets for which experimental evidence already existed. For example, DsrA of E. coli (2) was predicted to pair, in addition to rpoS mRNA, with the translation initiation region of dcuS and rbsA mRNAs. dcuS encodes the histidine kinase of the DcuS/R two-component system, involved in the control of C4-dicarboxylate usage, and rbsA encodes the ATPase subunit of the d-ribose ABC transporter. Assuming that DsrA would decrease the translation or the stability of these mRNAs, our predictions may explain previous observations, i.e. the overexpression of DsrA impairs E. coli growth when succinate or ribose are used as carbon source (Repoila, F. and Gottesmann, S. unpublished data). The RNAIII of S. aureus was also predicted to form a hybrid with a novel target the SA1000 mRNA, an interaction that has now been demonstrated experimentally ((5) and unpublished data).
LhrA, B and C in L. monocytogenes EGD-e strain have been described as Hfq-binding ncRNAs (15), suggesting that they could have mRNA targets. Our computational method did not predict statistically significant mRNA targets for Lhr ncRNAs. This might either be due to a real absence of mRNA targets or to the fact that our prediction method selects ncRNA–mRNA hybrids with strong pairing energies, but not other weaker interactions such as loop–loop contacts (kissing complex), as described for OxyS and fhlA mRNA in E. coli for instance (46).
mRNA targets were predicted, in addition to the antisense targets of RliD, RliE and RliH, for three of the nine novel ncRNAs, RliB, RliE, and RliI. Thus, in total we have predicted targets for five of our ncRNAs. No targets were predicted for RliA, C, F and G, indicating either that these ncRNAs may act on proteins rather than on mRNAs, or that interactions between these Rli ncRNAs and mRNA targets cannot be detected by our method. Out of the nine predicted mRNA targets for RliB, E and I, we tested duplex formation for RliB and lmo2104 mRNA, RliE and comEA-, comFA- and lmo0945 mRNAs, and RliI and lmo1035 mRNA. Our in vitro experiments demonstrate pairing between the ncRNAs and each predicted target tested, validating our in silico predictions. Unexpectedly, while the ▵G predicted for the complex between RliB and lmo2104-2105 was rather low (−44 kcal/mol, Figure 7A), the binding in vitro was not efficient. These data argue that structural constraints in both RNA partners prevented efficient pairing. It is to be noted that although the predicted hybrids Rli–mRNA are relatively long and stable (<−38 kcal/mol), none of the mRNA targets can be predicted by the previously reported alternative method (18) (see Figures 5–7). Together these data revealed that our computational method could be of a general use to find mRNA targets of ncRNAs.
Towards the mode of action and functions of rli genes
Taking into account both our in vitro gel shift assays and our in vivo data, several modes of action of our Rli ncRNAs can be distinguished.
RliD and RliH act as antisense RNAs
rliD and rliH overlap with and are transcribed in the opposite direction to pnpA and lmo1050 genes, respectively (Figure 2). More than 178 nt of RliD are perfectly complementary to the coding sequence of the PNPase, and the first 221 nt of RliH extend into the coding sequence of lmo1050, a transcription regulator similar to PocR in S. typhimurium. Since the 3′ ends of RliD and RliH also extend into the 5′ leader regions containing the ribosome-binding site of pnpA and lmo1050 mRNAs, these ncRNAs probably repress translation by sequestring the S.D. sequence. Interestingly as rliD in Listeria, sraG in E. coli is transcribed divergently from pnpA. rliD and sraG are not homologous but it is possible that they ensure similar functions on PNPase expression (30).
RliI and RliB act in trans
For RliB and RliI, we provided in vitro data showing the interaction of the ncRNAs and their predicted targets, lmo2104 and lmo1035 mRNAs, respectively. In vivo, when RliB and RliI were overexpressed, we observed a measurable change (either positive or negative) in the abundance of the corresponding targets, as compared to the empty plasmid vector used as negative control (Figures 5C and 6C). The empty vector increased the levels of mRNAs, but the effects on the respective targets were opposite when RliB or RliI were overexpressed from the same multicopy vector, highlighting the specific action of the ncRNAs. Although the effects observed in vivo for these two ncRNAs on their respective targets cannot be taken as a comprehensive physiological study, the combination of the data obtained in vitro and in vivo definitely support our in silico predictions. In addition, effects observed in vivo for RliB and RliI are reminiscent of observations reported for other ncRNAs acting on mRNA targets, increasing or decreasing the stability of their counterpart mRNAs (47–49).
Recently, it was demonstrated that mRNA degradation and translation involving ncRNAs in E. coli are independent events, suggesting that ncRNA–mRNA hybrids are not necessarily a substrate for degradation (50). The levels of lmo0512-0513 and lmo1172-1173 mRNAs, two predicted targets for RliB, were not modified by the overexpression of the ncRNA (not shown). This might either be due to the fact that these predicted targets are false positives, or that RliB modulates the translation process without degradation of the mRNAs.
RliB is remarkable by its five repeats of 29 nt, also entirely conserved in the two copies found in L. ivanovii, suggesting an important function of both the ncRNA and its repeats. Similar transcripts (CRISPR elements) generated on direct DNA repeats (21–47 bp in length), interspaced with unrelated sequences of similar length, have been described in many prokaryotes and archeae (51–53). CRISPR elements are flanked by genes coding for proteins involved in DNA and RNA metabolism (Cas proteins), reminiscent of the players involved in the eukaryotic RNA interference phenomenon (53). This observation led the authors to propose that the CRISPR–Cas system might constitute a gene arsenal for a mechanism of defense against phages and plasmids (53). Although this point should be further investigated, a PSI–BLAST analysis of protein-encoding genes in the vicinity of the rliB locus, did not show any particular homology to cas genes that have been defined so far (53).
In Enterobacteria, the CsrB and CsrC ncRNAs carry short repeats, 5′-CAGGA(U/A)CG-3′, that mimic a S.D. sequence and bind CsrA, an RNA-binding protein regulating genes involved in carbon storage and other functions (54). PSI–BLAST analysis did not detect any homolog to CsrA protein in L. monocytogenes. Similarly, repeats in RliB could be recognized and bound by a specific protein. We have shown a weak interaction of RliB with lmo2104-2105 mRNA, reinforcing the possibility of a protein partner involved in the interaction in vivo (Figure 7B).
RliE is a pleiotropic antisense ncRNA of com genes
RliE probably acts as an antisense on comC mRNA and pairs with comEA, comFA and lmo0945 mRNAs which are expressed from different loci. mRNA targets of RliE encode proteins similar to those of the competence machinery in B. subtilis (40). All four mRNA targets of RliE are polycistronic and their 5′ leaders may be sequestered in hybrids formed with the ncRNA, suggesting a global translation repressor effect of RliE on the entire set of genes (at least seven ORFs) carried by the four polycistronic mRNAs.
Although the Listeria genome carries numerous orthologs required for the early (i.e. codY, abrB, degU, spoOKA, B, C) and late (comC, comEA, comFA, comGA, mecA, clpC, clpP) steps of competence in B. subtilis (19,55), Listeria is not known to be competent. Regulatory genes controlling competence in B. subtilis are absent in Listeria, including comX and comQ, encoding a key pheromone that turns-on the competence system and the protein required for its proper expression and secretion, respectively. In addition, the two-component system responding to ComX, comA/P, and comS, encoding a small peptide essential for competence are absent (56). Moreover, comK, is disrupted by the insertion of the A118 cryptic phage in L. monocytogenes strain EGD-e and L. innocua (19). The absence of these genes in Listeria, in addition to the possible negative effect of RliE on comC, probably explains why this bacterium is not competent. However, the absence of a rliE homolog in B. subtilis indicates that a ncRNA-dependent regulation has evolved in Listeria. Whether this regulation is essential for competence control remains to be determined.
Is Hfq playing a role?
In E. coli, Hfq generally facilitates the pairing of ncRNAs, e.g. DsrA, RhyB, RprA, OxyS, Spot42, with their mRNA targets. It is also observed that levels of ncRNAs interacting with Hfq are usually decreased in a hfq-deficient strain as compared to the wild type (16). The level of Rli ncRNAs was not affected by the deletion of hfq in the EGD-e strain (not shown), even for RliB, E and I which are able to form hybrids with their counterpart mRNAs. This could reflect biological differences between E. coli and Listeria in the mode of action of Rli ncRNAs or in the involvement of Hfq. ncRNA–mRNA hybrids in the two bacteria may have different properties. Hybrids described for RliB, E and I are longer (>40 nt) than those involving Hfq found in E. coli (<30 nt) (16). This feature might permit to overcome the requirement of Hfq, as described for long and stable ncRNA–mRNA hybrids in E. coli for plasmidic systems or antisense ncRNAs (45). Alternatively, Hfq could be involved in the formation of Rli–mRNA duplexes, but the ncRNA would only affect translation of the mRNA, and the ncRNA–mRNA hybrids would not be substrate for degradation. Finally, we cannot rule out that a protein different from Hfq may play a similar role in the formation of Rli–mRNA hybrids.
In conclusion, we have identified nine novel ncRNAs in L. monocytogenes and provided a new computational approach to predict mRNA targets of ncRNAs in bacteria. This tool could be a major improvement in understanding the functions of regulatory ncRNAs. It is not yet known whether any of the rli genes modulates Listeria virulence and the identification of ncRNAs involved in virulence is now our major challenge.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR online
ACKNOWLEDGEMENTS
We thank M. Hamon, J. Johansson, G. Lindahl, P. Romby and A. Toledo-Arana for discussions. We are grateful to P. Romby for helpful comments on the manuscript and technical advices. This work was supported by Institut Pasteur (GPH9), ANR-05-MIIM-026-01, E.U. FP6-LSHM-CT-2005-018618. PM was granted by ‘La Fondation pour la Recherche Médicale’ and PC is an international research scholar from the Howard Hugues Medical Institute.
REFERENCES
- 1.Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 2.Repoila F, Majdalani N, Gottesman S. Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol. Microbiol. 2003;48:855–861. doi: 10.1046/j.1365-2958.2003.03454.x. [DOI] [PubMed] [Google Scholar]
- 3.Wassarman KM. Small RNAs in bacteria: diverse regulators of gene expression in response to environmental changes. Cell. 2002;109:141–144. doi: 10.1016/s0092-8674(02)00717-1. [DOI] [PubMed] [Google Scholar]
- 4.Johansson J, Cossart P. RNA-mediated control of virulence gene expression in bacterial pathogens. Trends Microbiol. 2003;11:280–285. doi: 10.1016/s0966-842x(03)00118-5. [DOI] [PubMed] [Google Scholar]
- 5.Romby P, Vandenesch F, Wagner EG. The role of RNAs in the regulation of virulence-gene expression. Curr. Opin. Microbiol. 2006;9:229–236. doi: 10.1016/j.mib.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pizarro-Cerda J, Cossart P. Subversion of cellular functions by Listeria monocytogenes. J. Pathol. 2006;208:215–223. doi: 10.1002/path.1888. [DOI] [PubMed] [Google Scholar]
- 8.Dussurget O, Pizarro-Cerda J, Cossart P. Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 2004;58:587–610. doi: 10.1146/annurev.micro.57.030502.090934. [DOI] [PubMed] [Google Scholar]
- 9.Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 10.Kim H, Marquis H, Boor KJ. SigmaB contributes to Listeria monocytogenes invasion by controlling expression of inlA and inlB. Microbiology. 2005;151:3215–3222. doi: 10.1099/mic.0.28070-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandin P, Fsihi H, Dussurget O, Vergassola M, Milohanic E, Toledo-Arana A, Lasa I, Johansson J, Cossart P. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol. Microbiol. 2005;57:1367–1380. doi: 10.1111/j.1365-2958.2005.04776.x. [DOI] [PubMed] [Google Scholar]
- 12.Williams T, Bauer S, Beier D, Kuhn M. Construction and characterization of Listeria monocytogenes mutants with in-frame deletions in the response regulator genes identified in the genome sequence. Infect. Immun. 2005;73:3152–3159. doi: 10.1128/IAI.73.5.3152-3159.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kalslipolitis BH. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 2004;186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 15.Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Sogaard-Andersen L, Kallipolitis BH. Identification of small Hfq-binding RNAs in Listeria monocytogenes. Rna. 2006;12:1383–1396. doi: 10.1261/rna.49706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 17.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, et al. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- 20.Vogel J, Sharma CM. How to find small non-coding RNAs in bacteria. Biol. Chem. 2005;386:1219–1238. doi: 10.1515/BC.2005.140. [DOI] [PubMed] [Google Scholar]
- 21.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.d’Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J. Mol. Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 24.Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 2005;33:D121–124. doi: 10.1093/nar/gki081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Durbin R, Eddy S, Krogh A, Mitchison G. Cambridge, UK: Cambridge University Press; 1998. Biological Sequence Analysis, Probabilistic Models of Proteins and Nucleic acids. [Google Scholar]
- 26.Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trieu-Cuot P, Carlier C, Poyart-Salmeron C, Courvalin P. A pair of mobilizable shuttle vectors conferring resistance to spectinomycin for molecular cloning in Escherichia coli and in gram-positive bacteria. Nucleic Acids Res. 1990;18:4296. doi: 10.1093/nar/18.14.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janabi N. Selective inhibition of cyclooxygenase-2 expression by 15-deoxy-delta(12,14)(12,14)-prostaglandin J(2) in activated human astrocytes, but not in human brain macrophages. J. Immunol. 2002;168:4747–4755. doi: 10.4049/jimmunol.168.9.4747. [DOI] [PubMed] [Google Scholar]
- 29.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 31.Axmann IM, Kensche P, Vogel J, Kohl S, Herzel H, Hess WR. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 2005;6:R73. doi: 10.1186/gb-2005-6-9-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc. Natl. Acad. Sci. U.S.A. 2005;102:14249–14254. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown JW. The Ribonuclease P Database. Nucleic Acids Res. 1999;27:314. doi: 10.1093/nar/27.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Withey JH, Friedman DI. A salvage pathway for protein structures: tmRNA and trans-translation. Annu. Rev. Microbiol. 2003;57:101–123. doi: 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- 35.Trotochaud AE, Wassarman KM. 6S RNA function enhances long-term cell survival. J. Bacteriol. 2004;186:4978–4985. doi: 10.1128/JB.186.15.4978-4985.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wassarman KM, Storz G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- 37.Lenz DH, Mok KC, Lilley BN, Kulkarni RV, Wingreen NS, Bassler BL. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell. 2004;118:69–82. doi: 10.1016/j.cell.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 38.Nelson KE, Fouts DE, Mongodin EF, Ravel J, DeBoy RT, Kolonay JF, Rasko DA, Angiuoli SV, Gill SR, Paulsen IT, et al. Whole genome comparisons of serotype 4b and 1/2a strains of the food-borne pathogen Listeria monocytogenes reveal new insights into the core genome components of this species. Nucleic Acids Res. 2004;32:2386–2395. doi: 10.1093/nar/gkh562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campos-Guillen J, Bralley P, Jones GH, Bechhofer DH, Olmedo-Alvarez G. Addition of poly(A) and heteropolymeric 3′ ends in Bacillus subtilis wild-type and polynucleotide phosphorylase-deficient strains. J. Bacteriol. 2005;187:4698–4706. doi: 10.1128/JB.187.14.4698-4706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen I, Dubnau D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2004;2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 41.Bobik TA, Ailion M, Roth JR. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J. Bacteriol. 1992;174:2253–2266. doi: 10.1128/jb.174.7.2253-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. U.S.A. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P. Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev. 2002;16:1696–1706. doi: 10.1101/gad.231702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner EG, Flardh K. Antisense RNAs everywhere? Trends Genet. 2002;18:223–226. doi: 10.1016/s0168-9525(02)02658-6. [DOI] [PubMed] [Google Scholar]
- 46.Argaman L, Altuvia S. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J. Mol. Biol. 2000;300:1101–1112. doi: 10.1006/jmbi.2000.3942. [DOI] [PubMed] [Google Scholar]
- 47.Lease RA, Belfort M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl. Acad. Sci. U.S.A. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Opdyke JA, Kang JG, Storz G. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 2004;186:6698–6705. doi: 10.1128/JB.186.20.6698-6705.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 50.Morita T, Mochizuki Y, Aiba H. Translational repression is sufficient for gene silencing by bacterial small noncoding RNAs in the absence of mRNA destruction. Proc. Natl. Acad. Sci. U.S.A. 2006;103:4858–4863. doi: 10.1073/pnas.0509638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J. Mol. Evol. 2006;62:718–729. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- 52.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit. Rev. Biochem. Mol. Biol. 2005;40:93–113. doi: 10.1080/10409230590918702. [DOI] [PubMed] [Google Scholar]
- 55.Borezee E, Msadek T, Durant L, Berche P. Identification in Listeria monocytogenes of MecA, a homologue of the Bacillus subtilis competence regulatory protein. J. Bacteriol. 2000;182:5931–5934. doi: 10.1128/jb.182.20.5931-5934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hamoen LW, Venema G, Kuipers OP. Controlling competence in Bacillus subtilis: shared use of regulators. Microbiology. 2003;149:9–17. doi: 10.1099/mic.0.26003-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.