Abstract
Pax6 transcription is under the control of two main promoters (P0 and P1), and these are autoregulated by Pax6. Additionally, Pax6 expression is under the control of the TGFβ superfamily, although the precise mechanisms of such regulation are not understood. The effect of TGFβ on Pax6 expression was studied in the FHL124 lens epithelial cell line and was found to cause up to a 50% reduction in Pax6 mRNA levels within 24 h. Analysis of luciferase reporters showed that Pax6 autoregulation of the P1 promoter, and its induction of a synthetic promoter encoding six paired domain-binding sites, were significantly repressed by both an activated TGFβ receptor and TGFβ ligand stimulation. Subsequently, a novel Pax6 binding site in P1 was shown to be necessary for autoregulation, indicating a direct influence of Pax6 protein on P1. In transfected cells, and endogenously in FHL124 cells, Pax6 co-immunoprecipitated with Smad3 following TGFβ receptor activation, while in GST pull-down experiments, the MH1 domain of Smad3 was observed binding the RED sub-domain of the Pax6 paired domain. Finally, in DNA adsorption assays, activated Smad3 inhibited Pax6 from binding the consensus paired domain recognition sequence. We hypothesize that the Pax6 autoregulatory loop is targeted for repression by the TGFβ/Smad pathway, and conclude that this involves diminished paired domain DNA-binding function resulting from a ligand-dependant interaction between Pax6 and Smad3.
INTRODUCTION
Pax6 is a member of the paired-type homeobox gene family, of which there are nine in total (1). A number of developmental abnormalities are attributable to mutations in Pax6 including Small Eye in mouse, Aniridia in humans and Eyeless in Drosophila, and overexpression can cause ectopic eye formation (2). Pax6 encodes a protein containing a paired domain, paired-type homeodomain and a C-terminal transactivation domain. The paired domain is 128 amino acids in length, and was first described in the Drosophila segmentation genes paired, gooseberry and gooseberry neuro (3), and is characteristic of all members of the Pax gene family. The bi-partite paired domain binds as a monomer to two half sites in adjacent major grooves in DNA comprising the core nonpalindromic sequence T{T/C}ACGC (4). The paired-type homeodomain binds preferentially to DNA as a dimer to a palindromic sequence comprising two TAAT half-sites surrounding a conserved central motif (5). Interestingly, the paired and homeodomains can interact directly and, based on co-immunoprecipitation studies with other homeodomain-containing proteins, it has been suggested that the DNA binding regions of Pax6 should also be viewed as important protein–protein interaction domains capable of both intramolecular and intermolecular interactions (6).
Pax6 transcription is under the control of at least two promoters (P0 and P1). Studies on the quail Pax6 promoters revealed that Pax6 protein is able to bind multiple sites in both P0 and P1 (7,8). Autoregulation was suggested based on mouse genetic experiments (9), and has since been observed with both of the human promoters (10), as well as several Pax6 enhancer sequences in mouse (11,12). Additional evidence for autoregulation comes from studies of Small eye mutant phenotypes in mice. One particular Small eye mutant allele, Sey, is the product of a single point mutation, and results in a protein that is truncated before the homeodomain (1). While this is sufficient to disrupt Pax6 function, it does not interfere with the detection of Pax6 mRNA by in situ hybridization. During the development of Sey homozygous mice, Pax6 mRNA is expressed normally prior to lens specification. However, expression is completely lost throughout the head surface ectoderm post-specification, when it would normally be confined to the presumptive lens placodes (13). An earlier study has also suggested the existence of Pax6 autoregulation in the developing forebrain (14).
The precise co-ordination of upstream signalling pathways in controlling Pax6 expression is not clear, although several pathways have been implicated including Wnts (15), FGFs (16), Notch (17) and members of the TGFβ superfamily (18–22). TGFβs control a broad range of normal biological activities including cell growth, bone development, cell migration, differentiation and apoptosis (23,24). TGFβs signal through serine/threonine kinase receptors that phosphorylate TGFβ/activin/BMP pathway restricted R-Smads (Smads 1, 2, 3, 5, 8). Receptors for activin/TGFβ can activate Smad2, Smad3 and Smad8, and receptors for BMPs activate Smad1 and Smad5. In all cases, the phosphorylated R-Smads then associate with a common-mediator or co-Smad (Smad4). These heteromeric complexes are translocated to the nucleus, where they regulate gene transcription by either association with DNA-binding proteins or direct binding to promoter sequences in target genes.
There is some circumstantial evidence in the literature for functional connections between TGFβs and Pax6, particularly in the context of eye and neural development. In BMP7-deficient mice, Pax6 expression disappears just prior to the time when the lens placode should appear, and this correlates with defects in eye development (19) suggesting that BMP7 functions upstream of Pax6 in controlling lens formation. In addition, studies involving the manipulation of BMP signalling in the chick neural tube have shown that BMP regulates the expression boundary of Pax6, and that this is essential for the generation of defined neural cell populations (19). Perhaps the most compelling evidence for a link is demonstrated by the treatment of isolated chick neural plate with Activin A resulting in complete loss of Pax6 expression in the neural tube and impeded motor neuron differentiation (18). More recent studies have also shown that Smad1+/− mice exhibit increased neural Pax6 expression (21).
In this study, we show for the first time that Pax6 expression can be directly controlled by the TGFβ/Smad signalling system, and also define the molecular basis of this novel regulatory mechanism. Pax6-stimulated activity of the Pax6 promoter is repressed by TGFβ signalling. In GST pull-down experiments, Pax6 interacts with Smad 1, 3, 4 and 5, but not Smad2, and the MH1 domain of Smad3 binds to the paired domain of Pax6 releasing it from its own promoter-binding site. Taken together, our data suggest a model in which TGFβ receptor activation represses Pax6 promoter activity by releasing Pax6 from autoregulating its own promoter.
MATERIALS AND METHODS
Cell culture and transfections
FHL124 cells were cultured in Eagles Minimal Essential Medium (Invitrogen) supplemented with penicillin, streptomycin and 5% (v/v) heat-inactivated fetal bovine serum (Gibco) as described (25). HEK-293 cells were maintained in Dulbecco's Modified Eagles Medium (Invitrogen) containing 4500 mg/ml glucose and supplemented with penicillin, streptomycin and 10% (v/v) heat-inactivated fetal bovine serum (Sigma). HEK-293 cells were transiently transfected by calcium phosphate precipitation of DNA. The quantities of transfected DNA were kept constant by adding an appropriate amount of empty vector pCMV1.
Quantitative RT-PCR
FHL 124 cells were seeded on to 35 mm dishes at 30,000 cells in 400 μl of 5%FCS-EMEM (Gibco, Invitrogen Ltd, Paisley, UK) and were maintained in 1.5 ml of 5%FCS-EMEM for 3 days. The medium was replaced with non-supplemented EMEM and cultured for a further 24 h before experimental conditions were applied. After 24 h under experimental conditions RNA was collected from the cells using RNeasy® mini kit (Qiagen Ltd, Crawley, UK). Five hundred nanograms of total RNA at a final concentration of 50 or 25 ng/μl was aliquoted into RNase-free thin-walled eppendorf tubes. Equal volumes of Random Primers (Promega, WI, USA) and 10 mM dNTP (Bioline, London, UK) were mixed together, and 2 μl of this mix was added to each of the diluted RNA samples. After brief vortex mixing and centrifuging for 30 s at 13,000 rpm, samples were placed in a Peltier Thermal Cycler-DNA Engine (MJ Research Inc., Reno, NV), and incubated at 65°C for 5 min, and then another 5 min at 4°C. A mixture containing: 40 U/μl of RNaseOUT Recombinant Ribonuclease inhibitor; 100 mM DTT and 5X First Strand Buffer (Invitrogen) was prepared in a ratio of 1:2:4, respectively, and 7 μl of this mixture was then added to each sample. Samples were centrifuged for 30 s at 13,000 rpm before incubation at 25°C for 10 minutes in the Peltier Thermal Cycler, followed by 42°C for 2 min. One microlitre of Superscript II (Invitrogen Ltd, Paisley, UK) was added, and reverse transcription was performed at 42°C for 50 min and then 70°C for 15 min. Final cDNA samples were diluted with sterile double distilled water to a final concentration of 5 ng/μl. The QRT-PCR was performed with an Opticon 2 DNA Engine (MJ Research Inc., Reno, NV). Primer oligonucleotide sequences specific for Pax6 were 5′-GAATCAGAGAAGACAGGCCA-3′ upstream and 5′-GTGTAGGTATCATAACTCCG-3′ downstream and for GAPDH were 5′-ACCACAGTCCATGCCATCAC-3′ upstream and 5′-TCCACCACCCTGTTGCTGTA-3′ downstream. Level of product was determined by SYBR® green (Finnzymes, Finland) which binds exclusively to double-stranded DNA resulting in a fluorescence emission proportional to the amount of the product. A 50 μl reaction mixture was prepared for each cDNA sample containing: 50 ng cDNA; SYBR® green 2x; 2 μM forward and reverse primers (Invitrogen) and double distilled water to total final volume. Serial dilutions of cDNA known to express the gene of interest were prepared to permit relative levels between test samples to be determined. QRT-PCR was performed using the following program: step 1—initial denaturation at 94°C 4 min; step 2—denaturation at 94°C for 20 s; step 3—annealing at 55°C for 30 s; step 4—extension at 72°C for 20 s; step 5—‘cut off’ for 10 s at 80°C (GAPDH) or 82°C (Pax6) to denature potential primer dimers, this was then followed by fluorescent dye measurement. Steps 2–5 were repeated for 35 cycles. Additional, melting curve analysis was performed to determine the quality of product.
Transcription reporter assays
The P6CON and human P1 luciferase reporter plasmids have been described elsewhere (26,27), while P1(▵PBS)-Luc is described below. HEK-293 cells were transiently transfected with these reporter constructs in combination with expression plasmids encoding Pax6 (pcDNA3.zPax6.1) and a constitutively activated TGFβ type I receptor (pCMV1-TGFβRI-T204D). Cells were lysed 48 h post-transfection and lysates were assayed for luciferase activity. FHL124 cells were transfected with Lipofectamine 2000 (Gibco) in serum-free media. Cells were incubated in 0.5% FCS-containing media overnight before stimulation with 5 ng/ml TGFβ and then assayed for luciferase activity.
DNA cloning and sequence analysis
The dominant negative paired domain expression construct PD (pCMV1.Pax6-PD-Flag) was generated as follows. Site-directed mutagenesis was used to insert an EcoRI restriction site into the coding sequence of pCMV1.xPax6-N-GFP downstream of the paired box using the following primers: 5′-GACAAGCTCAGGATGCTCGAGGGACAAACTGCAACT-3′ sense and 5′-AGTTGCAGTTTGTCCCTCGAGCATCCTGAGCTTGTC-3′ anti-sense. The paired box sequence was then excised by EcoRI restriction site, taking advantage of a pre-existing EcoRI site upstream of the paired box, and ligated into a pre-prepared expression vector, pCMV1.Flag.
The luciferase reporter P1(▵PBS)-Luc was generated by site-directed mutagenesis and restriction enzyme digestion of P1-Luc. First, EcoRI restriction sites were inserted immediately upstream and downstream of the putative paired domain-binding site in P1-Luc using the following primers: 5′-GCGAGCGGTGCATTTGAATTCTGCGGAGTGATTAGT-3′ sense and 5′-ACTAATCACTCCGCAGAATTCAAATGCACCGCTCGC -3′ anti-sense for the upstream site, and 5′-GGAGTGATTAGTGGGTTTGGAATTCGAACCGTGCTCGGCCTC-3′ sense and 5′-GAGGCCGAGCACGGTTCGAATTCCAAACCCACTAATCACTCC-3′ anti-sense for the downstream site. The plasmid was then digested with EcoRI to excise the intervening sequence before re-ligation.
All DNA sequence analyses and alignments were performed using the DNA Star Lasergene software package. Accession numbers for the human and Xenopus Pax6 P1 promoter sequences are U63833 and AY048575, respectively.
Immunoprecipitation and immunoblotting
HEK-293 cells were transiently transfected with expression plasmids encoding GFP-tagged Smads (pCMV1-Smad2-N-GFP, pCMV1-Smad3-N-GFP, pCMV1-Smad4-C-GFP) in combination with Flag-tagged Pax6 (p3xFlag-mPax6) and the constitutively activated TGFβ type I receptor. Cells were lysed in Lysis Buffer (50 mM HEPES pH7.5, 150 mM NaCl, 1 mM EDTA, 1% (v/v) Triton X-100, 10% (v/v) Glycerol) 48 h post-transfection and lysates were immunoprecipitated with anti-GFP antibody and protein A-sepharose. Immunoprecipitates were subjected to immunoblotting along with a fraction of each lysate for protein expression levels. For immunoprecipitation of endogenous proteins, FHL124 cells were serum-starved for 24 h before stimulation with 10 ng/ml of recombinant TGFβ1 for a further 24 h. Un-stimulated cells were maintained in serum-free conditions as a control. Cell lysates were immunoprecipitated with anti-Pax6 (H-295, Santa Cruz) antibody and protein A–sepharose. Immunoprecipitates and cell lysates were subjected to immunoblotting. Protein A-HRP conjugate was substituted for secondary antibody to prevent anti-Pax6 heavy chain from obscuring Pax6 and Smad3 signals in immunoprecipitates.
GST pull-down assays
GST fusion constructs of Smad1, 2, 3, 4, 5 and 7 or fragments of Smad3 were expressed in Escherichia coli and purified using glutathione-sepharose 4B beads (Amersham Pharmacia Biotech.). Equal amounts of GST or GST-Smads bound to glutathione-sepharose beads were incubated with lysates from HEK-293 cells transiently transfected with Pax6. Beads were washed five times in Wash Buffer (20 mM HEPES, 150 mM NaCl, 0.1% (v/v) Triton X-100, 10% (v/v) Glycerol), and interacting proteins were detected by immunoblotting. Expression of GST fusions was confirmed by Coomassie blue staining. Where appropriate, GST fusions of full-length Smad3, or fragments of Smad3, were purified from E. coli LE392 or E. coli BL21-Star(DE3)pLysS (Invitrogen) extracts using glutathione-sepharose beads (Amersham Pharmacia Biotech.). These were then incubated with pre-cleared Pax6, Pax6ΔHD and Pax6ΔPD synthesized by in vitro transcription and translation in the presence of [35S] methionine. Similarly, GST fusions of Pax6 paired domain or its individual sub-domains, PAI and RED, were incubated with [35S] methionine-labelled Smad3 and Smad4. These pull-down assays were performed exactly as described previously (6).
DNA binding assays
HEK-293 cells were transfected with expression plasmids encoding Flag-tagged Pax6 in combination with HA-tagged Smad3 (pCMV1-Smad3-N-HA) and the constitutively activated TGFβ type I receptor. Cells were lysed 48 hours post-transfection, and lysates were incubated for 2 h with 1 μg of 5′ biotinylated, double-stranded oligonucleotide corresponding to the P6CON paired domain-binding sequence using a protocol described previously (28). DNA–protein complexes were precipitated with streptavidin–agarose beads for 1 h and subjected to immunoblotting. For gel mobility shift assays, GST fusion proteins were purified from E.coli BL21-Star(DE3)pLysS extracts, and binding of GST-Pax6-PD and GST-Pax6-HD to 32P-labelled P1 promoter probes performed as described previously (29). The following PCR primers were used to generate probes: Full-length P1-F (5′-CCCGGGCTCGGGGGCCCTG-3′); Full-length/short P1-R (5′-GCCGGCGCCCGGCCTCGCCTCC-3′); Short P1-F (5′-ATTTGCATGTTGCGGAGTGATTAG-3′).
RESULTS
TGFβ represses endogenous Pax6 expression and Pax6-dependent promoter activity
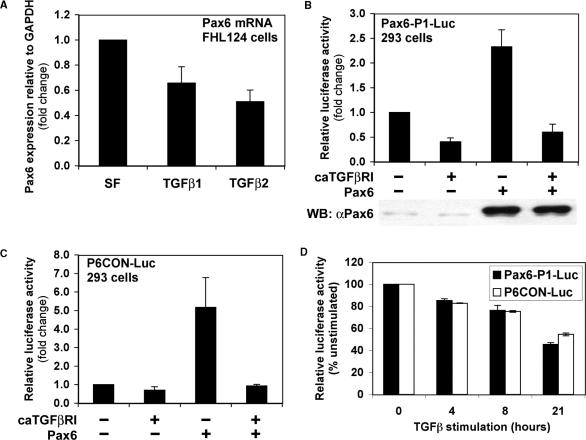
Initially we studied the effects of TGFβ signalling on Pax6 expression in the human lens FHL124 cell line. TGFβ1 and TGFβ2 caused significant reductions in endogenous Pax6 mRNA expression of 34 and 49%, respectively, within 24 h (Figure 1A). While both TGFβ isoforms yielded significant repressions of Pax6 expression, there was no significant difference in the extent of repression by the two isoforms. These data suggest that the Smad/TGFβ signalling pathway can regulate transcriptional activity at the Pax6 locus.
Figure 1.
TGFβ represses Pax6 expression and autoregulation. (A) TGFβ inhibits Pax6 expression in human lens epithelial cells. FHL124 cells were serum starved for 24 hours and stimulated with 10 ng/ml TGFβ1 or TGFβ2 for a further 24 h. Total RNA was extracted and reverse transcribed. Endogenous Pax6 expression was quantified by quantitative real-time PCR. Data were normalized with mGAPDH control and represent mean +/−SEM (n = 4). (B) Constitutively activated TGFβ receptor I inhibits expression and autoregulation of the Pax6 P1 promoter in reporter assays. HEK-293 cells were transiently transfected with 2 μg of P1-Luc in combination with 5 μg of caTGFβRI or Pax6. Cells were lysed 48 h post-transfection and luciferase assays were performed. The data represent mean +/−SEM (n = 9). (C) Constitutively activated TGFβ receptor I inhibits Pax6 protein function in reporter assays. HEK-293 cells were transiently transfected with 2 μg of P6CON-Luc in combination with 5 μg of caTGFβRI or Pax6. Cells were lysed 48 h post-transfection and luciferase assays were performed. The data represent mean +/−SEM (n = 4). (D) Time-course-dependent repression of P1-Luc and P6CON-Luc in FHL124 cells by TGFβ ligand stimulation. FHL124 cells were transfected with 1 μg of Pax6-P1-Luc or P6CON-Luc using Lipofectamine 2000, serum-starved overnight, and stimulated with 5 ng/ml TGFβ for the times indicated. Cells were lysed and luciferase assays were performed (n = 3).
Of all Pax6 promoters identified to date, the human P1 is the best characterized. Its core activity has been narrowed down to approximately 350 bp of sequence and includes the so-called Exon 1 enhancer (26). Reporter assays were performed with a Pax6 P1-Luc luciferase reporter plasmid comprising the minimal 350 bp of the Pax6 P1 promoter. HEK-293 human embryonic kidney cells were transfected with P1-Luc and co-transfected with or without a constitutively activated TGFβ type I receptor. Consistent with our analysis of the endogenous gene, P1 promoter activity dropped to 40% of its basal rate in the presence of activated receptor (Figure 1B). Autoregulation by Pax6 appears to be an evolutionary conserved behaviour of P1 promoters, and has been demonstrated in reporter assays for human, quail (8,10) and now Xenopus P1. In our experiments, we found that overexpression of Pax6 protein resulted in ∼2.5-fold induction of basal activity, confirming the autoregulatory potential of the human Pax6 P1 promoter (Figure 1B). Furthermore, co-expression of the activated TGFβ type I receptor, completely blocked this auto-induction revealing the dominance of TGFβ induced repression (Figure 1B). This effect could not be explained by a reduction in exogenous Pax6 expression by the activated TGFβ type I receptor based on western blotting of cell lysates with a specific Pax6 antibody (Figure 1B; lower panel). Interestingly, the same pattern of Pax6 autoregulation and TGFβ repression was observed for an equivalent reporter construct derived from the Xenopus P1 promoter (data not shown).
Given that TGFβ repression may be specifically targeting Pax6 autoregulation, the potential effects of TGFβ on Pax6 protein function were investigated. If the repression of P1 were due to impeded protein function, then other Pax6 responsive promoters might be expected to exhibit similar behaviours to P1. This issue was addressed with the synthetic Pax6 responsive promoter P6CON-Luc, containing 6 repeats of an optimized paired domain DNA binding sequence (30). Since this is a synthetic construct containing only Pax6 responsive elements, any repression by the TGFβ pathway must function independently of promoter specific cis-regulatory elements. HEK-293 cells were transfected with P6CON-Luc in combination with expression constructs encoding the constitutively activated TGFβ type I receptor and Pax6. Cells were lysed 48 h post-transfection, and lysates assayed for luciferase activity. Pax6 overexpression resulted in a 5-fold induction of P6CON activity, and the activated type I receptor exhibited the same dominant inhibition as with the P1 promoter (Figure 1C). These data suggest that TGFβ represses Pax6 gene expression by inhibiting Pax6 protein function, and thus autoregulation of its own promoter.
Next, we examined the effect of TGFβ ligand stimulation, rather than overexpressed activated TGFβ receptors, on P1-Luc and P6CON-Luc activity in transfected lens epithelial FHL124 cells in which Pax6 autoregulation is likely to play a more physiologically relevant role. The transcriptional activity of both P1-Luc and P6CON-Luc show a time-dependent repression reaching around 40% of the unstimulated activity after 21 h treatment with TGFβ (Figure 1D).
Pax6 binds to P1 and autoregulates promoter activity directly
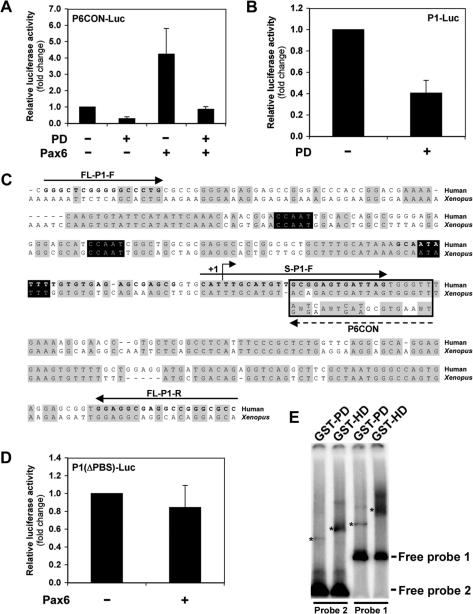
Given that the HEK-293 cells are known to express Pax6 endogenously (31), we attempted to determine whether P1-Luc basal activity included an autoregulatory component that might explain its repression by the activated TGFβ receptor. First, the Pax6 paired domain (PD) was isolated and cloned into an expression construct in an attempt to create a dominant negative peptide, since previous studies have reported that truncated Pax6 proteins act in dominant negative fashion (32,33). The efficacy of the PD peptide in blocking Pax6 protein function was determined in reporter assays whereby P6CON-Luc was co-transfected into HEK-293 cells in combination with PD and full-length Pax6. As shown in Figure 2A, PD was able to inhibit Pax6 activation of P6CON-Luc, recapitulating the pattern observed with the activated TGFβ receptor.
Figure 2.
Pax6 paired domain mediates direct autoregulation of promoter P1. (A) An isolated Pax6 paired domain (PD) inhibits the function of full-length Pax6 in dominant negative manner. HEK-293 cells were transiently transfected with 2 μg of P6CON-Luc in combination with 5 μg of Pax6 or PD. Cells were lysed 48 h post-transfection and luciferase assays were performed. The data represent mean +/−SEM (n = 3). (B) Dominant negative paired domain (PD) inhibits the basal activity of promoter P1 indicating an autoregulatory component. HEK-293 cells were transiently transfected with 2 μg of P1-Luc in combination with 5 μg of PD. Cells were lysed 48 h post-transfection and luciferase assays were performed. The data represent mean +/−SEM (n = 5). (C) Alignment of the human and Xenopus Pax6 P1 promoters reveals an evolutionary conserved putative paired domain-binding site. Sequences of the human and Xenopus Pax6 P1 promoters were aligned in the region corresponding to the reporter construct P1-Luc. This alignment was then aligned to the consensus-paired domain binding sequence, P6CON, which is boxed (Note: this is the reverse-complement P6CON sequence as the putative binding site is encoded 5′ to 3′ on the opposite strand). A broken arrow indicates orientation of the P6CON consensus. Nucleotides matching the majority sequence are shaded in grey, while the TATA and CCAAT boxes are shaded in black. Bold letters indicate the location of PCR primers used to generate DNA probes for gel-shift assays, and solid lines indicate primer orientation. Human transcription start site is indicated as +1. (D) Deletion of a putative paired domain binding site in promoter P1 (P1(▵PBS)-Luc) disrupts Pax6 autoregulation. HEK-293 cells were transiently transfected with 2 μg of P1(▵PBS)-Luc in combination with 5 μg of Pax6. Cells were lysed 48 h post-transfection and luciferase assays were performed. The data represent mean +/−SEM (n = 5). (E) Gel mobility shift assay of GST-Pax6-PD and GST-Pax6-HD binding to the P1 promoter. Equal amounts of GST-Pax6-PD and GST-Pax6-HD were used as described in the materials and methods. Probe 1 was amplified using the PCR primers FL-P1-F and FL-P1-R, and Probe 2 using S-P1-F and FL-P1-R as indicated in the P1 sequence shown in Figure 2C. The actual primer sequences used here are provided in the materials and methods. The migration of the major shifted bands is indicated as (*).
Next, the influence of the dominant negative paired domain on basal P1-Luc activity was assessed by co-transfection of HEK-293 cells with P1-Luc with or without PD, and subsequent reporter assay. The resulting data showed a 60% inhibition of basal P1-Luc activity in the presence of the dominant negative PD (Figure 2B), indicating that this expression includes a significant autoregulatory component. It is therefore likely that the repression of P1-Luc basal and Pax6-induced activity may be explained by a single mechanism for the TGFβ-repression of Pax6 autoregulation.
To confirm that Pax6 autoregulates P1-Luc directly, we attempted to identify a functional Pax6 binding site within P1. Since human and Xenopus P1 promoters were observed to behave identically in response to TGFβ receptor activation, the nucleotide sequences of these promoters were aligned to highlight evolutionary conserved nucleotides. A second alignment was then performed with the consensus-paired domain binding sequence P6CON (Figure 2C). Interestingly, P6CON aligned to a well-conserved region of the human and Xenopus P1 promoters, just downstream of the TATA box and transcription initiation site. P6CON aligned to human and Xenopus sequences with identities of 74 and 68% to the consensus paired domain-binding site, respectively.
Site-directed mutagenesis was then used to determine the functional significance of this putative paired domain-binding site (PBS). EcoRI restriction sites were inserted immediately upstream and downstream of this sequence in P1-Luc. The intervening putative binding site was then excised by EcoRI restriction enzyme digestion before P1-Luc was re-ligated, yielding P1(▵PBS)-Luc. The ability of Pax6 to autoregulate P1(▵PBS)-Luc was assessed following transfection of HEK-293 cells with the mutated reporter in combination with Pax6. Figure 2D shows that exogenous Pax6 is unable to autoregulate the P1 promoter following deletion of the putative paired domain binding site. These data suggest that Pax6 autoregulation of P1-Luc is direct and mediated through this novel paired domain-binding site. The direct interaction of Pax6 with the region of the P1 promoter encompassing this novel interaction site was then assessed by gel mobility shift assay. GST-Pax6-PD proteins comprising the paired domain bind to both the full-length P1 promoter and also very weakly, but significantly, to a shorter P1 probe that includes the putative paired domain-binding site (Figure 2E). Interestingly, GST-Pax6-HD proteins comprising the homeodomain are also bound to both the full-length and short P1 probes (Figure 2E). Equivalent gel shifted bands were not seen using GST proteins as a control (data not shown). The sites of homeodomain interaction with DNA are represented normally by a palindromic sequence comprising two TAAT half-sites separated by a conserved linker (5). There are no such consensus sites in the P1 promoter, and the significance of the homeodomain binding to P1 in the context of Pax6 autoregulation is not clear at present. Additionally, we cannot exclude the possibility that Pax6 interacts also with other regions in P1 that may explain the appearance of other slower migrating bands in the gel shift experiments. Most importantly, the data presented in Figure 2 show that the Pax6 paired domain can bind to the P1 promoter in a region that includes a predicted paired domain-binding site, the deletion of which completely abrogates the autoregulation of P1 by Pax6.
Pax6 interacts with Smad proteins
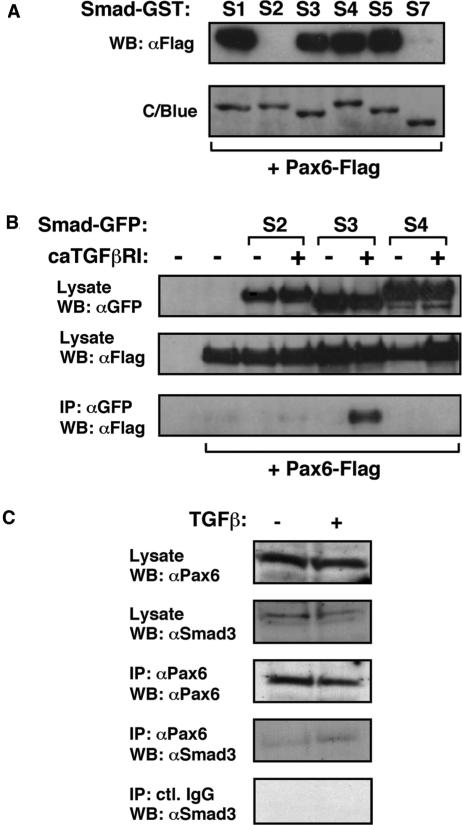
Upon ligand activation of the TGFβ receptor complex, the receptor activated Smad proteins (Smad2 and 3), together with the common mediator Smad (Smad4), translocate to the nucleus where they exert their influence on gene expression. To examine whether Smads are then able to interact with Pax6 once inside the nucleus, GST pull-down experiments were performed using a panel of GST-Smad proteins representing R-Smads, Co-Smad and I-Smads (R-Smad1, 2, 3, 5; Co-Smad4; I-Smad7), together with lysates from cells expressing Flag-tagged Pax6. As shown in Figure 3A, there are clear in vitro interactions between Pax6 and GST-Smad1, 3, 4 and 5, but not GST-Smad2 or 7. Since I-Smads are known to be the least structurally conserved Smad proteins, and given their role in the TGFβ pathway, an interaction between Smad7 and Pax6 was not expected. However, it appears from this assay that Smad2 is the only R-Smad that does not interact with Pax6. The failure to observe any interaction between these proteins might be explained by the unique structural features of Smad2, since the major isoform used in this assay contains an insert within its MH1 domain which disrupts that domain's ability to bind DNA (34).
Figure 3.
Interaction of Smads with Pax6. (A) Pax6 interacts with Smad1, 3, 4 and 5 in vitro. Lysates were prepared from HEK-293 cells that had been transiently transfected with p3xFlag-Pax6. These were then incubated with GST-Smad proteins bound onto glutathione beads. Following extensive washing, Pax6 associated with the Smad-GSTs was identified by elution of beads with SDS-Laemmli buffer, separation by 10% SDS-PAGE, and western blotting using a specific Pax6 antibody (Santa Cruz, cat. No. SC-20). GST alone was used as a control. The presence of the GST proteins was confirmed by staining gels with Coomassie Blue (C/Blue). (B) Pax6 interacts with Smad3 in the presence of constitutively activated TGFβ receptor I in vivo. HEK-293 cells were transfected as indicated. Pax6-Flag was immunoprecipitated with anti-FLAG antibody. Samples were separated by 10% SDS-PAGE and immunoblotted with anti-GFP antibody. (C) Pax6 and Smad3 interact endogenously in response to TGFβ stimulation of human lens epithelial cells.
Co-immunoprecipitation assays were performed to screen for in vivo interactions between Smad proteins and Pax6. HEK-293 cells were transfected with expression constructs encoding Flag-tagged Pax6 in combination with constitutively activated TGFβ type I receptor and GFP-tagged Smad2, 3 or 4, and cell lysates were immunoprecipitated with anti-Flag. None of the Smad proteins investigated were observed to interact with Pax6 in the absence of constitutively activated TGFβ receptor I (Figure 3B). Following TGFβ pathway activation however, Smad3 but not Smad2 or 4 was seen to interact with Pax6 (Figure 3B). These data serve to validate the interaction of Smad3, and non-interaction of Smad2 with Pax6, while invalidating the Smad4 interaction that was observed only in vitro. Most importantly these findings suggest that Smad3 interaction with Pax6 is dependent on TGFβ pathway activation in vivo.
We next sought to confirm that Pax6 interacts with Smad3 endogenously in FHL124 lens epithelial cells following stimulation with TGFβ ligand. Cells were serum-starved for 24 h and either stimulated with 10 ng/ml TGFβ1 for 8 h, or maintained in serum-free conditions for the same period. The cells were then lysed and lysates immunoprecipitated with anti-Pax6. Figure 3C demonstrates the endogenous interaction of Pax6 and Smad3 following TGFβ stimulation of lens epithelial cells, and immunoprecipitations with control immunoglobulins confirm that this interaction is specific. No detectable Smad2 was found in Pax6 immunoprecipitates even though FHL124 cells express reasonably high levels of Smad2 (data not shown). We have not been able to identify the presence of Smad4 in Smad3/Pax6 complexes, although the overall detectable levels of Smad4 are extremely low in FHL124 cells and the occurrence of this complex in vivo cannot be excluded. Additionally, we have not detected any DNA using ethidium bromide staining in Smad3/Pax6 complexes (data not shown).
The paired domain of Pax6 interacts with the MH1 domain of Smad3
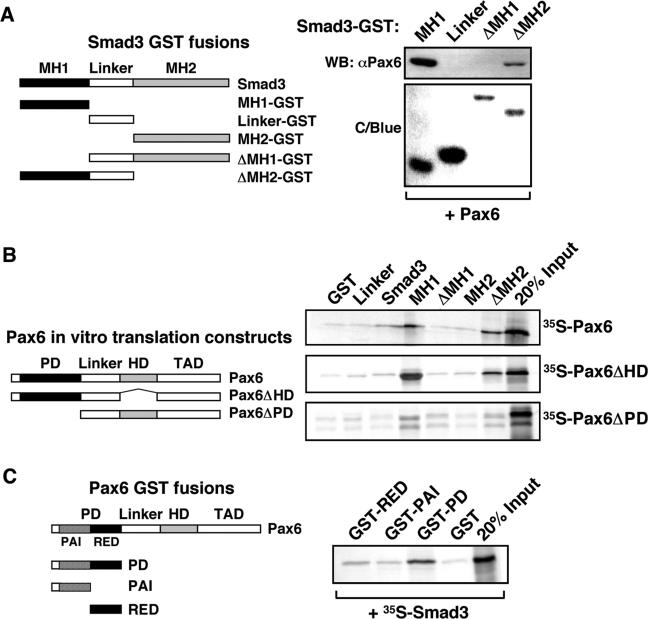
In order to investigate the specific domains involved in the observed Smad3/Pax6 interaction, further GST pull-down assays were performed using a panel of Smad3 protein fragments (Figure 4A), and lysates from cells expressing Flag-tagged Pax6. Pax6 clearly interacts with the isolated MH1 domain of Smad3 and a Smad3 fragment lacking the MH2 domain but retaining the linker region (Figure 4A). Moreover, no Pax6 interaction was observed with either the isolated linker region, or a Smad3 fragment lacking the MH1 domain. A similar analysis was performed in order to verify those data and determine the specific Pax6 domains required for Smad3 interaction. GST pull-down assays were performed using a panel of Smad3–GST protein fragments and a series of in vitro translated [35S]-methionine Pax6 constructs, either whole or possessing different Pax6 sub-domain deletions (Figure 4B). GST fusions with the Smad3 MH1 domain, but not the MH2 domain, were found to bind strongly with Pax6 constructs that include the paired domain and weakly to those including only the homeodomain (Figure 4B). GST fusions lacking the MH2 domain interacted more strongly than those containing it, suggesting intra-molecular regulation of the interaction within Smad3. We also found that the MH1 domain of Smad3 binds to the RED sub-region of the paired domain in Pax6 (Figure 4C). It is interesting to note that the strongest interaction is observed between isolated paired domain and MH1 domain. Intra-molecular regulation is known to occur between the paired and homeodomain of Pax6 (6,35), and between MH1 and MH2 domains of R-Smads (36). Such regulation is known to inhibit inter-molecular domain interactions between R-Smads, and thus formation of active Smad complexes in the absence of TGFβ receptor activation. It is possible therefore that intra-molecular inhibition within Smad3 may contribute to the TGFβ-dependence of Pax6 association.
Figure 4.
The MH1 domain of Smad3 interacts primarily with the paired domain of Pax6. (A) Lysates were prepared from HEK-293 cells that had been transiently transfected with pcDNA3-Pax6. These were then incubated with GST-Smad truncated proteins as indicated in the left-hand schematic diagram and, following extensive washing, associated proteins were identified by elution of beads with SDS-Laemmli buffer, separation by 10% SDS-PAGE, and western blotting using a specific Pax6 antibody. The presence of the GST proteins was confirmed by staining gels with Coomassie Blue (C/Blue). (B) Pax6 constructs used for in vitro translation are shown in the left-hand panel. GST pull-down assays were performed with full-length Smad3 and different domains of Smad3 fused to GST and immobilized on glutathione-agarose beads and Pax6, Pax6ΔHD and Pax6ΔPD produced by in vitro transcription and translation in the presence of [35S]-methionine. Ten-microliter portions of the in vitro translation reactions were preincubated with GST immobilized on glutathione-agarose beads before incubation with the GST fusion proteins. The GST beads, GST-Pax6ΔHD beads and GST-Pax6ΔPD beads were washed several times before they were boiled and run on a 10% SDS–polyacrylamide gel. Two microlitres of the in vitro translated proteins were run on the same gel to visualize the signal from 20% of the input as shown in the middle panel. (C) GST-Pax6 sub-paired domain constructs are shown in the left-hand schematic panel. The right panel shows the results of GST pull-down assays with the paired domain (PD) of Pax6 and the two sub-domains, PAI and RED fused to GST and immobilized on glutathione–agarose beads and Smad3 or Smad4 produced by in vitro transcription and translation in the presence of [35S]-methionine. Samples were prepared and separated as described above in Section B.
Smad3 prevents Pax6 paired domain from binding DNA
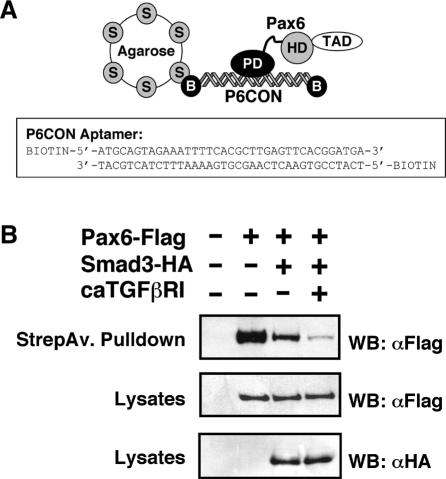
The interaction of Smad3 with the Pax6 paired domain also suggests an alternative mechanism for the repression of Pax6 function by the TGFβ signalling pathway. Since the paired domain is the dominant DNA binding domain of Pax6, it is possible that its association with Smad3 may inhibit Pax6 DNA binding, and so abrogate transactivation. DNA adsorption assays were performed to determine the potential influence of Smad3 over Pax6 DNA binding. We used an established DNA adsorption protocol (28), and the assay briefly consisted of binding a biotinylated P6CON aptamer with overexpressed Pax6 protein in cell lysates (Figure 5A). HEK-293 cells were transfected with expression constructs encoding Flag-tagged Pax6, HA-tagged Smad3 and constitutively activated TGFβ type I receptor. Streptavidin–biotin pull-down in the absence of P6CON aptamer yielded no Pax6 protein, while addition of aptamer resulted in efficient pull-down of Pax6 due to DNA binding (Figure 5B). When Pax6 was co-expressed with Smad3, a significant reduction was observed in the level of Pax6 DNA binding (Figure 5B). Moreover, Smad3 when expressed alone failed to co-precipitate with the P6CON (data not shown), suggesting that Pax6 paired-domain interactions with Smad3 and DNA are mutually exclusive.
Figure 5.
Smad3 prevents Pax6 paired domain from binding DNA. (A) Schematic representation of the experimental protocol for the DNA absorption assay together with the sequence of the biotinylated P6CON aptamer. (B) Biotinylated double-stranded P6CON oligonucleotide immobilized on streptavidin beads was incubated with lysates of HEK-293 cells transfected with the indicated expression plasmids. Pax6 bound to P6CON was analysed by immunoblotting using anti-Flag antibodies. The lower panels show the expression levels of Pax6-Flag and Smad3-HA proteins as analysed in immunoblots of the cell lysates.
DISCUSSION
This study establishes that the TGFβ pathway represses Pax6 expression by targeting the autoregulation of the Pax6 P1 promoter. Significantly, both P1-Luc and P6CON-Luc, whose activities are induced through the operation of Pax6 paired domain-binding elements, are identically repressed by TGFβ signalling, strongly suggesting that TGFβ targets Pax6 protein function rather than specific TGFβ response elements in either of the two promoters, since there are no Smad binding elements encoded in P6CON-Luc. We have not seen any significant effect of Pax6 overexpression on the activation of a Smad-dependent CAGA12-luciferase reporter by TGFβ (data not shown). Therefore, it is unlikely that Pax6/Smads can have inverse roles via Smad binding elements, although the structural basis for these differences are not clear at present.
Importantly, we have identified a novel paired domain-binding site in the P1 promoter, and shown that deletion of this sequence completely abrogates Pax6-stimulated P1 promoter activity. The basal activity of this mutant P1 promoter is still however partially repressed by TGFβ (data not shown), and it remains possible that the basal and Pax6-stimulated regulation of the P1 promoter will have additional layers of complexity that remain to be explored. Indeed, we have found Pax6 homeodomain interactions with the P1 promoter in vitro, for example, and other studies have defined a number of transcription factor binding sites that also have the potential to influence basal as well Pax6/Smad-regulated activity (26). Subsequently, we have explored the mechanism of Pax6 repression by TGFβ signalling and shown that it is due to specific Pax6/Smad protein interactions (Figure 6). Pax6 was found to interact with activated Smad3 following stimulation of TGFβ receptors with TGFβ ligand, or overexpression of a constitutively active TGFβ type I receptor. Furthermore, in vitro analyses of specific protein domains demonstrated the involvement of Smad3 MH1 domain and Pax6 paired domain in Pax6/Smad3 association. Using a simple DNA adsorption assay, we have also shown that Smad3 can release Pax6 from the P6CON consensus DNA-binding site. In vitro GST pull-down data implied an interaction between Pax6 and Smad4, which could not be validated by co-immunoprecipitation experiments, while Pax6 interactions with Smad1 and 5 of the BMP pathway are yet to be verified.
Figure 6.
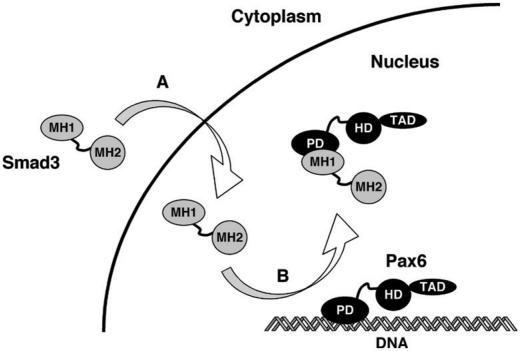
Summary of the mechanism for the repression of Pax6 function by Smads. Our results show that TGFβ represses Pax6 promoter activity by inducing the nuclear translocation of Smad3. (A) Once in the nucleus, Smad3 interacts with the RED sub-domain of the paired domain in Pax6 and releases Pax6 from its DNA binding site. (B) Thus, the Smad/TGFβ signalling pathway turns off Pax6 expression by preventing it from autoregulating its own promoter.
TGFβ signalling appears to be necessary for Pax6/Smad3 interaction, and this dependence may arise from two mechanistic components. First, as a transcription factor Pax6 is a predominantly nuclear protein, so its in vivo association with Smad3 would require nuclear shuttling of the R-Smad and therefore TGFβ signalling. Second, in vitro analysis indicated a possible role for the Smad3 MH2 domain in repressing Pax6 binding by the MH1 domain. Since the auto-inhibition between MH1 and MH2 domains is alleviated following C-terminal phosphorylation by an activated type I receptor (36), it is possible that TGFβ signalling also regulates Smad3 affinity for Pax6. While a small amount of endogenous Pax6/Smad3 interaction was observed in FHL124 cells in the absence of exogenous TGFβ ligand, it is possible that this may be attributable to endogenous autocrine signalling. We have not been able to see a significant reduction in this basal Pax6/Smad3 association in the presence of a specific type I TGFβ receptor inhibitor SB431542, however, FHL124 cells may express other functional type I TGFβ receptor isoforms that lack sensitivity to these inhibitors (37).
While early investigations of Smad-mediated transcriptional regulation largely concerned mechanisms of activation, an increasing number of reports are emerging which describe mechanisms of repression. Generally speaking, such mechanisms may be divided into those involving Smad/DNA binding and those that are independent of such associations. A recent study of the myogenic factor MEF2 provides an example of a Smad-mediated transcriptional repression independent of direct Smad/DNA binding. Smad3 MH2 domain was reported to interact with MEF2C while the latter is bound to its target DNA sequence (28). At the myogenin promoter, the physical association of Smad3 with MEF2C disrupts the transcription factor's recruitment of the coactivator GRIP-1, resulting in transcriptional repression. Another example of a Smad/DNA independent mechanism is that of Smad1/Hoxc-8 and the osteopontin promoter. BMP stimulation leads to the nuclear accumulation of Smad1 and its subsequent interaction with Hoxc-8 (38,39). However, rather than regulating co-factor recruitment, the Smad1/Hoxc-8 association disrupts Hoxc-8/DNA binding at target sequences in the osteopontin promoter. BMP stimulation of the Smad1/Hoxc-8 interaction results in the activation of the osteopontin promoter, since Hoxc-8 acts as a transcriptional repressor. Although this constitutes an activation of gene expression, this result is achieved through the targeted inhibition of transcription factor/DNA binding. Indeed, the basic mechanism closely resembles that of Smad3/Pax6 transcriptional repression reported here.
While Smad proteins have been reported to interact with many different homeodomain transcription factors, this is only the second study to demonstrate an interaction with a paired box transcription factor, and the first to do so endogenously. The only previous study concerned the class II paired box gene Pax8, and its interaction with Smad3 (40). Pax8/Smad3 interaction was demonstrated in vitro, as was its inhibitory effect on Pax8 DNA binding, however no endogenous interaction was reported. Another notable distinction between the current and previous studies is the demonstrated influence of TGFβ/Smad signalling on Pax6 autoregulation. This finding resulted from the analysis of Pax6 promoters, whereas Pax8 function was assayed only in relation to a downstream promoter. The current study raises at least one potential implication for Pax8 regulation since, in addition to Pax8 functional repression by TGFβ, there was a corresponding repression of Pax8 expression that could not be explained (40). Given that inhibition of Pax6 function leads to a repression of its autoregulation and thus expression, it is possible the same mechanism holds for Pax8. Although no other evidence exists that supports a case for Pax8 autoregulation, the TGFβ/Smad inhibition of Pax6 and Pax8, expression and function, are potentially equivalent and may represent a more general paradigm for the regulation of paired box transcription factors by the TGFβ superfamily. While Pax8 encodes a partial homeodomain, like all Pax genes it encodes a full-length paired domain. Moreover, Pax8, Pax5 and Pax2 all share a consensus paired domain-binding sequence which is closely related to that of Pax6 (27). Interestingly, other reports describe the inhibition of Pax gene expression by members of the TGFβ superfamily. Pax1 has a role in patterning the paraxial mesoderm, which forms the somites. BMP2 and 4 inhibit the expression of Pax1 in this tissue while the BMP antagonist noggin is required to abrogate this effect during normal development (41). Pax2 functions in renal tubule cells, where its expression may be inhibited by TGFβ1, although this repression is correlated with diminished mRNA stability (42). Pax3 and Pax7 are both involved in the development of skeletal muscle from the somites. Exposure of somites to ActivinA results in a loss of Pax3 expression while Pax7 is apparently unaffected (43). Since Pax3 and Pax7 are both assigned to class II of the paired box gene family, based partly on protein sequence homology (1), their differential response to the same TGFβ superfamily signal may not correspond to any gross structural divergence.
A dynamic change in Pax6 expression also underpins lens progenitor cell terminal differentiation, and mice devoid of Pax6 expression fail to form a lens (9). In the adult lens, Pax6 is required for maintenance of the lens epithelium (44), overexpression of Pax6 has been shown to suppress lens fibre differentiation by inhibiting the betaB1 crystallin promoter (45), and loss of Pax6 during differentiation leads to the expression of this lens fibre differentiation marker (46). A reduction in Pax6 expression has also been suggested to play a key role in the formation of anterior sub-capsular cataract, a condition which is also strongly associated with TGFβ (47). Therefore, TGFβ signalling could lower Pax6 expression and at the same time drive epithelial-mesenchymal transition to provide an important mechanism contributing to lens opacification and cataract formation.
In summary, this study contributes to our understanding of a new paradigm that identifies novel molecular connections linking Pax6 and the TGFβ signalling pathway, two cellular systems that are essential for normal tissue growth and differentiation, and also play pivotal roles in human disease. It is likely that the fine-tuning of the Pax6-P1 promoter in vivo is due to complex layers of both temporal and spatial regulation by multiple signalling pathways. Future work will also be needed to fully understand the roles of BMP-regulated R-Smads in the control of Pax6 expression and the general applicability of TGFβ superfamily members in coordinating the expression of other Pax family members.
ACKNOWLEDGEMENTS
The expert technical assistance of Turid Holm is greatly appreciated. This study was supported by grants to AC from the BBSRC and Wellcome Trust, and by grants to TJ from the Top Research programme of the Norwegian Research Council and the Norwegian Cancer Society. TG was supported by a Medical Research Council PhD Studentship. We thank Peter ten Dijke for providing Smad-GST plasmids, Grady Saunders for the human Pax6P1 luciferase reporter plasmid, John Reddan for the FHL124 cells, Ales Cvekl for the p3xFlag-Pax6 expression construct and P6CON-Luc reporter plasmid, and Dylan Sweetman for experimental help and advice.
Conflict of Interest Statement. None declared.
REFERENCES
- 1.Callaerts P, Halder G, Gehring WJ. PAX-6 in development and evolution. Annu. Rev. Neurosci. 1997;20:483–532. doi: 10.1146/annurev.neuro.20.1.483. [DOI] [PubMed] [Google Scholar]
- 2.Hanson I, Van Heyningen V. Pax6: more than meets the eye. Trends Genet. 1995;11:268–272. doi: 10.1016/s0168-9525(00)89073-3. [DOI] [PubMed] [Google Scholar]
- 3.Bopp D, Burri M, Baumgartner S, Frigerio G, Noll M. Conservation of a large protein domain in the segmentation gene paired and in functionally related genes of Drosophila. Cell. 1986;47:1033–1040. doi: 10.1016/0092-8674(86)90818-4. [DOI] [PubMed] [Google Scholar]
- 4.Xu HE, Rould MA, Xu W, Epstein JA, Maas RL, Pabo CO. Crystal structure of the human Pax6 paired domain-DNA complex reveals specific roles for the linker region and carboxy-terminal subdomain in DNA binding. Genes Dev. 1999;13:1263–1275. doi: 10.1101/gad.13.10.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson DS, Guenther B, Desplan C, Kuriyan J. High resolution crystal structure of a paired (Pax) class cooperative homeodomain dimer on DNA. Cell. 1995;82:709–719. doi: 10.1016/0092-8674(95)90468-9. [DOI] [PubMed] [Google Scholar]
- 6.Bruun JA, Thomassen EIS, Kristiansen K, Tylden G, Holm T, Mikkola I, Bjørkøy G, Johansen T. The third helix of the homeodomain of paired class homeodomain proteins acts as a recognition helix both for DNA and protein interactions. Nucleic Acids Res. 2005;33:2661–2675. doi: 10.1093/nar/gki562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plaza S, Dozier C, Saule S. Quail Pax-6 (Pax-QNR) encodes a transcription factor able to bind and trans-activate its own promoter. Cell Growth Differ. 1993;4:1041–1050. [PubMed] [Google Scholar]
- 8.Plaza S, Dozier C, Turque N, Saule S. Quail Pax-6 (Pax-QNR) mRNAs are expressed from two promoters used differentially during retina development and neuronal differentiation. Mol. Cell. Biol. 1995;15:3344–3353. doi: 10.1128/mcb.15.6.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grindley JC, Davidson DR, Hill RE. The role of Pax-6 in eye and nasal development. Development. 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- 10.Okladnova O, Syagailo YV, Mossner R, Riederer P, Lesch KP. Regulation of PAX-6 gene transcription: alternate promoter usage in human brain. Brain Res. Mol. Brain Res. 1998;60:177–192. doi: 10.1016/s0169-328x(98)00167-3. [DOI] [PubMed] [Google Scholar]
- 11.Aota S, Nakajima N, Sakamoto R, Watanabe S, Ibaraki N, Okazaki K. Pax6 autoregulation mediated by direct interaction of Pax6 protein with the head surface ectoderm-specific enhancer of the mouse Pax6 gene. Dev. Biol. 2003;257:1–13. doi: 10.1016/s0012-1606(03)00058-7. [DOI] [PubMed] [Google Scholar]
- 12.Kleinjan DA, Seawright A, Childs AJ, van Heyningen V. Conserved elements in Pax6 intron 7 involved in (auto)regulation and alternative transcription. Dev. Biol. 2004;265:462–77. doi: 10.1016/j.ydbio.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Ashery-Padan R, Marquardt T, Zhou X, Gruss P. Pax6 activity in the lens primordium is required for lens formation and for correct placement of a single retina in the eye. Genes Dev. 2000;14:2701–2711. doi: 10.1101/gad.184000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grindley JC, Hargett LK, Hill RE, Ross A, Hogan BL. Disruption of PAX6 function in mice homozygous for the Pax6Sey-1Neu mutation produces abnormalities in the early development and regionalization of the diencephalon. Mech. Dev. 1997;64:111–126. doi: 10.1016/s0925-4773(97)00055-5. [DOI] [PubMed] [Google Scholar]
- 15.Rasmussen JT, Deardorff MA, Tan C, Rao MS, Klein PS, Vetter ML. Regulation of eye development by frizzled signaling in Xenopus. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3861–3866. doi: 10.1073/pnas.071586298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faber SC, Dimanlig P, Makarenkova HP, Shirke S, Ko K, Lang RA. Fgf receptor signaling plays a role in lens induction. Development. 2001;128:4425–4438. doi: 10.1242/dev.128.22.4425. [DOI] [PubMed] [Google Scholar]
- 17.Onuma Y, Takahashi S, Asashima M, Kurata S, Gehring WJ. Conservation of Pax 6 function and upstream activation by Notch signaling in eye development of frogs and flies. Proc. Natl. Acad. Sci. U.S.A. 2002;99:2020–2025. doi: 10.1073/pnas.022626999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pituello F, Yamada G, Gruss P. Activin A inhibits Pax-6 expression and perturbs cell differentiation in the developing spinal cord in vitro. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6952–6956. doi: 10.1073/pnas.92.15.6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wawersik S, Purcell P, Rauchman M, Dudley AT, Robertson EJ, Maas R. BMP7 acts in murine lens placode development. Dev. Biol. 1999;207:176–88. doi: 10.1006/dbio.1998.9153. [DOI] [PubMed] [Google Scholar]
- 20.Timmer JR, Wang C, Niswander L. BMP signaling patterns the dorsal and intermediate neural tube via regulation of homeobox and helix-loop-helix transcription factors. Development. 2002;129:2459–2472. doi: 10.1242/dev.129.10.2459. [DOI] [PubMed] [Google Scholar]
- 21.Hester M, Thompson JC, Mills J, Liu Y, El-Hodiri HM, Weinstein M. Smad1 and Smad8 function similarly in mammalian central nervous system development. Mol. Cell. Biol. 2005;25:4683–4692. doi: 10.1128/MCB.25.11.4683-4692.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carpenter L, Zernicka-Goetz M. Directing pluripotent cell differentiation using ‘diced RNA’ in transient transfection. Genesis. 2004;40:157–163. doi: 10.1002/gene.20078. [DOI] [PubMed] [Google Scholar]
- 23.Hill CS. TGF-beta signalling pathways in early Xenopus development. Curr. Opin. Genet. Dev. 2001;11:533–540. doi: 10.1016/s0959-437x(00)00229-x. [DOI] [PubMed] [Google Scholar]
- 24.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 25.Wormstone IM, Tamiya S, Eldred JA, Lazaridis K, Chantry A, Reddan JR, Anderson I, Duncan G. Characterisation of TGF-beta2 signalling and function in a human lens cell line. Exp. Eye Res. 2004;78:705–714. doi: 10.1016/j.exer.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Zheng JB, Zhou YH, Maity T, Liao WS, Saunders GF. Activation of the human PAX6 gene through the exon 1 enhancer by transcription factors SEF and Sp1. Nucleic Acids Res. 2001;29:4070–4078. doi: 10.1093/nar/29.19.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Mol. Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D, Kang JS, Derynck R. TGF-beta-activated Smad3 represses MEF2-dependent transcription in myogenic differentiation. EMBO. J. 2004;23:1557–1566. doi: 10.1038/sj.emboj.7600179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikkola I, Bruun J-A, Holm T, Johansen T. Superactivation of Pax6-mediated transactivation from paired domain-binding sites by DNA-independent recruitment of different homeodomain proteins. J. Biol. Chem. 2001;276:4109–4118. doi: 10.1074/jbc.M008882200. [DOI] [PubMed] [Google Scholar]
- 30.Chauhan BK, Yang Y, Cveklova K, Cvekl A. Functional properties of natural human PAX6 and PAX6(5a) mutants. Invest. Ophthalmol. Vis. Sci. 2004;45:385–392. doi: 10.1167/iovs.03-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu ZP, Saunders GF. Transcriptional regulation of the human PAX6 gene promoter. J. Biol. Chem. 1997;272:3430–3436. doi: 10.1074/jbc.272.6.3430. [DOI] [PubMed] [Google Scholar]
- 32.Duncan MK, Cvekl A, Li X, Piatigorsky J. Truncated forms of Pax-6 disrupt lens morphology in transgenic mice. Invest. Ophthalmol. Vis. Sci. 2000;41:464–473. [PubMed] [Google Scholar]
- 33.Singh S, Tang HK, Lee JY, Saunders GF. Truncation mutations in the transactivation region of PAX6 result in dominant-negative mutants. J. Biol. Chem. 1998;273:21531–21541. doi: 10.1074/jbc.273.34.21531. [DOI] [PubMed] [Google Scholar]
- 34.Shi Y, Wang YF, Jayaraman L, Yang H, Massague J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- 35.Singh S, Stellrecht CM, Tang HK, Saunders GF. Modulation of PAX6 homeodomain function by the paired domain. J. Biol. Chem. 2000;275:17306–17313. doi: 10.1074/jbc.M000359200. [DOI] [PubMed] [Google Scholar]
- 36.Hata A, Lo RS, Wotton D, Lagna G, Massague J. Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4. Nature. 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- 37.Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- 38.Shi X, Yang X, Chen D, Chang Z, Cao X. Smad1 interacts with homeobox DNA-binding proteins in bone morphogenetic protein signaling. J. Biol. Chem. 1999;274:13711–13717. doi: 10.1074/jbc.274.19.13711. [DOI] [PubMed] [Google Scholar]
- 39.Yang X, Ji X, Shi X, Cao X. Smad1 domains interacting with Hoxc-8 induce osteoblast differentiation. J. Biol. Chem. 2000;275:1065–1072. doi: 10.1074/jbc.275.2.1065. [DOI] [PubMed] [Google Scholar]
- 40.Costamagna E, Garcia B, Santisteban P. The functional interaction between the paired domain transcription factor Pax8 and Smad3 is involved in transforming growth factor-beta repression of the sodium/iodide symporter gene. J. Biol. Chem. 2004;279:3439–3446. doi: 10.1074/jbc.M307138200. [DOI] [PubMed] [Google Scholar]
- 41.McMahon JA, Takada S, Zimmerman LB, Fan CM, Harland RM, McMahon AP. Noggin-mediated antagonism of BMP signaling is required for growth and patterning of the neural tube and somite. Genes Dev. 1998;12:1438–1452. doi: 10.1101/gad.12.10.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu S, Cieslinski DA, Funke AJ, Humes HD. Transforming growth factor-beta 1 regulates the expression of Pax-2, a developmental control gene, in renal tubule cells. Exp. Nephrol. 1997;5:295–300. [PubMed] [Google Scholar]
- 43.He L, Vichev K, Macharia R, Huang R, Christ B, Patel K, Amthor H. Activin A inhibits formation of skeletal muscle during chick development. Anat. Embryol. (Berl.) 2005;209:401–407. doi: 10.1007/s00429-005-0454-1. [DOI] [PubMed] [Google Scholar]
- 44.Cvekl A, Kashanchi F, Sax CM, Brady JN, Piatigorsky J. Transcriptional regulation of the mouse alpha A-crystallin gene: activation dependent on a cyclic AMP-responsive element (DE1/CRE) and a Pax-6-binding site. Mol. Cell Biol. 1995;15:653–660. doi: 10.1128/mcb.15.2.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Duncan MK, Haynes JI, II, Cvekl A, Piatigorsky J. Dual Roles for Pax6: a transcriptional repressor of lens fiber cell-specific-crystallin genes. Mol. Cell Biol. 1998;18:5579–5586. doi: 10.1128/mcb.18.9.5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Duncan MK, Xie L, David LL, Robinson ML, Taube JR, Cui W, Reneker LW. Ectopic Pax6 expression disturbs lens fiber cell differentiation. Invest. Ophthalmol. Vis. Sci. 2004;45:3589–3598. doi: 10.1167/iovs.04-0151. [DOI] [PubMed] [Google Scholar]
- 47.Lovicu FJ, Steven P, Saika S, McAvoy JW. Aberrant lens fiber differentiation in anterior subcapsular cataract formation: a process dependent on reduced levels of Pax6. Invest. Ophthalmol. Vis. Sci. 2004;45:1946–1953. doi: 10.1167/iovs.03-1206. [DOI] [PubMed] [Google Scholar]