Abstract
MAP kinase phosphatase-1 (MKP-1) controls nuclear MAP kinase activity with important consequences on cell growth or apoptosis. MKP-1 transcription is initiated constitutively but elongation is blocked within exon 1. It is unclear how induction of MKP-1 is controlled. Here, we report that the transcriptional elongation factors P-TEFb, DSIF and NELF regulate MKP-1 transcription in the pituitary GH4C1 cell line. Prior to stimulation, DSIF, NELF and RNA polymerase II (pol II) associate with the promoter-proximal region of the MKP-1 gene upstream of the elongation block site. Thyrotropin-releasing hormone (TRH) leads to recruitment of P-TEFb along the whole gene and a marked increase of DSIF and pol II downstream of the elongation block site, whereas NELF remains confined to the promoter-proximal region. 5,6-Dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) an inhibitor of P-TEFb eliminated TRH stimulation of MKP-1 transcription. DRB specifically inhibited TRH-induced recruitment of DSIF and P-TEFb to the MKP-1 gene. Furthermore, DRB treatment eliminated TRH-induced progression along the MKP-1 gene of pol II phosphorylated on Ser-2 of its CTD. These results indicate that P-TEFb is essential for gene-specific stimulated transcriptional elongation in mammalian cells via mechanisms which involve the activation of the DSIF–NELF complex and Ser-2 phosphorylation of pol II.
INTRODUCTION
MAP kinase phosphatase-1 (MKP-1) gene is an immediate early gene (IEG) induced by various extra-cellular stimulations. Expressed in the nucleus, MKP-1 controls the ERK MAP kinase, a key signaling enzyme which activates cell growth, cell differentiation and apoptosis (1). Recent reports show that MKP-1 is involved in cardiac hypertrophy and tumorigenesis as well as monocyte/macrophage chemotaxis (2–4). Furthermore, MKP-1 mRNA as well as protein is over-expressed at different stages of breast and prostate carcinoma (5–8). These findings underline the importance of MKP-1 for the cellular patho-physiology of many diseases. Hence, understanding how MKP-1 transcription is controlled may also be interesting for the medical field, in particular to understand emergence and progression of breast and prostate cancers.
Eukaryotic gene transcription requires, on the one hand, mechanisms which recruit RNA polymerase II (pol II) and the transcription factors needed to start transcription; on the other hand, complex mechanisms are needed to assure the output of correctly processed mRNA. Induction of most genes is achieved by stimulating mechanisms of transcription initiation. Studies on transcription control focus in general on the gene promoter, normally with the aim to identify the responsive elements which may explain induction of transcription. These promoter elements bind transcription factors needed to initiate gene transcription. The MKP-1 gene promoter comprises the calcium-cAMP response element CRE, E-box and GC-boxes. These response elements are targets of signaling via MAP kinase cascades, protein kinase C, cAMP, Ca2+, glucocorticoids and retinoic acids which are indeed controlling MKP-1 expression (9–17).
We have reported earlier that the response elements in the MKP-1 promoter favor initiation of transcription already in resting cells, and that transcription of the MKP-1 gene is mainly controlled at the level of transcriptional elongation (18). In resting cells, pol II transcribing the MKP-1 gene is arrested within 300 bp downstream from the transcription start site (18), unless extra-cellular stimuli trigger mechanisms permitting transcription to proceed. These observations suggest that the level of MKP-1 mRNA is mostly regulated via mechanisms which control elongation of transcripts, splicing, capping and polyadenylation. Although this is also the case for many other IEGs (19), it is at present largely unknown how signaling controls such mechanisms. Indeed, the control of elongation and of RNA processing has been considered so far mainly important to coordinate progressing elongation with splicing and capping of the nascent transcripts (20). Here, we address the new question of how intracellular signals enhance transcriptional elongation of the MKP-1 gene and thereby induce its expression.
The C-terminal domain (CTD) of a large subunit of pol II appears to be controlling elongation of transcripts, splicing, capping and polyadenylation (21–24). The CTD includes 52 repeated YSPTSPS motifs which are extensively phosphorylated and dephosphorylated at second and fifth serine (Ser-2 and Ser-5, respectively) during the transcription cycle of pol II (21–24). So far, various kinases which phosphorylate the CTD have been reported; among them positive elongation factor b (P-TEFb) has been studied most extensively.
In in vitro transcription systems, DRB sensitivity-inducing factor (DSIF) and negative elongation factor (NELF) can arrest elongation of transcripts by pol II soon after transcriptional initiation. When CTD of pol II is phosphorylated by P-TEFb, negative regulation by DSIF–NELF is overcome and elongation resumes (25–30). This mechanism established for in vitro transcription is reminiscent of transcriptional regulation involving a <block to elongation> in eukaryotic cells. Indeed, in Drosophila cells, NELF, DSIF and P-TEFb associate with promoter-proximal regions of immediately responsive heat shock genes, suggesting that these factors regulate transcriptional elongation machinery of IEGs (31–33). A decisive role in the p53 transcriptional program in vivo has been demonstrated for P-TEFb (34). NELF has been shown to regulate immediate early expression of the junB gene (35). DSIF being phosphorylated by P-TEFb on its C-terminal repeats (CTRs) is important to allow progression of pol II along the IEG c-fos demonstrated in vivo by Chromatin immunoprecipitation (ChIP) (36). How the three elongation factors operate jointly when regulating the transcription of a mammalian IEG in vivo remains to be addressed.
In this article, we examined whether the transcriptional elongation of the MKP-1 gene is also under the control of the DSIF–NELF complex and P-TEFb in vivo. We investigated how these factors are distributed on the MKP-1 gene prior to and during thyrotropin-releasing hormone (TRH) stimulation of pituitary cells, using ChIP assay and quantitative real-time PCR. Our results show that P-TEFb modulates the DSIF–NELF complex and mediates stimulated transcriptional elongation of the MKP-1 gene, presumably through Ser-2 phosphorylation of pol II CTD.
MATERIALS AND METHODS
Cell culture and stimulation
Rat pituitary GH4C1 cells were usually grown in Ham's F-10 Gluta Max medium (GIBCO) containing 2.5% (v/v) fetal bovine serum (FBS) and 15% (v/v) horse serum at 37°C in a humidified atmosphere of 5% CO2. For induction of MKP-1 transcription, GH4C1 cells were incubated in Ham's F-10 Gluta Max serum-free medium (SFM) for 24 h and then stimulated by TRH (Roche) at 100 nM for 0–48 min in the absence of serum. For inhibition of P-TEFb, GH4C1 cells were incubated with 30 μM 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB) (Sigma) for 2 h prior to TRH treatment.
RNA preparation and real-time RT-PCR analysis
Nascent transcripts were prepared as described earlier (37). Total RNA was extracted with an acid phenol/guanidinium reagent (TRI-Reagent; Molecular Research Center) according to the manufacturer's instructions. Nascent transcripts and total RNA were quantified by TaqMan RT-PCR (Applied Biosystems) as described earlier (18) using the following primers and TaqMan probes: MKP-1 exon 1; forward primer 5′-GGGACGCGCGGTGAAG-3′, reverse primer 5′-GATCTTGTGCGGTTTTTTGTGG-3′, TaqMan probe 5′-FAM (6-carboxyfluorescein)-CCTAAGTCCTCAAGTGCTCGCTGATCCTAATCT-TAMRA (6-carboxytetramethylrhodamine)-3′, MKP-1 exon 2; forward primer 5′-GAAGCGTTTTCGGCTTCCT-3′, reverse primer 5′-TCCGGATTCTGCACTGTCA-3′, TaqMan probe 5′-FAM-TCAGCCTCCCGCTGAGTACTAGTGTGC-TAMRA-3′, MKP-1 exon 4; forward primer 5′-CCCTGTTCACCCCACGAA-3′, reverse primer 5′-GCAGCTCGGAGAGGTTGTG-3′, TaqMan probe 5′-FAM-TGCCCTGAACTACCTTCAAAGCCCCA-TAMRA-3′, MKP-1 exon 1-2; forward primer 5′-CGCGCTCCACTCAAGTCTTC-3′, reverse primer 5′-GGTGGACTGTTTGCTGCACA-3′, TaqMan probe 5′-FAM-AGCCGAAAACGCTTCATATCCTCCTTGG-TAMRA-3′, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) exon 2-3; forward primer 5′-ATGGTGAAGGTCGGTGTGAAC-3′, reverse primer 5′-GAAGGCAGCCCTGGTAACC-3′. For normalization, amplification of 18S rRNA was performed under the standard condition by using 18S rRNA Predeveloped Assay Reagent (Applied Biosystems).
Chromatin immunoprecipitation (ChIP) assay
GH4C1 cells were grown on 5 × 107 cells in 175 cm2 flasks. Prior to stimulation, the cells were incubated with SFM for 24 h. Chromatin of cells stimulated by TRH for the indicated times was cross-linked by incubation for 1 h at 4°C with a final concentration of 1% formaldehyde. The cells were washed twice with ice-cold phosphate-buffered saline (PBS), then they were scrapped in PBS with 1 mM phenylmethylsulfonyl fluoride (PMSF) and collected by centrifugation. The cells were lysed in the cell lysis buffer (10 mM Tris pH 8.1, 1 mM EDTA, 0.5% NP-40, 1 mM PMSF, 50 mM NaF, 1 mM ortho-Vanadate, 1 μg/ml Aprotinin, 1 μg/ml Leupeptin) and then centrifuged to collect nuclei. The nuclei were further lysed in the nuclear lysis buffer (10 mM Tris pH 8.1, 1 mM EDTA, 0.5 M NaCl, 1% Triton, 0.5% deoxycholate, 0.5% Sarcosyl, 1 mM PMFS, 50 mM NaF, 1 mM ortho-Vanadate, 1 μg/ml Aprotinin, 1 μg/ml Leupeptin) and spun down. The pelleted chromatin was suspended in the sonication buffer (10 mM Tris pH 8.1, 1 mM EDTA, 100 mM NaCl) and fragmented with a Branson sonifier 250 to less than 1 kb on average. The amount of chromatin was estimated based on the DNA concentration measured by OD at 260 nm. The chromatin extracts (50 μg of DNA content) diluted in RIPA buffer (1% TritonX-100, 0.1% SDS, 0.1% deoxycholate, 140 mM NaCl, 1 mM EDTA, 10 mM Tris-HCl pH 8.0, 1mM PMSF) were pre-cleared with Protein A-Sepharose beads (Amersham-Pharmacia) for 1 h and then incubated with 5–10 μg of antibody at 4°C overnight. Protein A-Sepharose beads (40 μl, 50% slurry) were then added to the mixture for 3 h at 4°C. The beads were washed five times with RIPA buffer, one time with LiCl buffer (0.25 M LiCl, 0.5% NP-40, 0.5% deoxycholate, 1 mM EDTA, 10 mM Tris-HCl pH 8.0), and two times with TE buffer. The precipitated DNA–protein complexes were subjected to reverse cross-linking by overnight incubation at 65°C with proteinase K and 10% SDS. The DNA purified by phenol extraction and ethanol precipitation was dissolved in 8 mM NaOH and used as a template for quantitative real-time PCR. Real-time PCR was performed with 5 μl of DNA solution. PCR reactions were carried out in a final volume of 25 μl with 250 nM primer pairs and SYBR Green PCR Master Mix (Applied Biosystems) or with 250 nM primer pairs, 250 nM Taqman probes and Universal PCR master mix (Applied Biosystems). Real-time PCR was performed in triplicates in 96-well plates with ABPrism 7700 Sequence Detection System (Applied Biosystems). Each experiment was performed at least three times. Immuno-precipitated DNA was expressed as a percentage of input DNA. The percent input for <mock> ChIP (no primary antibody) was maximally 0.05% of input DNA in all the conditions examined. Antibodies used in this experiment were as follows: an anti-cyclin T1 polyclonal antibody (H-245) (Santa Cruz Biotechnology), an anti-Spt5 monoclonal antibody (BD Biosciences), an anti-NELF-A polyclonal antibody (A-20) (Santa Cruz Biotechnology), an anti-pol II polyclonal antibody (N-20) (Santa Cruz Biotechnology) and anti-pol II monoclonal antibodies (H5 and H14) (Convance).
Primers and TaqMan probes used in this experiment were as follows: MKP-1 exon 1; forward primer 5′-GGGACGCGCGGTGAAG-3′, reverse primer 5′-GATCTTGTGCGGTTTTTTGTGG-3′, TaqMan probe 5′-FAM-CCTAAGTCCTCAAGTGCTCGCTGATCCTAATCT-TAMRA-3′ MKP-1 exon 1b; forward primer 5′-CTTCTGGATTGTCGCTCCTTCT-3′, reverse primer 5′-CGTTCACTGAGCCCACGAT-3′, MKP-1 exon 4; forward primer 5′-CCCTGTTCACCCCACGAA-3′, reverse primer 5′-GCAGCTCGGAGAGGTTGTG-3′, TaqMan probe 5′-FAM-TGCCCTGAACTACCTTCAAAGCCCCA-TAMRA-3′, GAPDH 5′; forward primer 5′-CTCTCTGCTCCTCCCTGTTCTA-3′, reverse primer 5′-CTGGCACTGCACAAGAAGA-3′, GAPDH 3′; forward primer 5′-GGGCAGCCCAGAACATCA-3′, reverse primer 5′-CCGTTCAGCTCTGGGATGAC-3′, TaqMan probe 5′-FAM-CCCTGCATCCACTGGTGCTGCC-TAMRA-3′.
RESULTS
TRH induces rapidly a marked and sustained increase in the rate of MKP-1 transcription
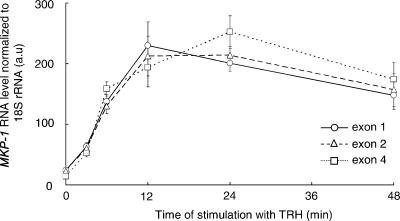
The induction of the MKP-1 gene by TRH is reflected by a progressive increase in transcription rate (Figure 1). Nascent transcripts reached maximal levels between 12 and 24 min. Elevated nascent transcript levels are sustained for at least 48 min. The kinetics of the increase in nascent transcripts indicates a very rapid acceleration of MKP-1 transcription rate. To monitor nascent transcripts by RT-PCR, random primers are used for reverse transcription, an approach which favors long transcripts. Indeed, the data in Figure 1 do not clearly indicate a higher abundance of the very short transcripts which would be anticipated if a block to elongation in exon 1 would stop transcription as was suggested from in vitro investigations (e.g. 18). In spite of this, the immediate onset of the rise in transcriptional rate is consistent with the proposal that MKP-1 transcription does not need to be initiated de novo.
Figure 1.
MKP-1 nascent transcripts after induction by TRH. Nascent transcripts of MKP-1 were purified from chromatin prepared from GH4C1 cells collected at various times after stimulation of 100 nM TRH. Transcripts which include MKP-1 exon 1, exon 2 and exon 4 sequences were quantified by real-time RT-PCR. A typical experiment repeated twice (mean ± SD in triplicates) is shown.
Dynamic redistribution of RNA polymerase II on the MKP-1 gene during stimulation of its transcription
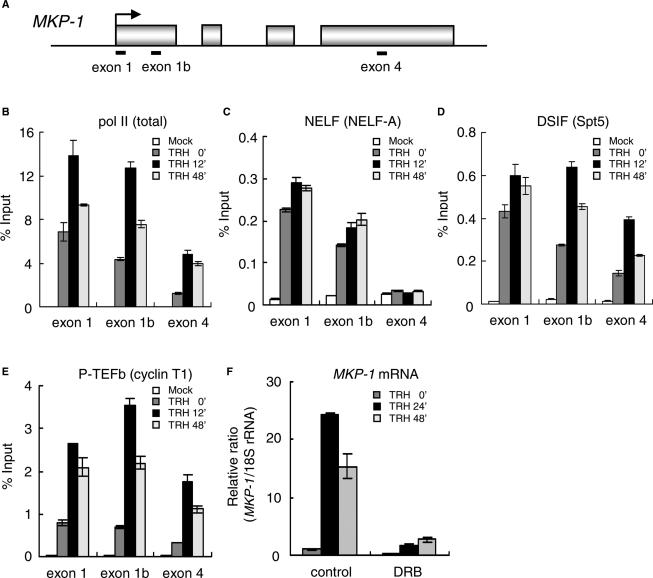
Density of pol II on the MKP-1 gene was quantified by ChIP using an antibody against the N-terminal region of Rbp-1, the largest subunit of pol II (Figure 2B). For this purpose, we prepared cross-linked and fragmented chromatin from GH4C1 cells collected at various times following TRH stimulation and immuno-precipitated the DNA fragments associated with pol II. DNA corresponding to the MKP-1 gene was then quantified by real-time PCR. Using three primer sets amplifying two exon 1 and an exon 4 regions (Figure 2A), this ChIP design yielded the relative density of pol II and of the elongation factors on three important sectors of the MKP-1 gene: the exon 1 primer set, centered at +58 representing the sector adjacent to the transcription start site upstream of the <block to elongation>; the exon 1b primer set, centered at +257 which is around the <block to elongation>; the exon 4 primer set, centered at +2076, representing the 3′ region of the MKP-1 gene.
Figure 2.
Association of pol II, NELF, DSIF and P-TEFb with the MKP-1 gene. (A) Rat MKP-1 genomic locus with the primer positions. (B–E) Distributions of pol II and elongation factors on the MKP-1 gene. ChIP assay was performed with an anti-pol II (N-20) (B), an anti-NELF-A (C), an anti-Spt5 (D) and an anti-cyclin T1 antibody (E) with chromatin prepared at various time points of TRH stimulation. Density on the MKP-1 gene was quantified by real-time PCR and presented as a percentage of input. A typical experiment (mean ± SD, n = 3) repeated three times is shown. (F) Suppression of MKP-1 transcription by DRB an inhibitor of P-TEFb. Cells were incubated with or without 30 μM DRB for 2 h prior to TRH stimulation. Total RNA was extracted 24 and 48 min after TRH, and transcripts of the MKP-1 were quantified by real-time PCR. A typical experiment repeated three times (mean ± SD in triplicates) is shown.
In resting cells, pol II is already strongly associated with exon 1 and to a lesser extent with exon 1b and least with exon 4. During acute TRH induction (12 min) pol II is strongly recruited to all the three sectors tested. The strongest relative increase in pol II (4-fold) is seen on exon 4. In the stimulated steady state, the elevated transcription rate (Figure 1) corresponds to a marked presence of pol II on exon 4 maintained for 48 min. These dynamic changes in pol II density along the MKP-1 gene during the different phases of TRH induction suggest that initiation of transcription and elongation of transcripts are independently regulated. Furthermore, the marked presence of pol II on exon 1 prior to any stimulation is consistent with the proposed <block to elongation> in the middle of exon 1 arresting pol II in resting cells (18).
Differential association of NELF, DSIF and P-TEFb with the pol II complex transcribing the MKP-1 gene in vivo
To address the role of negative (NELF, DSIF) and positive (P-TEFb) transcription factors in the induction of the MKP-1 gene, we determined the in vivo association of NELF, DSIF and P-TEFb with the MKP-1 gene during stimulation of its transcription. Density of the elongation factors was assessed with ChIP using antibodies against NELF (NELF-A), DSIF (Spt5) and P-TEFb (cyclin T1).
First, we focused on the negative elongation factor NELF which consists of four subunits (A, B, C or D and E), all required for NELF to function as a negative elongation factor (28–30). We examined the association of NELF by using an anti-NELF-A antibody (Figure 2C). In resting cells, NELF was associated with the MKP-1 gene at exon 1 and also exon 1b regions. NELF association with exon 1 increased only very slightly during TRH stimulation. There was no specific association of NELF with the exon 4 of the MKP-1 gene (compare specific versus mock in Figure 2C) regardless of TRH stimulation. Thus, NELF is never recruited to the 3′ region and functions mainly at the promoter-proximal regions of the MKP-1 gene.
DSIF which consists of Spt5 and Spt4 is reported to cooperate with NELF as a negative regulator for the arrest of pol II in vitro (28–30). Notably however, the distribution of DSIF assessed by ChIP using an anti-Spt5 antibody was markedly different from that of NELF (Figure 2C versus 2D). In resting cells, DSIF was present at exon 1 and exon 1b and to a lesser extent even at exon 4. In contrast to NELF the distribution of which did not change after TRH stimulation, DSIF density changed markedly and dynamically during TRH stimulation. On exon 1b and exon 4, DSIF density was more than doubled at the peak of transcriptional rate (12 min). Even at the steady state of sustained stimulated transcription (48 min), DSIF on exon 1b and exon 4 was still significantly increased over resting levels. Taken together, these results lead us to postulate that NELF and DSIF act as a complex at the promoter-proximal region to arrest pol II elongation in resting cells; TRH stimulation separates the complex. Strikingly, the dynamics of redistribution of DSIF on the MKP-1 gene were very similar to that of total pol II (Figure 2B versus 2D), suggesting that following its dissociation from NELF, DSIF remains associated with pol II on the transcribed MKP-1 gene during transcriptional elongation.
P-TEFb which consists of cyclin T1 and the cyclin-dependent kinase 9 (CDK9) plays a key role during the stimulation of elongation. Indeed, in vitro it has been shown to phosphorylate both the CTD of pol II and the CTRs of Spt5 eliminating thereby the negative action of the DSIF–NELF complex on transcriptional elongation by pol II (36, 38–45). The distribution of P-TEFb on the transcribed MKP-1 gene was assessed by ChIP assay with an anti-cyclin T1 antibody (Figure 2E). A small amount of P-TEFb was present on all regions (exon 1, exon1b and exon 4) prior to TRH stimulation. TRH caused a marked recruitment to all parts of the gene. P-TEFb was recruited most abundantly at the peak of transcriptional rate (12 min); subsequently P-TEFb density was reduced but its levels at the steady-state rate of transcription (48 min) remained significantly elevated over the initial levels prior to TRH stimulation. Densities of pol II and P-TEFb change with similar dynamics (Figure 2E versus 2B). This is consistent with a continuous association of pol II and P-TEFb during stimulated transcription of MKP-1. The data on P-TEFb recruitment are strongly suggesting that P-TEFb plays its role to favor transcriptional elongation also in intact cells in vivo.
The CDK9 inhibitor DRB represses MKP-1 transcription
To examine whether CDK9 activity of P-TEFb is required for the induction of MKP-1 transcription in vivo, protein kinase activity of CDK9 was inhibited by DRB. As reported earlier (18) and expected from Figure 1, MKP-1 mRNA levels were dramatically increased by TRH stimulation: 25- and 15-fold over basal at 24 and 48 min after stimulation, respectively (Figure 2F). However, in GH4C1 cells treated with DRB prior to TRH stimulation, the induction of MKP-1 mRNA was almost completely suppressed such that the levels in the presence of DRB 24 min after TRH stimulation were only 10% of the levels reached in cells in which CDK9 was not inhibited. This result clearly shows that CDK9 in P-TEFb is essential for the induced transcription of MKP-1 gene.
A gene-specific mechanism controls stimulated recruitment of elongation factors to the MKP-1 gene
To examine whether TRH stimulated specifically the recruitment of elongation factors to the MKP-1 gene, we assessed densities of pol II and P-TEFb also on the housekeeping gene GAPDH. P-TEFb densities on GAPDH were found lower, unaffected by TRH stimulation and insensitive to inhibition of CDK-9 by DRB (see supplementary data). However, the low levels of P-TEFb correspond to similarly low levels of pol II. Thus mechanisms which control transcription elongation involving elongation factors such as P-TEFb are likely common to all genes. However, the rapid and reversible recruitment of elongation factors upon cell stimulation occurs specifically on induced genes such as MKP-1.
DRB inhibits the association of P-TEFb and DSIF, but not of NELF to the MKP-1 gene
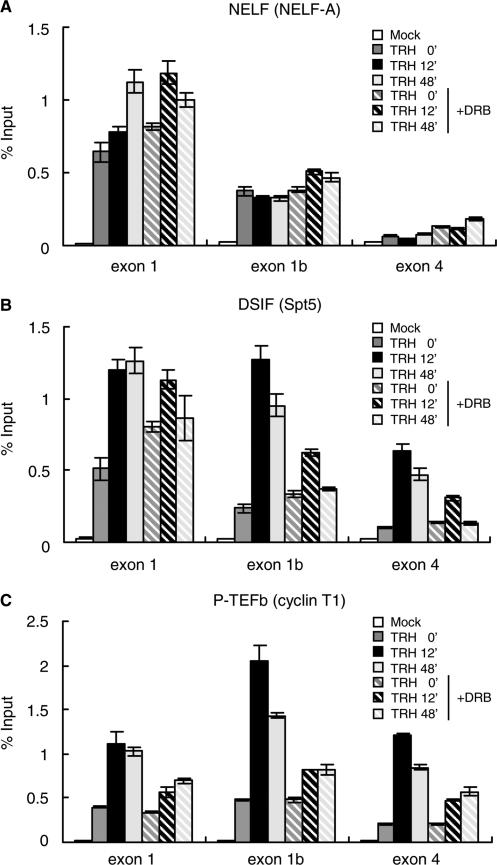
To assess the role of CDK9 activity, we further investigated the density of NELF, DSIF and P-TEFb on the MKP-1 gene during TRH stimulation under DRB treatment. DRB had no significant effect on the distribution of NELF which functions at the promoter-proximal region (Figure 3A). In contrast, the association of DSIF with the MKP-1 gene was very sensitive to DRB (Figure 3B). While the density of DSIF on exon 1 was essentially unaffected by DRB treatment neither in basal nor in stimulated conditions, DSIF densities at exon 1b and exon 4 after TRH stimulation were reduced to less than 50% of control by DRB. CDK9 activity is thus not important for the recruitment of DSIF to exon 1 of MKP-1; however the marked TRH-induced recruitment of DSIF to exon 1b and exon 4 which is associated with stimulated transcriptional elongation depends upon CDK9 activity.
Figure 3.
DRB inhibits recruitment of P-TEFb and dynamic redistribution of DSIF on the MKP-1 gene. Densities of NELF, DSIF and P-TEFb on the MKP-1 gene were assessed at various time points of TRH stimulation in the absence (filled bars) or presence (hatched bars) of DRB. ChIP assay was performed with an anti-NELF-A (A), an anti-Spt5 (B) and an anti-cyclin T1 antibody (C). Density on the MKP-1 gene was analyzed by real-time PCR and presented as a percentage of input. A typical experiment (mean ± SD, n = 3) repeated three times is shown.
DRB also significantly decreased TRH-induced recruitment of P-TEFb to the whole of the MKP-1 gene. Whereas P-TEFb densities in resting cells were not affected by DRB, the recruitment induced by the stimulation was reduced to less than 30% of control by DRB (Figure 3C). Therefore, kinase activity of CDK9 is essential for the induced recruitment of the whole P-TEFb complex to the MKP-1 gene. DRB by inhibiting CDK9 inhibits the association of P-TEFb with pol II and thereby the progression of pol II as well as the progression of DSIF with pol II and P-TEFb to the downstream parts of the MKP-1 gene.
Dynamics of phosphorylation of Ser-2 of pol II CTD upon TRH stimulation are linked to transcriptional elongation
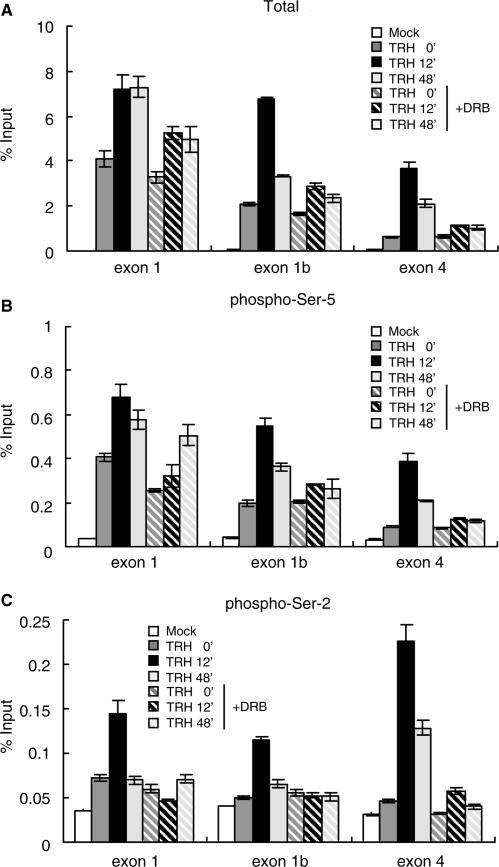
To study the effects of CTD phosphorylation by P-TEFb in the context of MKP-1 transcriptional elongation, we investigated distribution of CTD-phosphorylated pol II on the MKP-1 gene in the presence or absence of DRB. As shown in Figure 4A, in resting cells condition, the association of total pol II was unaffected by DRB. At the peak of transcriptional rate 12 min after TRH stimulation, the recruitment of total pol II at exon 1 region was suppressed only slightly by DRB (∼20% less than the condition without DRB treatment). In contrast, the association of total pol II with the exon 1b and exon 4 regions was markedly reduced by DRB to around one-third of the corresponding controls, indicating that transcriptional elongation by pol II induced by TRH stimulation was attenuated in the presence of DRB.
Figure 4.
Phosphorylation by P-TEFb of Serine-2 in the CTD pol II is essential for transcriptional elongation. The density of total and CTD-phosphorylated pol II on the MKP-1 gene was assessed at various time points of TRH stimulation in the absence (filled bars) or presence (hatched bars) of the DRB. ChIP assay was performed with specific anti-pol II antibodies: (A) an anti-pol II antibody (N-20), which recognizes N-terminal region of pol II; (B) an anti-pol II antibody (H14), directed against phospho-Ser-5 CTD; and (C) an anti-pol II antibody (H5), directed against phospho-Ser-2 CTD. Density on the MKP-1 gene was quantified by real-time PCR and is presented as a percentage of input. A typical experiment (mean ± SD, n = 3) repeated three times is shown.
The distribution of pol II phosphorylated on the Ser-5 residues of the CTD was assessed by ChIP using the H14 antibody which recognizes the phospho-Ser-5 CTD heptad repeats (Figure 4B). The distribution of Ser-5-phosphorylated CTD was almost identical to that of total pol II regardless of DRB treatment. This result suggests that the decreased association of Ser-5-phosphorylated CTD along the gene following DRB treatment reflects strictly the reduction of total pol II. Consistent with this, the constitutive association of pol II Ser-5-phosphorylated on its CTD with the region upstream of the <block to elongation> (exon 1) should not be significantly affected by DRB, which is indeed the case.
In contrast, the distribution of Ser-2-phosphorylated CTD was quite different from that of total pol II (Figure 4C versus 4A). ChIP assay using a phosphorylated Ser-2-specific antibody showed that pol-II-associated exon 1 is not Ser-2-phosphorylated prior to stimulation (Figure 4C, compare signal to mock). At the peak of transcriptional rate 12 min after TRH, Ser-2 phosphorylation was substantial on pol II situated along the whole of the MKP-1 gene, but most abundant at the exon 4 region. DRB prevented completely the Ser-2-phosphorylation of pol II associated with the MKP-1 gene. The essential role of P-TEFb for MKP-1 transcriptional elongation (Figure 2F), and the dynamics of Ser-2 phosphorylation of pol II transcribing the MKP-1 gene (Figure 4) are strong indicators for an essential role of Ser-2 phosphorylation for stimulated transcriptional elongation on the MKP-1 gene in vivo. Consistent with earlier studies in yeast attesting to the importance of Ser-2 phosphorylation for elongation (40), this suggests that P-TEFb phosphorylation of Ser-2 is a key control element for pol II transcription of MKP-1 and probably other genes which are regulated at the elongation steps in mammalian cells.
DISCUSSION
Biochemical data shows that the DSIF–NELF complex can induce the arrest of pol II, and P-TEFb allows pol II to override such negative regulation in vitro (25–30). However, it is less clear how and for what genes the DSIF–NELF complex functions in vivo. The association of NELF, DSIF and P-TEFb with pol II transcribing heat shock genes in Drosophila has been demonstrated (31–33). More recently, Yamada et al. showed that Spt5 phosphorylation is important for EGF activation of c-fos transcription (36). Here, we provide the first evidence that the DSIF–NELF complex regulates the elongation by pol II of the MKP-1 gene in mammalian cells. Our data strongly support a central role of P-TEFb as a positive elongation factor acting by modulating the function of the DSIF–NELF complex causing its dissociation and the association of DSIF with pol II during stimulated transcriptional elongation in vivo.
Association of the DSIF–NELF complex and P-TEFb at the promoter-proximal region
The MKP-1 promoter functions as a constitutive promoter, and MKP-1 transcription is presumably controlled at the level of transcriptional elongation (17,18,46). In the absence of stimulation, MKP-1 transcription is prematurely arrested within the first exon. This finding led to the hypothesis of a <block to elongation>, a notion that has remained without any molecular equivalent so far. Here, we asked which factors regulate positively and negatively the transcriptional elongation of the MKP-1 gene? Since pol II cannot progress on some model genes in vitro when it is paused by the negative elongation factors, DSIF and NELF (26,28–30), these were prime candidate negative regulators. Indeed, the DSIF–NELF complex associated with MKP-1 gene in basal condition in vivo. The limited association of P-TEFb with the MKP-1 gene in basal conditions (Figure 2E) and a powerful and gene-specific inhibition of induced MKP-1 transcription by DRB (Figures 2F and 3) are in agreement with a model whereby the DSIF–NELF complex arrests pol II on the MKP-1 gene preventing elongation as long as P-TEFb is either not activated, or inhibited by DRB (26,28–30).
Nuclear run-on experiments have localized the presumed <block to elongation> at around 300 bp downstream from the transcription start site of the MKP-1 gene (18); given the bias of in vitro elongation of transcripts, this technique may not correspond to the most frequent arrest position of pol II (19). Indeed, our ChIP data indicated that prior to stimulation, the DSIF–NELF complex is found preferentially in the promoter-proximal region (Figures 2 and 3). Therefore, the inhibitory DSIF–NELF complex would function at the promoter-proximal region in vivo similar to its behavior in vitro (28). Upon TRH stimulation, P-TEFb is recruited to the promoter-proximal region (Figures 2 and 3), suggesting that P-TEFb enables pol II to override the arrest by the DSIF–NELF complex. P-TEFb and DSIF–NELF acting upstream leaves the question open whether the <block to elongation> contains any regulatory sequences.
A dual role of DSIF
DSIF does not only functions as a negative elongation factor. Spt4 and Spt5 are essential for transcription and cellular viability in yeast (47–50). DSIF localizes at a large number of transcriptionally active chromosomal sites on polytene chromosomes in Drosophila (31–33). DSIF is necessary for appropriate RNA splicing and transcriptional termination (51,52). Additionally, DSIF prevents premature RNA release by cooperating with human immunodeficiency virus (HIV) type 1 Tat in vitro (53). Thus DSIF also functions positively during transcriptional elongation. Spt5 is phosphorylated by P-TEFb on its CTR (36,39,42). Phosphorylation of the threonine 4 residues has been shown to be critical for processive transcriptional elongation. Over-expression of exogenous Spt5 mutated at the threonines in the CTR reduces EGF induced c-fos gene transcription by pol II in vivo (36). Since c-fos transcription is controlled at the elongation steps (46) this strongly suggests that after phosphorylation by P-TEFb of Spt5, DSIF becomes a positive elongation factor. We show here that endogenous DSIF migrates along the MKP-1 gene after TRH stimulation. TRH-induced association of DSIF with downstream parts of the MKP-1 gene was suppressed by DRB (Figure 3), whereas the presence of DSIF on the promoter-proximal region was not affected by the blockade of P-TEFb. Thus DSIF plays a dual role for elongation switching from a negative to a positive function as a consequence of the P-TEFb-dependent phosphorylation of the CTR of Spt5.
The dynamic redistribution of DSIF along the MKP-1 gene is in marked contrast to the static presence of NELF on the MKP-1 promoter-proximal regions. NELF is neither affected by TRH stimulation, nor by the inhibition of CDK9 by DRB. While it has been reported that NELF-E is phosphorylated by P-TEFb (54), our data underline that NELF functions only as a negative regulator at the promoter-proximal region on the MKP-1 gene irrespective of the action of P-TEFb. The data of our in vivo study is consistent with the model proposed earlier derived mainly from in vitro data (36) in which the DSIF–NELF complex would function as a negative regulator at the promoter-proximal region in basal condition, whereas DSIF detached from NELF and phosphorylated on its CTR would favor stimulated transcription.
During stimulated transcriptional elongation, the C-terminal domain of pol II is phosphorylated by P-TEFb on Serine-2 residues
Phosphorylation of the CTD of pol II is of key importance for the control of RNA production and maturation. The CTD is a lengthy extension consisting of multiple repeats of a seven amino acid motif which includes five potential phosphate acceptor sites. CTD is an essential element for most of the steps leading to mature mRNA. Specific phosphorylation of the CTD characterizes the pol II in various stages of progression from the promoter to the end of the transcribed gene. In view of the complexity of potential CTD phosphorylation pattern, the term ‘CTD-code’ has been coined (55) to illustrate that each of the multiple functions assigned to the CTD could require its specific pattern of multiple phosphorylation. Here, we concentrated on the phosphorylation of the serines in position 2 and 5 (Ser-2 and Ser-5) and its relation to stimulated elongation involving CDK9, the kinase in P-TEFb. Our data show very clearly that in the context of stimulated elongation in vivo, CDK9 phosphorylates mainly the Ser-2 residues on pol II which transcribes the MKP-1 gene (Figure 4). Indeed, we find Ser-2 phosphorylated pol II on the downstream parts of the MKP-1 gene only during stimulated transcriptional elongation, but neither in the basal state nor when CDK9 is inhibited by DRB. In contrast, Ser-5 phosphorylation is found prior to stimulation on the pol II on exon 1 of the MKP-1 gene in a manner independent of DRB. Distribution pattern of Ser-5-phosphorylated pol II change in parallel to the distribution pattern of total pol II upon stimulation with TRH; as DRB inhibits elongation both total pol II and Ser-5 phosphorylated pol II are reduced in parallel downstream of the <block to elongation>. In marked contrast, DRB inhibition of CDK9 completely prevents Ser-2 phosphorylation, reducing ChIP with the H5 antibody to the level of mock IP.
Taken together, our data are thus consistent with the proposal made earlier that phosphorylation of Ser-5 and Ser-2 of CTD are responsible for pol II initiation and elongation, respectively (20–24). Note that these findings do not define the <CTD code> which will <unlock> blocked elongation, but they strongly suggest that phospho-Ser-2 is a necessary feature.
Continuous regulatory input by P-TEFb associated with elongating pol II
The precise mechanisms which link signaling cascades to elongation control via the three factors P-TEFb, DSIF and NELF remain to be established. Possibilities for gene-specific recruitment of elongation factors (e.g. 34) have been recently reviewed (56) and include interactions with RNA-binding proteins or with DNA-binding transcription factors such as NFκB (57) or the estrogen receptor (58).
The TRH-induced association of P-TEFb with exon 1b and exon 4 regions suggests that P-TEFb regulates pol II elongation along the MKP-1 gene by migrating together with pol II after its recruitment to the promoter-proximal region. Against a hypothetical background of constitutive dephosphorylation of both pol II CTD (23) and Spt5 CTR the continuous presence of P-TEFb in the transcribing complex would assure a continuous regulatory input and as a result finely tuned transcriptional rates. Alternatively, P-TEFb may be required for the recruitment and phosphorylation of proteins involved in RNA splicing, capping and polyadenylation.
CONCLUSIONS
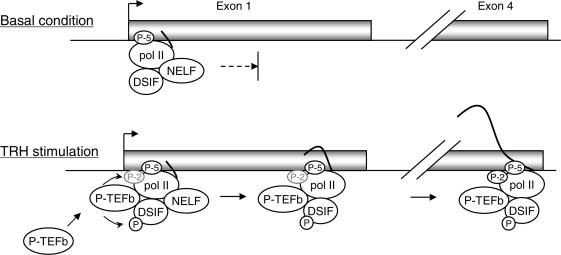
In Figure 5, we present a model of how transcriptional elongation of the MKP-1 gene involves the elongation factors NELF, DSIF and P-TEFb. In basal condition, the DSIF–NELF complex arrests pol II elongation at the promoter-proximal region. TRH stimulation results in the recruitment of P-TEFb which will phosphorylate CTR of Spt5 and Ser-2 of pol II CTD, resulting in the functional change of DSIF and release of pol II from the arrest. P-TEFb and DSIF detached from NELF move together with pol II probably to promote further elongation of transcripts and processing of nascent RNA into mature mRNA.
Figure 5.
A model for the mechanisms by which the DSIF–NELF complex and P-TEFb control MKP-1 transcription. In basal condition, the DSIF–NELF complex associates with pol II arrested in the promoter-proximal region. TRH stimulation induces recruitment of P-TEFb to the pol II complex resulting in phosphorylation of the CTR of Spt5 and of the CTD of pol II. This leads to the dissociation of the DSIF–NELF complex and the progression of pol II elongation. P-TEFb and DSIF remain associated with pol II during elongation of the transcript, possibly contributing also to the control of RNA processing and maturation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
ACKNOWLEDGEMENTS
This investigation was supported in parts by The Mochida Memorial Foundation for Medical and Pharmaceutical Research, by the Swiss National Science Foundation grant 3100A0-102147 and by the Fondation pour Recherches Médicales. Funding to pay the Open Access publication charge was provided by the Foudation pour Recherdies Médicales.
Conflict of interest statement. None declared.
REFERENCES
- 1.Camps M, Nichols A, Arkinstall S. Dual specificity phosphatases: a gene family for control of MAP kinase function. FASEB J. 1999;14:6–16. [PubMed] [Google Scholar]
- 2.Bueno OF, De Windt LJ, Lim HW, Tymitz KM, Witt SA, Kimball TR, Molkentin JD. The dual-specificity phosphatase MKP-1 limits the cardiac hypertrophic response in vitro and in vivo. Circ. Res. 2001;88:88–96. doi: 10.1161/01.res.88.1.88. [DOI] [PubMed] [Google Scholar]
- 3.Grimshaw MJ, Balkwill FR. Inhibition of monocyte and macrophage chemotaxis by hypoxia and inflammation–a potential mechanism. Eur. J. Immunol. 2001;31:480–489. doi: 10.1002/1521-4141(200102)31:2<480::aid-immu480>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 4.Wang HY, Cheng Z, Malbon CC. Overexpression of mitogen-activated protein kinase phosphatases MKP1, MKP2 in human breast cancer. Cancer Lett. 2003;191:229–237. doi: 10.1016/s0304-3835(02)00612-2. [DOI] [PubMed] [Google Scholar]
- 5.Leav I, Galluzzi CM, Ziar J, Stork PJ, Ho SM, Loda M. Mitogen-activated protein kinase and mitogen-activated kinase phosphatase-1 expression in the Noble rat model of sex hormone-induced prostatic dysplasia and carcinoma. Lab. Invest. 1996;75:361–370. [PubMed] [Google Scholar]
- 6.Loda M, Capodieci P, Mishra R, Yao H, Corless C, Grigioni W, Wang Y, Magi-Galluzzi C, Stork PJ. Expression of mitogen-activated protein kinase phosphatase-1 in the early phases of human epithelial carcinogenesis. Am. J. Pathol. 1996;149:1553–1564. [PMC free article] [PubMed] [Google Scholar]
- 7.Magi-Galluzzi C, Mishra R, Fiorentino M, Montironi R, Yao H, Capodieci P, Wishnow K, Kaplan I, Stork PJ, Loda M. Mitogen-activated protein kinase phosphatase 1 is overexpressed in prostate cancers and is inversely related to apoptosis. Lab. Invest. 1997;76:37–51. [PubMed] [Google Scholar]
- 8.Magi-Galluzzi C, Montironi R, Cangi MG, Wishnow K, Loda M. Mitogen-activated protein kinases and apoptosis in PIN. Virchows Arch. 1998;432:407–413. doi: 10.1007/s004280050184. [DOI] [PubMed] [Google Scholar]
- 9.Charles CH, Abler AS, Lau LF. cDNA sequence of a growth factor-inducible immediate early gene and characterization of its encoded protein. Oncogene. 1992;7:187–190. [PubMed] [Google Scholar]
- 10.Keyse SM, Emslie EA. Oxidative stress and heat shock induce a human gene encoding a protein-tyrosine phosphatase. Nature. 1992;359:644–647. doi: 10.1038/359644a0. [DOI] [PubMed] [Google Scholar]
- 11.Kwak SP, Hakes DJ, Martell KJ, Dixon JE. Isolation and characterization of a human dual specificity protein-tyrosine phosphatase gene. J. Biol. Chem. 1994;269:3596–3604. [PubMed] [Google Scholar]
- 12.Guo YL, Kang B, Williamson JR. Inhibition of the expression of mitogen-activated protein phosphatase-1 potentiates apoptosis induced by tumor necrosis factor-alpha in rat mesangial cells. J. Biol. Chem. 1998;273:10362–10366. doi: 10.1074/jbc.273.17.10362. [DOI] [PubMed] [Google Scholar]
- 13.Laderoute KR, Mendonca HL, Calaoagan JM, Knapp AM, Giaccia AJ, Stork PJ. Mitogen-activated protein kinase phosphatase-1 (MKP-1) expression is induced by low oxygen conditions found in solid tumor microenvironments. A candidate MKP for the inactivation of hypoxia-inducible stress-activated protein kinase/c-Jun N-terminal protein kinase activity. J. Biol. Chem. 1999;274:12890–12897. doi: 10.1074/jbc.274.18.12890. [DOI] [PubMed] [Google Scholar]
- 14.Sommer A, Burkhardt H, Keyse SM, Luscher B. Synergistic activation of the mkp-1 gene by protein kinase A signaling and USF, but not c-Myc. FEBS Lett. 2000;474:146–150. doi: 10.1016/s0014-5793(00)01566-0. [DOI] [PubMed] [Google Scholar]
- 15.Kassel O, Sancono A, Kratzschmar J, Kreft B, Stassen M, Cato AC. Glucocorticoids inhibit MAP kinase via increased expression and decreased degradation of MKP-1. EMBO J. 2001;20:7108–7116. doi: 10.1093/emboj/20.24.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu Q, Konta T, Furusu A, Nakayama K, Lucio-Cazana J, Fine LG, Kitamura M. Transcriptional induction of mitogen-activated protein kinase phosphatase 1 by retinoids. Selective roles of nuclear receptors and contribution to the antiapoptotic effect. J. Biol. Chem. 2002;277:41693–41700. doi: 10.1074/jbc.M207095200. [DOI] [PubMed] [Google Scholar]
- 17.Ryser S, Massiha A, Piuz I, Schlegel W. Stimulated initiation of mitogen-activated protein kinase phosphatase-1 (MKP-1) gene transcription involves the synergistic action of multiple cis-acting elements in the proximal promoter. Biochem. J. 2004;378:473–484. doi: 10.1042/BJ20031022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryser S, Tortola S, van Haasteren G, Muda M, Li S, Schlegel W. MAP kinase phosphatase-1 gene transcription in rat neuroendocrine cells is modulated by a calcium-sensitive block to elongation in the first exon. J. Biol. Chem. 2001;276:33319–33327. doi: 10.1074/jbc.M102326200. [DOI] [PubMed] [Google Scholar]
- 19.Krumm A, Meulia T, Groudine M. Common mechanisms for the control of eukaryotic transcriptional elongation. Bioessays. 1993;15:659–665. doi: 10.1002/bies.950151005. [DOI] [PubMed] [Google Scholar]
- 20.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–512. doi: 10.1016/s0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 21.Conaway JW, Shilatifard A, Dvir A, Conaway RC. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 2000;25:375–380. doi: 10.1016/s0968-0004(00)01615-7. [DOI] [PubMed] [Google Scholar]
- 22.Kobor MS, Greenblatt J. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta. 2002;1577:261–275. doi: 10.1016/s0167-4781(02)00457-8. [DOI] [PubMed] [Google Scholar]
- 23.Palancade B, Bensaude O. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur. J. Biochem. 2003;270:3859–3870. doi: 10.1046/j.1432-1033.2003.03794.x. [DOI] [PubMed] [Google Scholar]
- 24.Meinhart A, Kamenski T, Hoeppner S, Baumli S, Cramer PA. A structural perspective of CTD function. Genes Dev. 2005;19:1401–1415. doi: 10.1101/gad.1318105. [DOI] [PubMed] [Google Scholar]
- 25.Wada T, Takagi T, Yamaguchi Y, Ferdous A, Imai T, Hirose S, Sugimoto S, Yano K, Hartzog GA, et al. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 1998;12:343–356. doi: 10.1101/gad.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 1998;17:7395–7403. doi: 10.1093/emboj/17.24.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi Y, Wada T, Handa H. Interplay between positive and negative elongation factors: drawing a new view of DRB. Genes Cells. 1998;3:9–15. doi: 10.1046/j.1365-2443.1998.00162.x. [DOI] [PubMed] [Google Scholar]
- 28.Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell. 1999;97:41–51. doi: 10.1016/s0092-8674(00)80713-8. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi Y, Inukai N, Narita T, Wada T, Handa H. Evidence that negative elongation factor represses transcription elongation through binding to a DRB sensitivity-inducing factor/RNA polymerase II complex and RNA. Mol. Cell. Biol. 2002;22:2918–2927. doi: 10.1128/MCB.22.9.2918-2927.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narita T, Yamaguchi Y, Yano K, Sugimoto S, Chanarat S, Wada T, Kim DK, Hasegawa J, Omori M, et al. Human transcription elongation factor NELF: identification of novel subunits and reconstitution of the functionally active complex. Mol. Cell. Biol. 2003;23:1863–1873. doi: 10.1128/MCB.23.6.1863-1873.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Andrulis ED, Guzman E, Doring P, Werner J, Lis JT. High-resolution localization of Drosophila Spt5 and Spt6 at heat shock genes in vivo: roles in promoter proximal pausing and transcription elongation. Genes Dev. 2000;14:2635–2649. doi: 10.1101/gad.844200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol. Cell. Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu CH, Yamaguchi Y, Benjamin LR, Horvat-Gordon M, Washinsky J, Enerly E, Larsson J, Lambertsson A, Handa H, et al. NELF and DSIF cause promoter proximal pausing on the hsp70 promoter in Drosophila. Genes Dev. 2003;17:1402–1414. doi: 10.1101/gad.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gomes NP, Bjerke G, Llorente B, Szostek SA, Emerson BM, Espinosa JM. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 2006;20:601–612. doi: 10.1101/gad.1398206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aida M, Chen Y, Nakajima K, Yamaguchi Y, Wada T, Handa H. Transcriptional pausing caused by NELF plays a dual role in regulating immediate-early expression of the junB gene. Mol. Cell. Biol. 2006;26:6094–6104. doi: 10.1128/MCB.02366-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yamada T, Yamaguchi Y, Inukai N, Okamoto S, Mura T, Handa H. P-TEFb-mediated phosphorylation of hSpt5 C-terminal repeats is critical for processive transcription elongation. Mol. Cell. 2006;21:227–237. doi: 10.1016/j.molcel.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 37.Wuarin J, Schibler U. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol. Cell. Biol. 1994;14:7219–7225. doi: 10.1128/mcb.14.11.7219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 39.Ivanov D, Kwak YT, Guo J, Gaynor RB. Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol. Cell. Biol. 2000;20:2970–2983. doi: 10.1128/mcb.20.9.2970-2983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, Brady JN. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol. Cell. Biol. 2000;20:5077–5086. doi: 10.1128/mcb.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lavoie SB, Albert AL, Handa H, Vincent M, Bensaude O. The peptidyl-prolyl isomerase Pin1 interacts with hSpt5 phosphorylated by Cdk9. J. Mol. Biol. 2001;312:675–685. doi: 10.1006/jmbi.2001.4991. [DOI] [PubMed] [Google Scholar]
- 43.Ramanathan Y, Rajpara SM, Reza SM, Lees E, Shuman S, Mathews MB, Pe’ery T. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 2001;276:10913–10929. doi: 10.1074/jbc.M010975200. [DOI] [PubMed] [Google Scholar]
- 44.Kim YK, Bourgeois CF, Isel C, Churcher MJ, Karn J. Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol. Cell. Biol. 2002;22:4622–4637. doi: 10.1128/MCB.22.13.4622-4637.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shim EY, Walker AK, Shi Y, Blackwell TK. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 2002;16:2135–2146. doi: 10.1101/gad.999002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ryser S, Tortola S, Schlegel W. Map kinase phosphatase-1 gene expression and regulation in neuroendocrine cells. J. Recept. Signal Transduct. Res. 2002;22:17–29. doi: 10.1081/rrs-120014586. [DOI] [PubMed] [Google Scholar]
- 47.Swanson MS, Malone EA, Winston F. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell Biol. 1991;11:3009–3019. doi: 10.1128/mcb.11.6.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Malone EA, Fassler JS, Winston F. Molecular and genetic characterization of SPT4, a gene important for transcription initiation in Saccharomyces cerevisiae. Mol. Gen. Genet. 1993;237:449–459. doi: 10.1007/BF00279450. [DOI] [PubMed] [Google Scholar]
- 49.Compagnone-Post PA, Osley MA. Mutations in the SPT4, SPT5, and SPT6 genes alter transcription of a subset of histone genes in Saccharomyces cerevisiae. Genetics. 1996;143:1543–1554. doi: 10.1093/genetics/143.4.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rondon AG, Garcia-Rubio M, Gonzalez-Barrera S, Aguilera A. Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J. 2003;22:612–620. doi: 10.1093/emboj/cdg047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui Y, Denis CL. In vivo evidence that defects in the transcriptional elongation factors RPB2, TFIIS, and SPT5 enhance upstream poly(A) site utilization. Mol. Cell. Biol. 2003;23:7887–7901. doi: 10.1128/MCB.23.21.7887-7901.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lindstrom DL, Squazzo SL, Muster N, Burckin TA, Wachter KC, Emigh CA, McCleery JA, Yates JR, III, Hartzog GA. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 2003;23:1368–1378. doi: 10.1128/MCB.23.4.1368-1378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bourgeois CF, Kim YK, Churcher MJ, West MJ, Karn J. Spt5 cooperates with human immunodeficiency virus type 1 Tat by preventing premature RNA release at terminator sequences. Mol. Cell. Biol. 2002;22:1079–1093. doi: 10.1128/MCB.22.4.1079-1093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM. Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol. Cell. Biol. 2004;24:787–795. doi: 10.1128/MCB.24.2.787-795.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buratowski S. The CTD code. Nat. Struct. Biol. 2003;10:679–680. doi: 10.1038/nsb0903-679. [DOI] [PubMed] [Google Scholar]
- 56.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 57.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 58.Aiyar SE, Sun JL, Blair AL, Moskaluk CA, Lu YZ, Ye QN, Yamaguchi Y, Mukherjee A, Ren DM, et al. Attenuation of estrogen receptor alpha-mediated transcription through estrogen-stimulated recruitment of a negative elongation factor. Genes Dev. 2004;18:2134–2146. doi: 10.1101/gad.1214104. [DOI] [PMC free article] [PubMed] [Google Scholar]