Abstract
Hfq protein is vital for the function of many non-coding small (s)RNAs in bacteria but the mechanism by which Hfq facilitates the function of sRNA is still debated. We developed a fluorescence resonance energy transfer assay to probe how Hfq modulates the interaction between a sRNA, DsrA, and its regulatory target mRNA, rpoS. The relevant RNA fragments were labelled so that changes in intra- and intermolecular RNA structures can be monitored in real time. Our data show that Hfq promotes the strand exchange reaction in which the internal structure of rpoS is replaced by pairing with DsrA such that the Shine-Dalgarno sequence of the mRNA becomes exposed. Hfq appears to carry out strand exchange by inducing rapid association of DsrA and a premelted rpoS and by aiding in the slow disruption of the rpoS secondary structure. Unexpectedly, Hfq also disrupts a preformed complex between rpoS and DsrA. While it may not be a frequent event in vivo, this melting activity may have implications in the reversal of sRNA-based regulation. Overall, our data suggests that Hfq not only promotes strand exchange by binding rapidly to both DsrA and rpoS but also possesses RNA chaperoning properties that facilitates dynamic RNA–RNA interactions.
INTRODUCTION
RNA molecules serve as important regulatory factors in a variety of cellular processes. Examples include small interfering and micro RNA in eukaryotes and non-coding small RNA (sRNA) and riboswitches in prokaryotes. Protein–RNA interactions form the basis of RNA-based regulation of gene expression, but our understanding of its details at the molecular level is still poor. Fluorescence resonance energy transfer (FRET) is a powerful tool to study intermolecular interactions and intramolecular conformational changes, and therefore it serves as a desirable technique to investigate the regulatory mechanisms involving RNA–protein interactions. Here, we have developed FRET assays to probe how E. coli Hfq protein modulates the interaction between a sRNA and its target mRNA.
Hfq was originally discovered in E. coli as a host factor for Q-β-replicase (1), but analysis of hfq mutations suggested its involvement in many other metabolic pathways. Indeed, Hfq is now known to be a pleiotropic regulator that modulates the stability as well as translational activity of several mRNAs (2–4). The role of Hfq in translational regulation has been the focus of much attention lately since many sRNAs have been identified to require Hfq for their activity. For example, Hfq is involved in the post-transcriptional regulation of the rpoS gene (encoding the σs RNA polymerase subunit involved in bacterial stress response) (5). Hfq modulates rpoS expression by altering the binding of sRNAs such as DsrA, OxyS and RprA (6–8). Other examples of protein pathways regulated by Hfq-mediated coupling of a regulatory sRNA to a target mRNA include OxyS … fhlA, and Spot42 … galK, where mRNA targets are italicized (reviewed in ref. (9)). Although it is likely that only a fraction of sRNA molecules that require Hfq as an additional factor have been identified, it is already clear that many of these sRNAs act by base-pairing with mRNA (10–12).
Hfq binds both sRNA and its corresponding mRNA simultaneously, forming a ternary complex and it was suggested that the enhanced local concentration of the RNA leads to final annealing of the two RNA which then exposes the mRNA coding sequence for translation (13,14). It has also been proposed that Hfq acts by changing the RNA structure (15,16). Furthermore, Hfq was shown to rescue a group I intron splicing intermediate trapped in a misfolded form in vivo (17) and hence qualifies as a RNA chaperone (18). It is likely that RNA chaperone activities of Hfq are also involved in translational regulation by sRNA (15,19).
Our current molecular understanding of Hfq activity largely stems from recent findings that it is a Sm-like protein. The most thoroughly characterized Sm proteins are those that form the cores of eukaryotic spliceosomal ribonucleoproteins implicated in pre-mRNA splicing. The Sm domain is highly conserved across many species and its wide phylogenetic distribution underlines its likely role in early evolution of RNA metabolism (20–22). In the case of Hfq, the relationship between eubacterial protein and Sm topology was first suggested by weak sequence conservation and subsequently confirmed by crystallographic structures of S. aureus, E. coli and P. aeruginosa Hfq proteins (23–26). Eukaryotic and archaeal Sm proteins form closed rings composed of seven monomers (identical or different) (27–29), whereas Hfq forms homo-hexameric rings. The toroidal-shaped Hfq ring is ∼70 Å in diameter (7,30), with a central cationic pore that forms the RNA-binding site. The structure of S. aureus Hfq-AU5G revealed RNA bound to one side of Hfq in a circular manner with each nucleotide stacking on the side chain of an aromatic residue (Tyr or Phe 42 for S. aureus and E. coli Hfq, respectively) (24,25). Furthermore, recent work has also provided compelling evidence that Hfq has multiple RNA binding sites (14,31).
In this study, we have focused on DsrA … Hfq … rpoS interactions to examine the chaperoning role of Hfq in promoting intermolecular base-pairing. It is generally accepted that, in the absence of stress conditions, the translation of rpoS mRNA is repressed by a stem region upstream of the AUG start codon that sequesters the Shine-Dalgarno sequence. The current model proposes that stress results in stimulation of base pairing between rpoS and DsrA, at least in part by increasing the synthesis of DsrA. DsrA … rpoS pairing allows disruption of the stem in rpoS and makes its ribosome binding site accessible (Figure 1a). Nuclease footprinting experiments indicated that Hfq recognizes several sites in rpoS mRNA without changing its secondary structure in the region that inhibits translation (13) and that Hfq does not alter DsrA secondary structure (32). In addition, results of native gel electromobility shift assays (EMSA) indicated that Hfq accelerates DsrA … rpoS annealing by a factor of two (13). Finally, it was proposed that binding of Hfq to DsrA stimulates annealing of rpoS to DsrA, followed by Hfq release from the sRNA … mRNA complex (13). In these in vitro assays, however, it was not clear how the RNA chaperone activities of Hfq are related to the enhanced DsrA … rpoS interaction.
Figure 1.
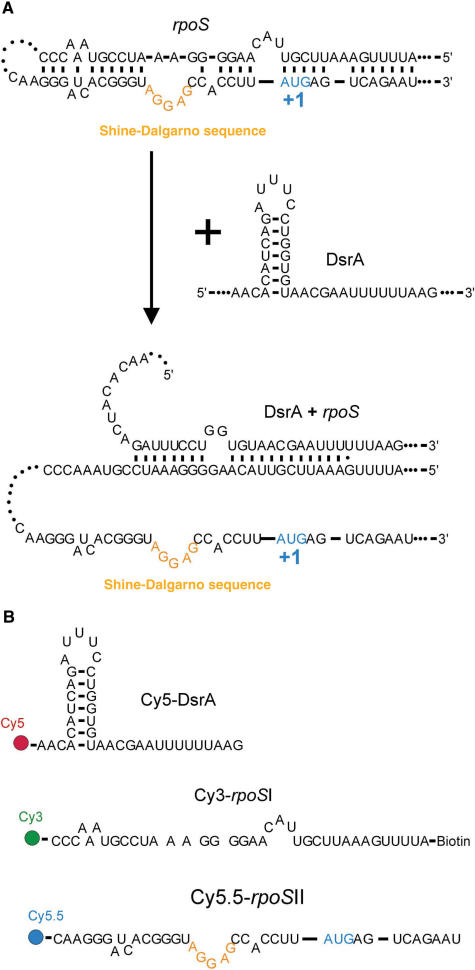
Translation regulation of rpoS by DsrA and FRET assay. (A) Proposed regulation mechanism of rpoS translation by DsrA. Base pairing between DsrA and rpoS 5′ UTR releases the rpoS segment containing the Shine–Dalgarno sequence. (B) RNA constructs for FRET measurements. DsrA, rpoSI and rpoSII are labelled with Cy5, Cy3 (and biotin) and Cy5.5, respectively.
Here, we have developed real-time FRET assays to monitor the influence of Hfq on DsrA … rpoS annealing. Our studies show that Hfq not only binds rapidly to both DsrA and pre-melted rpoS sequence but also acts as a RNA chaperone which modulates the RNA secondary structures in a sequence-specific manner. A model is proposed for how Hfq helps in annealing DsrA and rpoS.
METHODS
Unless otherwise specified, all enzymes and chemicals were either from Sigma or Merck-Biochemicals.
RNA sequences
We used the following RNA sequences for FRET measurements (5′→3′): Cy5-DsrA, Cy5-AACACAUCAGAUUUCCUGGUGUAACGAAUUUUUUAAG; Cy3-rpoSI, biotin-AUUUUGAAAUUCGUUACAAGGGGAAAUCCGUAAACCC-Cy3; Cy5.5-rpoSII, Cy5.5-CAAGGGAUCACGGGUAGGAGCCACCUUAUGAGUCAGAAU. Cy5-DsrA and Cy3-rpoSI were purchased from Dharmacon and deprotected according to the manufacturer's recommendations. For Cy5.5-rpoSII, we labelled an amine-reactive RNA (IDTDNA) with NHS ester Cy5.5 following the scheme below.
Cy5.5 dye labelling
Here, 3.2 µl of RNA (4.2 mM in H2O), 5.6 µl of H2O, and 11.2 µl of amine-reactive Cy5.5 dissolved in DMSO (18 µg/µl) was added to 60 µl of sodium tetraborate buffer (0.1 M, pH 8.5), and incubated overnight at 4°C. Labelled RNA was recovered by ethanol precipitation.
RNA annealing
For RNA duplex melting experiments, two different RNA complexes were annealed. For rpoSI+II, we mixed 10 µl of Cy3-rpoSI (40 µM in T50 (10 mM Tris pH 8.0, 50 mM NaCl)) and 10 µl Cy5.5-rpoSII (100 µM in T50) and slowly cooled the mixture from 90°C to room temperature. After the sample reached room temperature (∼4 h), we incubated it for another four hours at 4°C. For rpoSI+DsrA construct, we mixed 2 µl of Cy3-rpoSI (40 µM in H2O), 7 μl of Cy5-DsrA (52 µM in H2O), and 1 µl of NaCl (5 M) and followed the same annealing protocol as used for rpoSI+II.
Fluorescence measurements
Fluorescence emission spectra were measured in T50 buffer using a Varian Eclipse fluorospectrophotometer. While the sample was excited at 500 nm, fluorescence emission for Cy3, Cy5 and Cy5.5 were collected at 565, 670 and 702 nm, respectively. To increase the stability of RNA secondary structures and mimic the low temperatures that might be prevalent during effective rpoS translation, all measurements were done at 15°C. Fluorescence kinetic time traces were analysed using SigmaPlot (Systat) and Origin (Microcal).
Electro-mobility shift assay (EMSA)
The bandshift assay was performed with the same concentrations of labelled RNA and Hfq as described for FRET studies. After incubation, samples were loaded onto a 0.75 mm thick, 20% polyacrylamide (29:1) gel containing 150 mM Tris-HCl (pH 8.0). The gel was subjected to electrophoresis using a Mini-PROTEAN (Bio-Rad) apparatus in 25 mM Tris base and 250 mM glycine at 20°C for 4 h at 70 V. The gels were scanned using a Typhoon imager (Amersham Biosciences, USA). For quantification of annealed and melted RNA, we analysed the Cy3 label using the ImageJ software (http://rsb.info.nih.gov/ij/).
Purification of Hfq
Hfq was purified from the BL21(DE3) E. coli strain transformed with the plasmid pTE607 as previously described (33). UV absorption spectrum of Hfq indicated that Hfq is RNA and ATP free.
RESULTS
RNA constructs for FRET studies
To investigate how Hfq affects the interaction between the DsrA (sRNA) and its target site on the rpoS (mRNA), we reduced the size of the interacting RNAs to three short fragments of 37 nt each containing all the sequences involved in intra- and intermolecular base pairing governing the translational regulation of rpoS (Figure 1b) (13). The DsrA fragment was labelled with Cy5 (acceptor I) and a rpoS fragment, containing sequences upstream of the ribosome-binding site which is partially complementary to DsrA, was labelled with Cy3 (donor); they are referred to as Cy5-DsrA and Cy3-rpoSI, respectively. The third fragment containing the Shine–Dalgarno sequence and the start codon of rpoS harbours a Cy5.5 label (acceptor II) and is named Cy5.5-rpoSII. Annealing of Cy3-rpoSI and Cy5.5-rpoSII mimics the stem of the secondary structure that masks the Shine-Dalgarno sequence of rpoS. All three RNA fragments were labelled at their 5′ or 3′ extremities so that annealing of Cy5-DsrA to Cy3-rpoSI and Cy5.5-rpoSII to Cy3-rpoSI results in high FRET (see Methods and Figure 1).
Hfq stimulates the annealing of rpoS and DsrA
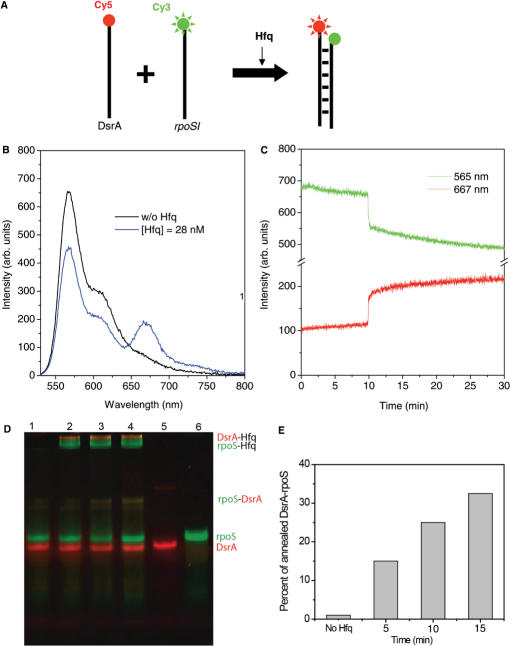
To test whether Hfq promotes DsrA … rpoS annealing (Figure 2a), we prepared 25 nM of Cy3-rpoSI and 50 nM of Cy5-DsrA in T50 buffer at 15°C. In the absence of Hfq, the emission spectrum showed negligible Cy5 signal (Figure 2b, black line), which did not change significantly for 5 min (Figure 2c), indicating that spontaneous annealing is slow under these conditions. We did observe significant spontaneous annealing at higher temperatures and higher RNA concentrations (Supplementary Figure S1). Addition of Hfq (28 nM in hexamer) at t = 9.5 min caused an abrupt increase in the Cy5 signal and a corresponding decrease in the Cy3 signal, indicating that Hfq facilitates rapid association of Cy5-DsrA and Cy3-rpoSI (Figure 2c). This large increase in FRET, which occurred in less than 20 s, was followed by a more gradual increase in FRET (lifetime of ∼9 min). The emission spectrum obtained 25 min after Hfq addition (Figure 2b, blue line) also shows a clear increase in the Cy5 signal and a decrease in the Cy3 signal. This result was confirmed by electromobility shift assay (EMSA) which showed that 15% of rpoSI is in the annealed form after the first 5 min with the reaction proceeding further with a lifetime of >10 min, similar to the slow phase in the FRET data (Figure 2d and e). The Cy5-DsrA lane shows an additional larger molecular-weight species that we attribute to the DsrA dimer as reported previously (13). Based on these data, we interpret the rapid FRET increase as the ternary complex formation via simultaneous binding of Hfq to rpoSI and DsrA and assign the subsequent slow FRET increase to the eventual annealing of the two RNA molecules. A similar ternary complex has been reported previously with the annealing process occurring over several minutes (13).
Figure 2.
Hfq helps annealing of DsrA and rpoS. (A) Scheme to examine the annealing of DsrA and rpoSI mediated by Hfq. (B) Emission spectra of 25 nM Cy3-rpoSI and 50 nM Cy5-DsrA in T50 buffer without Hfq or after incubating the sample for 10 min without Hfq (black line), and then with 28 nM of Hfq (blue line). (B) Emission intensity time trace of Cy3 at 565 nm (green trace) and Cy5 at 667 nm (red trace). Hfq (28 nM) added at 10 min results in an abrupt increase (<20s) in FRET followed by a slow increase (average annealing time = 9 min). (D) EMSA experiment confirms the annealing reaction. Green and red bands correspond to the fluorescence scans with Cy3 and Cy5 filters respectively. Lane 1: Cy3-rpoSI (25 nM) and Cy5-DsrA (50 nM) in the absence of Hfq, Lanes 2–4: Cy3-rpoSI and Cy5-DsrA in the presence of Hfq (28 nM) after incubation for 5, 10 and 15 min at 15°C, Lane 5: Cy5-DsrA and Lane 6: Cy3-rpoSI. (E) The amount of Cy3-RNA in the annealed form (indicated in the gel image) is quantified and shown in the plot.
Hfq melts the rpoSI+DsrA duplex
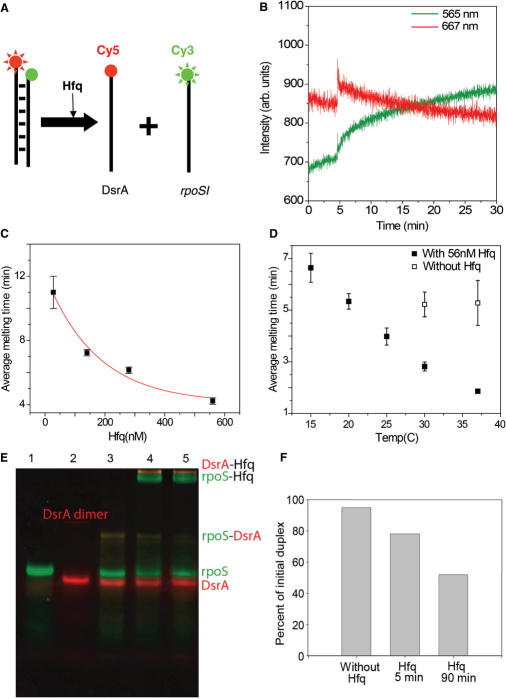
Next, we examined if Hfq can promote the reverse reaction, i.e. melting of the rpoSI-DsrA duplex (Figure 3a). We pre-annealed Cy3-rpoSI and Cy5-DsrA (See Methods for details), termed here as rpoSI+DsrA. Addition of 28 nM Hfq to 25 nM rpoSI+DsrA (in T50; 15°C) has two clear effects on the fluorescence signals (Figure 3b). First, we observed an abrupt increase in both Cy3 and Cy5 signals upon the addition of Hfq. The increase in both fluorescence signals, typically 20–30% (Supplementary Figure S2a), reflects an interaction of the protein with the cyanine dyes which blocks a non-radiative decay pathway (34,35). We cannot distinguish whether this effect is due to Hfq binding to the individual strands or to the annealed rpoSI+DsrA since the EMSA experiments suggest that there is a substantial fraction of RNA in the single-stranded form even after thermal annealing (Figure 3e). Second, the abrupt increase of fluorescence is followed by a gradual decrease in the Cy5 signal and an accompanying increase in the Cy3 signal, which indicates that Hfq melts rpoSI+DsrA over a time scale of minutes. The lifetime of FRET signal change decreased with increasing Hfq concentration and was ∼4 min at saturation (560 nM Hfq), (Figure 3c). The melting activity of Hfq was enhanced with increase in temperature with the lifetime of 1.7 min at 37°C for 56 nM Hfq (Figure 3d). Spontaneous melting of the rpoSI+DsrA duplex in the absence of Hfq was not observed at our standard temperature of 15°C but was observed at 37°C and was about 4-fold slower than the Hfq-induced melting (Figure 3d). This disruption of rpoSI+DsrA duplex by Hfq over a time scale of minutes was also confirmed by EMSA performed under the same conditions (Figure 3e and f). In contrast, we found that the melting of DsrA loop I domain by Hfq to be extremely slow even under saturating conditions (up to 2 μM Hfq; Kd = 320 nM) with lifetimes >30 min (data not shown), suggesting that melting of rpoSI+DsrA by Hfq is a specific activity of the protein rather than a result of non-specific binding of the protein to RNA duplexes.
Figure 3.
Unwinding of DsrA+rpoS complex. (A) Schematic representation of rpoSI+DsrA melting by Hfq. (B) 25 nM rpoSI+DsrA complex was prepared in T50 buffer. The graph shows the intensity time traces of Cy3 at 565 nm (green line) and Cy5 at 667 nm (red lines). Hfq (28 nM) was added at 5 min. Increase in intensities of both dyes is followed by gradual decrease in FRET (average melting time = 11 min). (C) Average melting time as a function of Hfq concentration is plotted and fitted to an exponential decay function (red line). The fit decays to a value of 4.1 min for saturating concentrations of Hfq. (D) Average melting time as function of temperature in the presence of 56 nM Hfq (solid squares) and in the absence of Hfq (hollow squares) (E) An EMSA experiment that shows the melting reaction. Green and red bands correspond to the fluorescence scans with Cy3 and Cy5 filters respectively. Lane 1: Cy3-rpoSI (25 nM); Lane 2: Cy5-DsrA; Lane 3: Thermally annealed DsrA+rpoSI (25 nM); Lane 4–5: DsrA+rpoSI with 28 nM Hfq after 5 and 90 min. (F) The amount of Cy3-RNA in the annealed form is quantified and shown as a bar graph.
Hfq melts the stem structure of rpoS mRNA
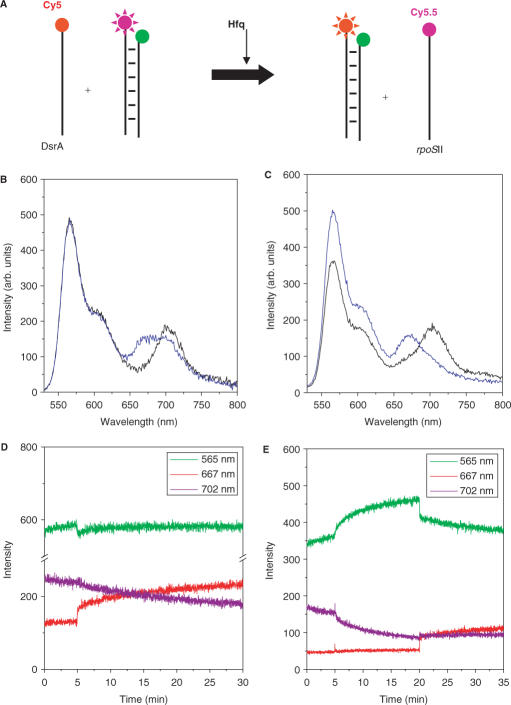
We examined if Hfq has the ability to disrupt the rpoS stem structure which would enhance the eventual annealing with DsrA (Figure 4a). For this measurement, we annealed Cy3-rpoSI and Cy5.5-rpoSII as described in Methods. The annealed RNA complex, named rpoSI+II, mimics the secondary structure of rpoS. Before adding Hfq, the emission spectrum shows significant Cy5.5 signal (Figure 4b, black line) due to intact rpoSI+II (25 nM in T50 at 15°C). Cy3 and Cy5.5 signals did not change significantly for 5 min (Figure 4c) indicating that rpoSI+II is stable under these conditions. However, when 28 nM Hfq was added, the Cy5.5 signal gradually decreased with a concomitant Cy3 signal increase over a time scale of minutes (Figure 4c). The emission spectrum obtained 25 min after Hfq addition (Figure 4b, blue line) also shows a clear decrease in FRET. These results demonstrate that Hfq can induce melting of the rpoS stem, independent of DsrA. The average melting time obtained by fitting the FRET decrease to an exponential decay decreases with Hfq concentration and begins to saturate at about 4 min at concentrations above 300 nM Hfq (Figure 4d). This saturation is consistent with the formation of a complex between Hfq and rpoS with a Kd around 150 nM as shown by previous affinity measurements (13) and our anisotropy measurements (Supplementary Figure S2b). Therefore, binding of Hfq to rpoS is slow, taking many minutes, under our standard conditions (28 nM Hfq hexamer) and hence the slower melting observed in Figure 4c.
Figure 4.
Hfq melts the stem structure of rpoS but does not promote annealing. (A) Scheme to show Hfq assisted melting of rpoS. (B) Emission spectra of 25 mM rpoSI+II or after incubating the sample for 5 min without Hfq (black line), and then for 25 min with 28 nM Hfq (blue line). (C) Intensity time trace of Cy3 (green lines) and Cy5.5 (red purple lines); Hfq (28 nM) was added at 5 min. Addition of Hfq results in gradual FRET decrease (average lifetime ∼15 min). (D) Average melting times were measured as a function of Hfq concentration (as plotted) and it decreases to ∼4 min in saturating concentrations of Hfq. (E) Hfq does not promote annealing of rpoSI and rpoSII. (F) Emission spectra of the mixture of 25 nM Cy3-rpoSI and 62.5 nM Cy5.5-rpoSII in T50 buffer without (black lines), after incubating the sample for 5 min without Hfq, and then with 28 nM of Hfq (blue lines).
Hfq does not anneal the rpoS stem
In contrast to Hfq's ability to promote rpoSI…DsrA annealing, we did not observe any increase in FRET when Cy3-rpoSI and Cy5.5-rpoSII were incubated in the presence of Hfq under the same condition (Figure 4e and f), despite the fact that Hfq is able to bind both rpoSI and rpoSII as observed by EMSA and anisotropy measurements (Supplementary Figure S2c and b). For this measurement, we incubated 25 nM Cy3-rpoS and 62.5 nM Cy5.5-rpoSII at 15°C for 25 min without Hfq (black curve) and with 28 nM of Hfq (blue curve). Therefore, the annealing activity of Hfq is specific to DsrA and rpoSI.
Strand exchange reaction
We then probed the effect of Hfq on the interaction between DsrA and rpoS (rpoSI+II) as shown schematically in Figure 5a. First, we mixed 25 nM rpoSI+II duplex and 65 nM Cy5-DsrA in T50 buffer at 15°C to investigate whether Cy5-DsrA can anneal by itself to Cy3-rpoSI and release Cy5.5-rpoSII. After 30 min incubation at 15°C, we measured the emission spectrum (Figure 5b). Although Cy3 and Cy5.5 emission is clear, Cy5 signal is negligible, indicating that spontaneous interaction between DsrA and rpoS does not occur effectively under our experimental conditions (black line; Figure 5b). However, incubation with 28 nM Hfq for 25 min results in a decreased Cy5.5 signal and an increase in Cy5 emission (blue line; Figure 5b). A larger change in fluorescence signals is seen with 280 nM Hfq (Figure 5c). The data show that Hfq enhances both the disruption of rpoS internal structure and annealing of DsrA with the upstream strand of the rpoS stem, thus in effect carrying out a strand exchange reaction.
Figure 5.
Strand exchange reaction promoted by Hfq. (A) Schematic of the strand exchange reaction mediated by Hfq. (B) Emission spectra of the mixture of 25 nM of the rpoSI+II and 62.5 nM of Cy5-DsrA in T50 buffer without Hfq. Comparison of the emission spectra of the mixture 15 min after adding 28 nM Hfq (blue lines) and before adding Hfq (black lines) shows a shift in emission spectrum from Cy5.5 (707 nm) to Cy5 (667 nm). (C) Comparison of the emission spectra before (black line) and after addition of 282 nM Hfq (blue line). (D) Intensity time traces at Cy3, Cy5 and Cy5.5 emission wavelengths where 28 nM Hfq is added at t = 5 min to a solution containing rpoSI+II and Cy5-DsrA. (E) Intensity times traces at Cy3, Cy5 and Cy5.5 emission wavelengths where Hfq is added at t = 5 min and Cy5-DsrA is added at t = 20 min to a solution containing rpoSI+II.
Kinetic data on the strand exchange reaction provide additional details (Figure 5d and e). When Hfq was added to a mixture of DsrA and rpoSI+II at t = 5 min, we observed an abrupt decrease in Cy3 signal and a simultaneous increase in Cy5 signal which we attribute to the rapid association of free single-stranded DsrA and rpoSI as we have shown in Figure 2c. This was followed by a slow decrease in the Cy5.5 signal and a slow increase in the Cy5 signal without significant changes in the Cy3 signal indicating that DsrA gradually replaces rpoSII to pair up with rpoSI. The decay lifetime of the Cy5.5 signal decrease is ∼13 min, similar to the time scale of melting of rpoSI+II at the same Hfq concentration. Figure 5e shows fluorescence signals versus time from rpoSI+II where Hfq (28.5 nM) is added at t = 5 min followed by DsrA addition at t = 20 min. Slow melting of rpoSI+II was observed upon Hfq addition similar to what has been seen in Figure 4c. When DsrA was added, we observed a very rapid increase in FRET from Cy3 to Cy5.5 followed by a slower FRET increase as we have seen from the experiment using rpoSI and DsrA only. This experiment suggests that the strand exchange reaction can also proceed by slow melting of the rpoS duplex followed by rapid association of DsrA to melted rpoS.
DISCUSSION
We have examined how Hfq affects the interaction between a non-coding sRNA, DsrA and its regulation target mRNA, rpoS using real-time FRET assays. In order to focus on the specific parts of the system that are involved in translational regulation of rpoS by DsrA, we limited our investigation to small fragments of DsrA and rpoS. In particular, we omitted the P3 site in rpoS and loop II domain in DsrA both of which contact Hfq strongly according to footprinting experiments (13). However, our constructs conserved all the known intra- and inter-strand base-pairing as well as uridine-rich regions important for Hfq binding (13,16).
We demonstrated that Hfq assists in annealing of pre-melted DsrA and rpoSI. FRET measurements showed a rapid FRET increase within a few seconds after Hfq addition due to the binding of both DsrA and rpoSI to Hfq and formation of a partially annealed complex (allowing energy transfer between the RNAs that are in close proximity). This rapid binding step is followed by a slower FRET increase signifying the subsequent RNA annealing. This is consistent with previous observations of a partially annealed intermediate DsrA … rpoS complex and only two fold enhancements in annealing in the presence of Hfq at 30°C (13) and our own EMSA studies. This implies that actual rates of Hfq-mediated annealing are much slower than RNA binding and are in the time scale of minutes at low temperatures.
Our real-time FRET analysis also showed that Hfq melts rpoSI+II complex over several minutes. This activity could be important in promoting the final strand exchange since annealing of DsrA and rpoS requires pre-melting of the rpoS secondary structure. This observation is contrary to previous nuclease footprinting experiments which reported that Hfq recognized several sites on the rpoS mRNA without changing the secondary structure in the region that inhibits translation (13). However, the RNA constructs used in the two experiments are different. For example, our fluorescent construct does not contain the P3 site and the two strands of rpoS are not covalently linked in our study. Our current result is analogous to that of sodB (iron superoxide dismutase) mRNA where Hfq binding to the mRNA does alter its structure by partially opening a loop (15). We propose that both the rates of denaturation of rpoS secondary structure and complete annealing of DsrA … rpoS are slow at low temperatures (15°C) and will manifest in a slow formation rate (order of 0.1 min−1) of the final DsrA … rpoS duplex. In fact, temperature-dependent rates of DsrA … rpoS hybridization determined in a previous report range from 1 min−1 at 42°C to 0.01 min−1 at 8°C that agree well with our estimates (13).
Surprisingly, Hfq was also capable of melting the rpoS+DsrA complex with the average reaction time of ∼4 min. The net effect is that Hfq can accelerate both annealing and melting reactions depending on initial conditions. The time scales of DsrA … rpoS annealing and melting promoted by Hfq are similar, in the range of several minutes, suggesting that both of these processes might be competing to achieve equilibrium.
Note that the experiments were performed at 15°C in order to mimic the physiological function of the mRNA rpoS which codes for a factor required for cold shock response. At this temperature, the protein could promote these annealing and melting reactions only 30–75 times per generation time of the bacteria; therefore, some additional co-factors might also be involved in one or both of these Hfq activities in vivo. For example, ribosomal protein S1 has been already shown to bind to DsrA and rpoS (36) and was suggested to interact with Hfq and RNA polymerase (37). This also raises an interesting possibility of a coupling between transcription and Hfq-mediated translational regulation. Consequences of such coupled transcription–translational systems need to be examined in the context of role of co-factors in Hfq activity.
The Hfq protein preparation used in this study is free of RNA and ATP (as judged from absorbance spectrum, see Methods). Thus, both annealing and melting activities are independent of ATP. The hydrolysis of ATP powers the unwinding activity of RNA helicases (38), which promotes disruption of RNA duplexes. By comparison, RNA chaperones have two distinct activities, secondary structure melting and strand annealing, which do not necessarily require NTP hydrolysis. Although Hfq was reported to have an ATPase activity (37), such an activity does not appear to be necessary for its RNA chaperone functions in sequence-specific RNA annealing or melting we observed here.
CONCLUSION
Our results confirm that Hfq rapidly associates with small RNA regulator, DsrA, and its target rpoS mRNA simultaneously increasing their local concentration. We also show that Hfq is a chaperone that can change the conformation of rpoS making it accessible to DsrA. We suggest that Hfq accelerates the arrival of the dynamic equilibrium between annealing and dissociation of RNA strands, and spectroscopic assays described here were able to track these processes in real time. Two primary results must be emphasized: (i) Hfq dramatically accelerates the association of DsrA and rpoS but eventual annealing of DsrA … rpoS is slower and (ii) Hfq melts the rpoSI+II complex over several minutes. Taking these results into account, we propose a model for the function of Hfq in the translational regulation of rpoS by DsrA (Figure 6). Hfq binds rapidly to both rpoS and DsrA and melts the stem of rpoS (average reaction time ∼4 min), which appears to be irreversible. Increased local concentration of both RNAs and melting of rpoS facilitates the annealing between rpoS and DsrA (average reaction time ∼9 min) (13). Finally, Hfq dissociates from RNA duplex due to its low binding affinity (13). We have also shown that at least one step in the reaction is reversible so that Hfq facilitates the melting of DsrA and rpoS (average reaction time ∼4 min). Such an activity of Hfq may be useful in the reversal of post-transcriptional regulation (for example, by re-sequestering the Shine–Dalgarno sequence and blocking the ribosome binding site). The overall process is rather slow and it may be that additional co-factors might be assisting this translational regulatory machinery in vivo.
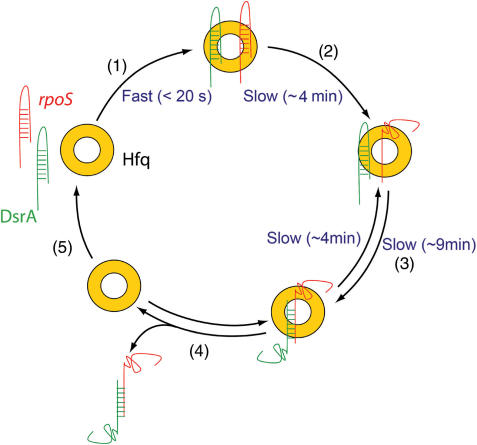
Figure 6.
A model for Hfq function in the translational regulation of rpoS by DsrA. (1) Hfq binds rapidly to both rpoS and DsrA. (2) After binding to rpoS, it slowly melts the stem region of rpoS. (3) Combined with the increased local concentration of both RNAs, this unwinding event could accelerate the annealing between rpoS and DsrA. Melting of the annealed DsrA … rpoS also takes place when bound to Hfq. (4) Hfq releases the DsrA … rpoS duplex and (5) it is free to get recycled.
ACKNOWLEDGEMENTS
This work was supported by CNRS (UPR 9073), University Denis Diderot-Paris 7 and by the National Institutes of Health (GM 065367). We are very grateful to A. Zhang (NIH) and J. Plumbridge (IBPC) for reading the manuscript and to J. Le Derout and J. Banroques for their help in performing EMSA and gel imaging. T.H. is an investigator with the Howard Hughes Medical Institute. Funding to pay the Open Access publication charge was provided by the Howard Hughes Medical Institute.
REFERENCES
- 1.Franze de Fernandez MT, Hayward WS, August JT. Bacterial proteins required for replication of phage Qβ ribonucleic acid. J. Biol. Chem. 1972;247:824–821. [PubMed] [Google Scholar]
- 2.Vecerek B, Moll I, Blasi U. Translational autocontrol of the Escherichia coli hfq RNA chaperone gene. RNA. 2005;11:976–984. doi: 10.1261/rna.2360205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hajnsdorf E, Régnier P. Host factor HFq of Escherichia coli stimulates elongation of poly(A) tails by poly(A)polymerase I. Proc. Natl. Acad. Sc. U.S.A. 2000;97:1501–1505. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rasmussen AA, Eriksen M, Gilany K, Udesen C, Franch T, Petersen C, Valentin-Hansen P. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol. 2005;58:1421–1429. doi: 10.1111/j.1365-2958.2005.04911.x. [DOI] [PubMed] [Google Scholar]
- 5.Muffler A, Traulsen DD, Fischer D, Lange R, Hengge-Aronis R. The RNA-binding protein HF-1 plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the σS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 1997;179:297–300. doi: 10.1128/jb.179.1.297-300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sledjeski DD, Whitman C, Zhang A. HFq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases oxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 8.Masse E, Majdalani N, Gottesman S. Regulatory roles for small RNA in bacteria. Current Opinion in Microbiology. 2003;6:120–124. doi: 10.1016/s1369-5274(03)00027-4. [DOI] [PubMed] [Google Scholar]
- 9.Storz G. An expanding universe of noncoding RNAs. Science. 2002;296:1260–1263. doi: 10.1126/science.1072249. [DOI] [PubMed] [Google Scholar]
- 10.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol. Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 11.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 12.Kawamoto H, Koide Y, Morita T, Aiba H. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 2006;61:1013–1022. doi: 10.1111/j.1365-2958.2006.05288.x. [DOI] [PubMed] [Google Scholar]
- 13.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rolle K, Zywicki M, Wyszko E, Barciszewska MZ, Barciszewski J. Evaluation of the dynamic structure of DsrA RNA from E. coli and its functional consequences. J. Biochem. (Tokyo) 2006;139:431–438. doi: 10.1093/jb/mvj045. [DOI] [PubMed] [Google Scholar]
- 17.Moll I, Leitsch D, Steinhauser T, Blasi U. RNA chaperone activity of the Sm-like Hfq Protein. EMBO Reports. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Herschlag D. RNA chaperones and the RNA folding problem. J. Biol. Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 19.Rajkowitsch L, Semrad K, Mayer O, Schroeder R. Assays for the RNA chaperone activity of proteins. Biochem. Soc. Trans. 2005;33:450–456. doi: 10.1042/BST0330450. [DOI] [PubMed] [Google Scholar]
- 20.Seraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. Embo J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He W, Parker R. Functions of Lsm proteins in mRNA degradation and splicing. Curr. Opin. Cell Biol. 2000;12:346–350. doi: 10.1016/s0955-0674(00)00098-3. [DOI] [PubMed] [Google Scholar]
- 22.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature. 1999;401:177–180. doi: 10.1038/43694. [DOI] [PubMed] [Google Scholar]
- 23.Arluison V, Derreumaux P, Allemand F, Folichon M, Hajnsdorf E, Regnier P. structural modelling of the Sm-like protein Hfq from Escherichia coli. J. Mol. Biol. 2002;320:705–712. doi: 10.1016/s0022-2836(02)00548-x. [DOI] [PubMed] [Google Scholar]
- 24.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq- RNA complex: a bacterial Sm-like protein. Embo J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sauter C, Basquin J, Suck D. Sm-like proteins in Eubacteria: the crystal structure of the Hfq protein from Escherichia coli. Nucleic Acids Res. 2003;31:4091–4098. doi: 10.1093/nar/gkg480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nikulin A, Stolboushkina E, Perederina A, Vassilieva I, Blaesi U, Moll I, Kachalova G, Yokoyama S, Vassylyev D, Garber M, et al. Structure of Pseudomonas aeruginosa Hfq protein. Acta Crystallogr D Biol Crystallogr. 2005;61:141–146. doi: 10.1107/S0907444904030008. [DOI] [PubMed] [Google Scholar]
- 27.Kambach C, Walke S, Young R, Avis JM, de la Fortelle E, Raker VA, Luhrmann R, Li J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 28.Thore S, Mayer C, Sauter C, Weeks S, Suck D. Crystal structures of the pyrococcus abyssi Sm core and its complex with RNA: common features of RNA-binding in Archaea and Eukarya. J. Biol. Chem. 2003;29:29. doi: 10.1074/jbc.M207685200. [DOI] [PubMed] [Google Scholar]
- 29.Mura C, Kozhukhovsky A, Gingery M, Phillips M, Eisenberg D. The oligomerization and ligand-binding properties of Sm-like archaeal proteins (SmAPs) Protein Sci. 2003;12:832–847. doi: 10.1110/ps.0224703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. Hfq: A bacterial Sm-like protein that mediates RNA–RNA Interaction. Molecular Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 31.Sun X, Wartell RM. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry. 2006;45:4875–4887. doi: 10.1021/bi0523613. [DOI] [PubMed] [Google Scholar]
- 32.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arluison V, Mura C, Guzman MR, Liquier J, Pellegrini O, Gingery M, Regnier P, Marco S. Three-dimensional structures of fibrillar Sm proteins: Hfq and other Sm-like proteins. J. Mol. Biol. 2006;356:86–96. doi: 10.1016/j.jmb.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Mikelsons L, Carra C, Shaw M, Schweitzer C, Scaiano JC. Experimental and theoretical study of the interaction of single-stranded DNA homopolymers and a monomethine cyanine dye: nature of specific binding. Photochem. Photobiol. Sci. 2005;4:798–802. doi: 10.1039/b508720a. [DOI] [PubMed] [Google Scholar]
- 35.Widengren J, Schwille P. Characterization of photoinduced isomerization and back-isomerization of the cyanine dye Cy5 by fluorescence correlation spectroscopy. J. Phys. Chem. A. 2000;104:6416–6428. [Google Scholar]
- 36.Koleva RI, Austin CA, Kowaleski JM, Neems DS, Wang L, Vary CPH, Schlax PJ. Interactions of ribosomal protein S1 with DsrA and rpoS mRNA. Biochem. Biophys. Res. Commun. 2006;348:662–668. doi: 10.1016/j.bbrc.2006.07.102. [DOI] [PubMed] [Google Scholar]
- 37.Sukhodolets MV, Garges S. Interaction of Escherichia coli RNA polymerase with the ribosomal protein S1 and the Sm-like ATPase Hfq. Biochemistry. 2003;42:8022–8034. doi: 10.1021/bi020638i. [DOI] [PubMed] [Google Scholar]
- 38.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]