Abstract
Development of methods that will allow exogenous imposition of inheritable gene-specific methylation patterns has potential application in both therapeutics and in basic research. An ongoing approach is the use of targeted DNA methyltransferases, which consist of a fusion between gene-targeted zinc-finger proteins and prokaryotic DNA cytosine methyltransferases. These enzymes however have so far demonstrated significant and unacceptable levels of non-targeted methylation. We now report the development of second-generation targeted methyltransferase enzymes comprising enhanced zinc-finger arrays coupled to methyltransferase mutants that are functionally dominated by their zinc-finger component. Both in vitro plasmid methylation studies and a novel bacterial assay reveal a high degree of target-specific methylation by these enzymes. Furthermore, we demonstrate for the first time transient expression of targeted cytosine methyltransferase in mammalian cells resulting in the specific methylation of a chromosomal locus. Importantly, the resultant methylation pattern is inherited through successive cell divisions.
INTRODUCTION
The link between DNA methylation and gene expression has come under intense scrutiny over recent years and a number of groundbreaking discoveries have been made, linking DNA methylation, histone methylation and/or deacetylation to the formation or modulation of higher order, repressive chromatin structures through various protein–DNA interactions (1–5). Moreover, high levels of DNA methylation are often found in the promoters of transcriptionally silent genes, especially in cancer. Such observations have led to the proposition that methylation is a key mechanism for controlling gene expression in the cell. The ability to impose DNA methylation at specific sites in the genome as a potential means of controlling heritable gene expression for both therapeutic and research applications is therefore a desirable goal.
Whether methylation is the cause or consequence of transcriptional silencing in all cases is still unclear. This is largely because it has not been possible to investigate the effects of de novo methylation in the cell as it occurs or simulate it in a meaningful way and much of our understanding of this epigenetic process therefore comes from post-event observations. Attempts to examine the consequences of de novo methylation on gene expression, methylation spread and transcriptional elongation have mainly involved transient transfection of in vitro methylated reporter constructs or, more elegantly, by Cre/LoxP-mediated genomic placement of in vitro methylated DNA (6). Even the latter approach however is imperfect, since it cannot divorce methylation mediated effects from the action of the integration/repair processes involved in DNA placement. Reports have described the use of siRNA targeted to the EF1A, erbB2, IL-2 and E Cadherin promoters, which resulted in the localized methylation and transcriptional down regulation of the gene promoters (7–9). Recruitment of DNA or histone methyltransferases to target loci via siRNA interaction with DNA or nascent transcripts are two postulated routes by which this effect may occur. However, the exact mechanism by which this effect is implemented remains unclear at present, as does the global applicability of this approach, since other studies have failed to detect similar RNA-directed methylation of targeted genes (10,11). Moreover, additional evidence suggests that short double-stranded RNA can induce transcriptional silencing in the absence of DNA methylation (12). The targeted methylation of the GSTP1 and IGF2 promoters using single-stranded methylated oligodeoxynucleotides complementary to those promoter regions has also been reported, although again the mechanism remains unclear (13,14). Given the sporadic outcome of these methodologies, their application in the generation of targeted methylation currently appears limited.
A more robust approach to delivering targeted cytosine methylation is the use of site-biased DNA methyltransferase (Mtase) enzymes, which potentially can deliver both CpG and non-CpG methylation directly at the target locus. The use of zinc fingers as the targeting component for a number of proteins with diverse function has been extensively described, allowing the progression of this class of molecule into the therapeutic arena (15). The first report of a targeted Mtase enzyme described a fusion between the CpG-specific Mtase M.SssI and the Zif268 three zinc-finger protein or a three zinc-finger protein specific for the p53-binding site found in the p21WAF/CIP1 gene (16). When similar enzymes were recently assayed in a yeast model, although enhanced methylation was observed at the target region, levels of CpG methylation at non-targeted sites were equivalent to those obtained using non-targeted wild-type Mtase enzyme (17). We have also similarly shown that other CpG Mtases can be targeted to specific sites on DNA, but that non-specific methylation is still a significant problem (18).
We now report the construction and evaluation of targeted Mtases with substantially enhanced targeted methylation properties. Using rational mutagenesis we have modified the catalytic/binding attributes of the Mtase components of two different targeted Mtases, M.HpaII and M.HhaI. These enzymes recognize the sequences 5′-CCGG-3′ and 5′-GCGC-3′, respectively, and the core methylation site is the cytosine in the sequence 5′-CpG-3′. We demonstrate that these enzymes are highly site-specific, producing a significantly reduced background methylation in the absence of a targetable site, in comparison with targeted wild-type enzymes. Expression of a targeted HpaII enzyme in mammalian cells results in specific de novo methylation of a target locus. The persistence of the methylation pattern over time shows that the de novo methylation pattern is inherited through successive cell divisions. Furthermore, the de novo methylation remains confined to the initial target region with little observable spread to flanking regions.
MATERIALS AND METHODS
Constructs
The pGex vector pG4Zf harbouring a four zinc-finger protein (4aZf) recognizing the sequence 5′-GACGCAGAAGCC-3′ has been described previously (19). The four zinc-finger coding DNA fragment was excised from this vector via NdeI and BamHI digestion and cloned into pG3ZfHhaI and pG3ZfHpaII vectors [harbouring three zinc-finger HhaI or HpaII wild-type and mutant fusion sequences, which have been described previously (18)], which had been similarly restricted, thus replacing the original three zinc-finger sequence. Gene sequences coding for the four zinc-finger Mtase fusion proteins were cloned into the modified pLysE vector described below, as NdeI/NotI fragments. The four zinc-finger gene for the protein 4bZf, designed to recognize the target sequence 5′-GACGCGGCGCTC-3′, was constructed using published zinc-finger recognition codes and assembled directly via ligation of oligonucleotides coding for each individual zinc-finger sequence. The recognition code (α-helical residues −1, 1, 2, 3, 4, 5, 6) used was ASDDLTQ (CTC subsite), RSDDLQR (GCG) and DRSNLTR (GAC). The four zinc-finger gene coding for the 4cZf protein was designed to recognize the sequence; 5′-TGGGGAGTGGCG-3′, and was similarly constructed [RSDELTR (GCG), RSDALSR (GTG), DSAHLTR (GGA) and RSDHLST (TGG)]. The zinc-finger backbone sequence used was that described for the anti-p190BcrAbl 3Zf protein (20). A six zinc-finger gene coding for two repeats of the zif268 three zinc-finger gene was constructed by ligation of two PCR products, each coding for one set of three zinc fingers, using the following primers: primers used were 1A and 1B for one PCR; 5′-gcgccagatcttggaacgcccatatgcttgccctgtcgagtcctg-3′, 5′-gcgcggatccgcccccgtccttctgtcttaaatggattttggtatgcctcttgcgtt-3′, and primers 2A and 2B for the second PCR; 5′-gcgcgagatctgaacgcccatatgcttgccctgtcgagtcctgcgat-3′ and 5′-gcgcggatccgtccttctgtcttaaatggattttggtatgcc-3′. PCR products were restricted with BamHI and BglII and sequentially cloned into BamHI restricted pGex4T-1 vector (Amersham Biosciences). The assembled six zinc-finger gene was PCR amplified from this vector using primer 1B above, and primer 6R 5′-gcgcattaatggatctgatcgaaggtcgtgggatcttggaa-3′, restricted with BamHI and AseI (underlined in the above sequences) and ligated onto the modified pLysE vectors described below, replacing the existing zinc-finger gene sequences present in targeted methylase constructs, excised as NdeI/BamHI fragments.
Mutagenesis of Mtase enzymes was carried out using the ‘quickchange’ protocol (Stratagene, CA) according to the manufacturer's instructions, using pGex-based clones as templates. Primers for mutagenesis were supplied by MWG (Germany) or Sigma (UK) and were gel purified before use. Primer sequences (only one strand is given) were as follows: HhaI-null mutant (stop codon introduced), 5′-ggaaaacaaaaaggattctagaacagcagaggtacgctc-3′; HhaI R(165)G, 5′-gaaaacaaatcatatagatcccttcccttttctgtg-3′; HhaI Q(237)G, 5′-cttggtattgtaggaaaaggtgggggcggagaacgaatttatagtactagaggcattgca-3′; HhaI Q(237)V, 5′-cttggtattgtaggaaaaggtggggtgggagaacgaatttatagtactagaggcattgca-3′; HhaI Q(237)I, 5′-cttggtattgtaggaaaaggtgggatcggagaacgaatttatagtactagaggcattgca-3′; HhaI Q(237)E, 5′-cttggtattgtaggaaaaggtggggaaggagaacgaatttatagtactagaggcattgca-3′; HpaII F(38)H, 5′-tttacttttatcgatttacatgcaggtattggtgga-3′ and HpaII R(189)G, 5′-ccacaaaatagagaagggatctatattgtaggcttt-3′. The HpaII-null construct was obtained by restriction of wild-type vector with MunI, followed by filling in the overhangs and religating the vector, thus generating a frameshift within the Mtase coding sequence.
Target site vectors were constructed in the following manner. Annealed oligonucleotides (with single A-base overhangs) harbouring single HpaII and HhaI sites (boldface) and flanked by the 6a and 4a zinc-finger recognition sites (underlined) (5′-agttccgcccacgctcgcccacgctacgctcttaaccggcgcctggttaggctgtgggacgcagaagcctcctc-3′) or by the 4c and 4b zinc-finger recognition sites, respectively (5′-tggccacgccactccccaggctccattccaggccggcgcaatgctagactcgacgcggcgctcgttgcca-3′) were cloned directly into pTopo2.1. These oligonucleotides (designated Ta and Tb/c, respectively) were also used as probes in band shift assays. These vectors were used directly in in vitro methylation assays. The insert and flanking sequence were then amplified by PCR using the specific primers described below and cloned back into pTopo2.1. Primer sequences used to generate the HpaII target site vector were 5′-gtatgttgtgtggaattgtgagcggat-3′ and 5′-ggcgcagcggtcgggctgaacgggggg-3′; and to generate the HhaI target site vector were 5′-cgcaattaatgtgagttagctcactcatta-3′ and 5′-ctgttgggaagggcgatcggtgcgggcctc-3′. These plasmids were used in bacterial-based assays. DNA fragments generated by this PCR were also used in initial methyltransferases assays and were designated as either ‘ZfM’ or ‘M’, depending on which zinc-finger recognition sites they contained in the context of the assay being performed.
Vectors expressing targeted Mtase enzymes for in situ methyltransferase specificity analysis were based on the pLysE vector (Novagen), harbouring a pTac promoter. The complementary oligonucleotides: (5′–3′) gatctgttgacaattaatcatcggctcgtataatgtgtggaattgtgagcggataacaatttcacatatgctaaagctttgagcggccgctag; agctctagcggccgctcaaagctttagcatatgtgaaattgttatccgctcacaattccacacattatacgagccgatgattaattgtcaaca, were annealed generating BamHI and HindIII overhangs, and ligated into the appropriate restriction sites in pLysE vector. Internal NdeI and NotI sites are underlined and were used for subsequently cloning genes for zinc-finger–Mtase fusion proteins into the modified pLysE expression vector, from pGex vectors described previously. Target site vector (Targ1) for mammalian experiments was made by oligonucleotide insertion into the HindIII site of pBLB-Bcr-Abl (21). Targeted methyltransferases were expressed in mammalian cells from the CMV promoter, as fusions with the SV40 nuclear localization sequence and the FLAG epitope tag using the pFLAG CMV-2 vector (Sigma), the latter was used for protein detection in western analysis.
Reagents
Restriction enzymes, T4 ligase, T4 kinase and high-purity dNTPs were purchased from NEB (UK). General chemicals were from Sigma. Taq polymerase was from Promega, poly(dI)–(dC) from Pharmacia Biotech and [γ-32P]ATP (>11 TBq/mmol) was purchased from ICN.
Cell culture
HeLa cells were grown in DMEM supplemented with 10% foetal calf serum. All transfections were performed according to the BES CaPO4 method (22). Stable cell lines were generated by co-transfection of the Targ1 target site vector and a vector that expressing puromycin at a ratio of 10:1, respectively, using a total of 3 μg DNA on cells seeded at 5 × 105 cells per 5 cm dish. Single clonal colonies were assessed for reporter gene expression. One clone was chosen for further transfection studies for which Southern blot analysis indicated as having a single integration event. This target clonal cell line was co-transfected with expression or control vectors, together with a vector expressing the low density truncated nerve growth factor receptor (LNGFR) at a ratio of 4:1, using 8 μg of methylase expression vector and a total of 12 μg DNA on 2 × 106 cells. After 72 h, cells were isolated by affinity capture using anti-LNGFR-biotin/streptavidin coupled magnetic beads (Dynal) according to the manufacturer's instructions. This was done to enhance the ratio of transfected to non-transfected cells, with respect to methyltransferase expression vectors, in order to assist downstream DNA methylation analysis. For expression studies, equivalent transfections were performed and RNA or protein was collected downstream.
DNA-binding and methylation assays
Escherichia coli ER1647 [F− fhuA2 Δ(lacZ)rl supE44 trp31 mcrA1272::Tn10(Tetr) his-1 rpsL104(Strr) xyl-7 mtl-2 metB1 Δ(mcrC-mrr)102::Tn10(Tetr) recD1014] was used for the expression of GST–four zinc finger HpaII and GST-HpaII fusion proteins. Protein purification and EMSA analysis were performed as described previously (18,19). Protein concentrations were quantified and balanced against a known BSA standard after separation by SDS–PAGE and gel staining with coumassie blue. Apparent dissociation constants were determined from protein titration EMSAs, and were calculated as the protein concentration at 50% DNA–protein complex formation using Graphpad Prizm software, essentially as described previously (18). In vitro plasmid methylation assays involved incubation of purified protein with plasmid in binding buffer (20 mM HEPES, pH 7.5, 1 mM dithiothreitol, 20 μM ZnSO4, 5 mM MgCl2, 50 mM KCl, 5% glycerol, 0.1% NP-40 and 0.1 mg/ml BSA) containing 100 μM S-adenosylmethionine for 1 h at 37°C. The reaction was stopped by heating at 80°C for 20 min and then digested with 20 U of HpaII restriction enzyme overnight at 37°C. Products were analysed on a 2% agarose gel run in TBE buffer.
Bacterial ‘in situ’ targeted methylation assay
E.coli Top10 cells (Invitrogen) [F− mcrA Δ(mrr-hsdsRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7696 galU galK rpsL (strr) endA1 nupG] were transformed with combinations of relevant target and expression vectors. Colonies were grown overnight in the presence of ampicillin and chloramphenicol and plasmids subsequently isolated by standard alkaline lysis methodology (22) and subjected to restriction with the relevant restriction enzymes. Restriction products were analysed by agarose gel electrophoresis.
Arbitrary primed PCR analysis
Genomic DNA (2 μg) was isolated from E.coli expressing targeted HpaII or HhaI methyltransferase enzymes or from untransformed E.coli using GenElute bacterial genomic DNA isolation kit (Sigma), and subjected to overnight digestion with >30 U of relevant restriction enzyme. After heat killing the enzyme at 70°C for 20 min and visually confirming the digestion of genomic DNA, 200 ng of the digest was used in a PCR employing the following primer sequences (5′–3′): tggccgccga, ntgggcggga, ctgacgttg, attgtgcttg. Cycling conditions were as follows: 95°C for 30 s, 35°C for 2 min, 72°C for 2 min, for 40 cycles, after an initial pre-melt incubation at 95°C for 2 min. Products were analysed on a 4% acrylamide–8 M urea gel and visualized by staining with Sybr green (Sigma). Differences in methylation between genomic samples were interpreted both from the relative intensities and band distribution of the resultant PCR profiles.
RT–PCR
RNA was extracted using the Trizol methodology (Invitrogen) according to the manufacturer's instructions. Poly(A) RNA was isolated using ‘Micro to Midi’ columns (Invitrogen) and cDNA generated using the ‘Cloned AMV first strand synthesis kit’ (Invitrogen) according to the manufacturer's instructions. PCR was performed using the following parameters: 94°C 30s, 55°C 30s, 72°C 45s for increasing cycle numbers. Primers used were either specific for the targeted Mtase gene (5′-CTTACCTGTAGCTTGAATAGCC-3′; 5′-CGTGAACGTAATCTTGTAATTG) or for the β-Actin gene (5′-GTGGGCCGCTCTAGGCACCAA-3′; 5′-CTCTTTGATGTCACGCACGATTTC-3′). PCR products were analysed on a 1% agarose gel.
Bisulphite sequencing
Genomic DNA isolated from HeLa cells was subjected to bisulphite modification using the CpGenome kit (Chemicon) according to the manufacturer's instructions. Primers used for 1st and 2nd round PCR, designed to amplify modified DNA and sequence the top strand, but were not biased towards methylation, have the following sequence: 1st round forward 5′-ggtattagagtagattgtattgagagtg-3′, reverse 5′-ctatcccatatcaccaactcaccatctttc-3′; 2nd round forward 5′-ggatgtgttgtaagg(c/t)gattaagttggg-3′, reverse 5′-caactaactaaaatacctcaaaatattc-3′. PCR products were cloned into PCRTopo2.1 cloning vector (Invitrogen). Only sequences shown to be converted (>90%, based on relative numbers of unconverted Cs in the context of non-CpG sequences) were used in subsequent analysis of methylation status. Sequencing was carried out by Lark Technologies (UK) and MRC Geneservice (UK).
RESULTS
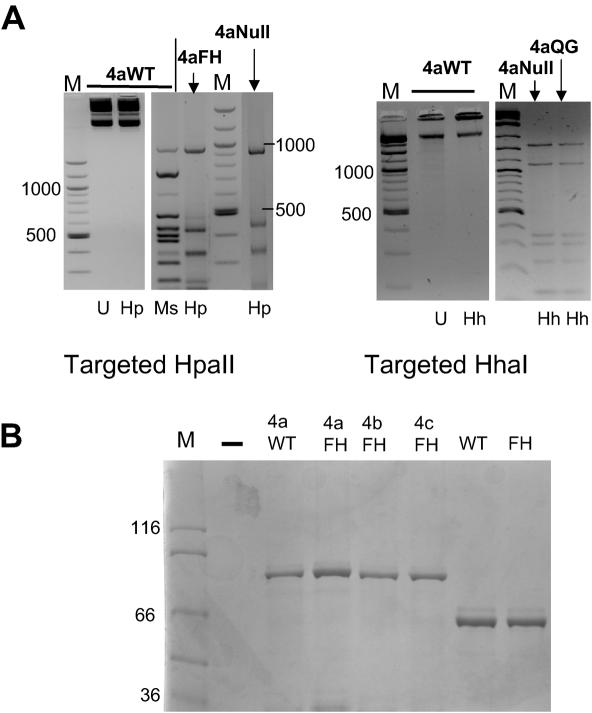
We have previously shown that three zinc-finger–HpaII and HhaI wild-type enzymes bind preferentially in vitro to a target site composed of adjacent zinc-finger and Mtase recognition sites. However, these enzymes also displayed significant non-specific methyltransferase activity, not immediately apparent in in vitro studies, but noticeable when these enzymes were expressed in E.coli (18). Previously we have demonstrated enhanced binding attributes of an array-extended four zinc-finger derivative (henceforth designated 4a) of the progenitor three zinc-finger protein component of this enzyme, which recognizes the sequence 5′-GACGCAGAAGCC-3′ (19,21). We therefore initially examined whether this four zinc-finger component would enhance targeted methylation by wild-type Mtase enzymes and reduce non-specific background methylation. An initial assay of the 4aWT HpaII and 4aWT HhaI Mtase enzymes reveal that the coding vectors are methylated when isolated from bacteria, as judged by insensitivity to HpaII or HhaI restriction, respectively (Figure 1A, left-hand side of each panel), despite having no zinc-finger recognition site within the plasmid sequences.
Figure 1.
(A) Methylation sensitive restriction analysis of expression plasmids coding for wild-type and mutant targeted methyltransferases. Expression plasmids coding for 4aWT, 4aFH and 4aNull HpaII based enzymes and 4aWT, 4aQG and 4aNull HhaI based enzymes were isolated from E.coli and subjected to restriction with methylation sensitive and/or insensitive isoschizomer restriction enzymes (left and right panels). The middle panel shows methylation-sensitive restriction digestion of higher copy number GST fusion expression plasmids coding for HpaII wild-type and FH mutants and as fusions with the 4a zinc-finger gene. Digestion products are shown. U, uncut; Hp, HpaII; Ms, MspI; Hh, HhaI restriction enzymes. (B) SDS–PAGE analysis of purified recombinant zinc-finger–Mtase fusion proteins, used for in vitro binding and methylation specificity studies.
Mutagenesis and zinc-finger arrays
In order to overcome the high levels of non-targeted methylation observed for wild-type-based enzymes generally, we introduced mutations to the Mtase component of the fusion enzymes. The rationale for this was that either a reduction of the catalytic activity or binding affinity of the Mtase component would allow the zinc-finger component to dominate protein–DNA interactions, giving lower levels of background (non-targeted) methylation. A highly conserved sequence homology exists between cytosine 5-Mtases, located within 10 specific homology blocks (23) and the contributions made by these conserved elements have been largely elucidated, especially for the HhaI Mtase (24–29). For example, F18 in the conserved element F18xGxG in M.HhaI helps to position the adenine ring of the co-factor S-adenosylmethionine in the protein binding pocket, while residue Q237, which surprisingly is not conserved between cytosine Mtases, helps to stabilize the protein–DNA structure by taking the place of the extruded cytosine base. Such mutational analyses have provided the basis of a rational mutagenesis approach for the enhancement of targeted Mtase function, focused predominantly on targeted HpaII F35H (corresponding to F18 in HhaI) and HhaIQ237 to glycine (G), valine (V) and isoleucine (I) mutant Mtase enzymes.
An initial assessment of the plasmids coding for the 4aF35H HpaII (henceforth referred to as 4aFH) and 4aQ237G HhaI (henceforth referred to as 4aQG) Mtase proteins revealed that they were digested completely by HpaII and HhaI restriction enzymes, respectively, giving patterns similar to plasmids coding for null activity enzymes (Figure 1A). This was therefore in stark contrast to the respective targeted wild-type enzymes and provided an initial indication that mutant enzymes were not functional in the absence of a zinc-finger recognition site. In addition to mutagenesis of the Mtase component, two additional four zinc-finger proteins and a six zinc-finger protein were constructed with varying specificities, designed using published zinc-finger–DNA recognition database information (see Materials and Methods for details). These zinc-finger proteins were constructed both to test zinc-finger recognition code validity and to generate targeted Mtase enzymes with differing affinities for their target sites, so that the role of zinc-finger DNA binding in targeted methyltransferase specificity could be further assessed.
DNA-binding and methylation assays
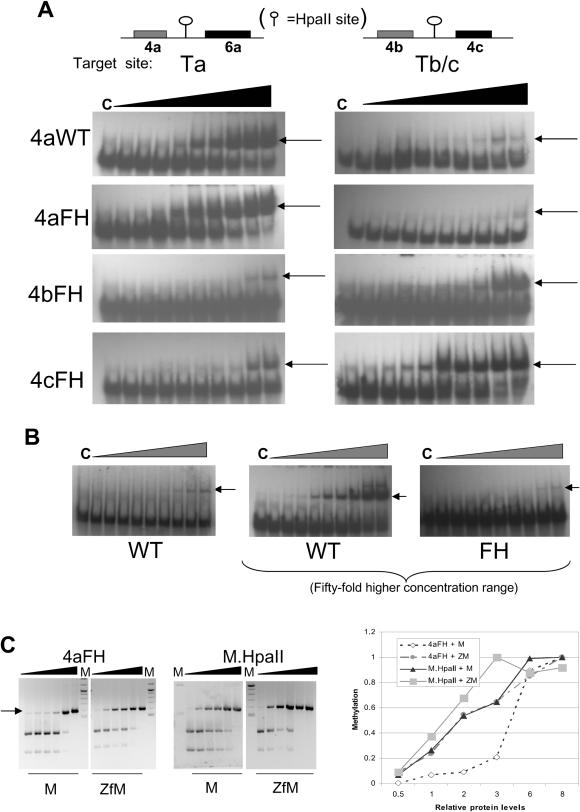
Initial studies focused on purified GST fusion proteins comprising various 4Zf proteins, fused to the HpaII FH Mtase mutant. In addition to the 4a zinc-finger protein, 4b and 4c zinc-finger proteins were designed to recognize the sequences 5′–3′ GACGCGGCGCTC and TGGGGAGTGGCG, respectively. Purified zinc-finger HpaII Mtase proteins (see Figure 1B) were used in gel shift assays to assess the relative affinities of the zinc-finger component of each fusion protein. The results of titrative EMSAs are shown in Figure 2A and B. Probe Ta comprises the 4a zinc-finger and HpaII recognition sites, separated by 14 bp. Probe Tb/c comprises a single HpaII site flanked on either side by a site for the 4b and 4c zinc-finger proteins, respectively. Results confirm high-affinity binding of each fusion protein only to probes harbouring the relevant recognition site, but not to probes harbouring just the Mtase site, which bound weakly. Analysis of these binding data revealed that the 4bFH Mtase protein binds most weakly to its target site (apparent Kd ≈ 38 nM); the 4cFH Mtase protein binds most strongly (≈3 nM). The 4aFH Mtase bound to its target site with intermediate strength (Kd ≈ 11 nM) and similar to the 4a wild-type paII enzyme (≈9 nM). All enzymes bound to just HpaII site-containing probe with a very low affinity that was not quantifiable under the conditions used, compared with that seen for target site probe. Taken together, these results confirm previous observations made for three zinc-finger Mtase fusions binding to target oligonucleotides that binding is dominated by zinc-finger–DNA interactions (18). Non-zinc-finger fusion wild-type HpaII (Figure 2B, left-hand panel) bound significantly more weakly to its target site (Kd ≈ 170 nM, to HpaII DNA) than the targeted enzymes to their target sites, but only slightly stronger to that seen for targeted enzymes binding to HpaII site DNA. Much higher protein levels were also used in this assay so that non-zinc-finger wild-type and FH mutant protein binding could be directly compared (Figure 2B, middle and right-hand panels). Binding by the non-zinc-finger FH mutant was considerably weaker than that seen for the wild-type non-zinc-finger enzyme and could not be quantified. It was also significantly weaker than the observed binding of 4aFH to HpaII site probe, with similar levels of complex formation observed at non-fusion FH protein levels 50 fold higher than that of the targeted enzyme, suggesting that the fusion protein as a whole has an enhanced, albeit relatively weak, non-specific interaction with DNA compared with just the Mtase component alone. As we have noted before, however, differences in the activity of these enzymes when expressed in bacteria, for example (see Figure 1A), do not generally reflect the EMSA-binding data, either as a consequence of differences in catalytic rate and processive activity once bound to DNA or due to local concentration and time-scale effects.
Figure 2.
(A) EMSA analysis of purified recombinant four zinc-finger–HpaII (F35H) mutant Mtase fusion proteins (the 4a, 4b and 4c zinc-finger components of each protein are described in the text), compared with recombinant non-zinc-finger fusion M.HpaII wild-type and FH proteins. Recombinant protein was titrated against either a target site or non-target site containing probe, at 0.1, 0.3, 0.5, 1, 3, 5, 10, 30 and 50 fmol. Lane C is probe only. The target sites are given under the relevant panels. (B) Binding of M.HpaII wild-type protein to HpaII site DNA at equivalent concentration range to (A) is shown in the left-hand panel. Non-zinc-finger fusion M.HpaII wild-type and FH proteins were also used at 50-fold higher levels, so that some comparative evidence of binding by the HpaII FH protein to HpaII site DNA could be seen (middle and right-hand panels). (The probe used was Tb/c). (C) Methylation analysis of 4aFH versus M.HpaII protein for target and non-target site containing DNA fragments. Increasing amounts of each protein (25, 50, 100, 150, 250 and 350 fmol) were incubated with DNA fragments (1.0 μg) containing just an HpaII site (M), or an HpaII site and flanking 4a zinc-finger recognition site (ZfM). Following incubation, the reaction was stopped and DNA subjected to restriction with R.HpaII and products resolved by electrophoresis. The intensity of the uncut band was measured for each lane (AIDA software, raytest, GmbH) and normalized against the end point intensity. These data are presented in graphical form in the right-hand panel.
The methyltransferase characteristics of these proteins were examined using in vitro and in vivo assays. Initial analysis focused on the ability of the 4aFH protein to specifically methylate at a simple target site, in comparison with the non-targeted wild-type enzyme. Incubation of increasing amounts of 4aFH Mtase or non-targeted M.HpaII enzyme (NEB) with ZfM or M DNA, followed by a HpaII restriction enzyme digest of these fragments, revealed the relative Mtase activity of each protein for these different substrates. The results are shown in Figure 2C. The non-targeted HpaII wild-type enzyme was, as expected, unable to discriminate between targeted and non-targeted HpaII site containing DNA fragments, as evidenced by the similar resistance of these DNA fragments to cleavage, as Mtase protein levels are increased. The 4aFH Mtase enzyme, however, clearly shows a preference for DNA which contains both zinc-finger and adjacent HpaII sites, compared with DNA containing just a HpaII site, denoted by the relatively early appearance of DNA resistant to HpaII cleavage (arrowed in Figure 2C). Presentation of these data in graphical form confirms similar gradients for the non-targeted wild-type enzyme binding to either DNA fragment. A similar curve is seen for 4aFH methylation of target site DNA. Methylation of DNA containing just a HpaII site by this enzyme was however significantly lower at lower levels of enzyme concentration, but with a rapid increase in methylation at higher protein concentrations.
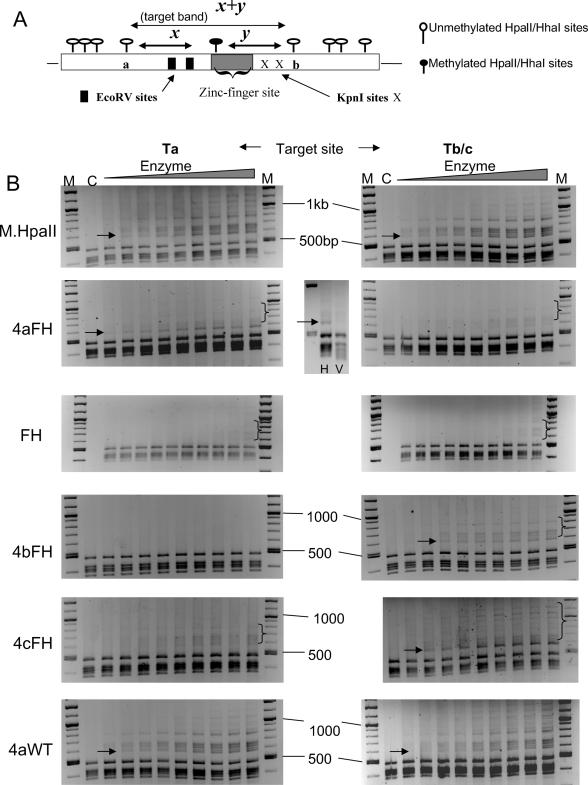
We next examined the ability of purified proteins to methylate at a targeted site against a complex background of HpaII sites, using a plasmid-based assay. The plasmids used contained a unique targetable sequence, comprising adjacent zinc-finger and HpaII recognition sites, based essentially on the Ta and Tb/c probes used previously. An additional 23 ‘background’ HpaII sites exist on these plasmids within a 3.9 kb sequence and thus presents a significant test of targeting by these enzymes. Methylation at the target site, followed by HpaII restriction of the plasmid, should generate a unique restriction pattern indicative of targeted methylation and give rise to a dominant unique band of 560 bp in size (see Figure 3A for schematic description of this rationale). The results of this assay are shown in Figure 3B. Purified four zinc-finger FH and 4aWT HpaII Mtase proteins, as well as non-targeted HpaII wild-type Mtase protein were incubated at increasing concentrations with plasmids harbouring a targetable HpaII site either for the 4aFH Mtase (plasmid Ta, left panels) or 4bFH and 4cFH Mtase proteins (plasmid Tb/c, right panels). After the reaction was stopped, the plasmids were restricted with rHpaII. Immediately apparent in some instances is the appearance of a dominant intense band which is of the predicted size, shown using arrows in the relevant panels. This occurs only when the plasmid contains the appropriate target recognition site. This is most noticeable for the 4cFH and 4aFH protein which also show low background methylation. The 4bFH enzyme gives the weakest evidence of targeted methylation and displays a non-specific background methylation similar to that seen for targeted and non-targeted wild-type enzyme. Generally, all the targeted FH proteins displayed an approximately equivalent low level of non-specific methylation in the absence of a target site. This was more apparent when higher concentrations of protein were used but again, was proportionally and significantly less than that seen for the targeted wild-type enzyme (data not shown). Confirmation that the 560 bp band was indeed derived from targeted methylation was shown by further restriction of plasmids with EcoRV. This site is uniquely present immediately flanking the zinc-finger recognition sites in each plasmid (see Figure 3A) and the targeted methylation-derived band is cleaved as expected (shown for the 4aFH protein in Figure 3, central panel). The identity of this band was also initially confirmed by sequencing. Significantly, the 4aWT protein gave a methylation profile similar to that of non-targeted HpaII enzyme under these conditions, although a small degree of targeted methylation was observed against a high background of non-specific events.
Figure 3.
(A) Schematic diagram of the basis for plasmid-based and in situ bacterial assays for targeted methylation. A detailed diagram of the target site (which is embedded in the target site vector) is shown. Following either incubation of plasmid with recombinant targeted methyltransferases or isolation from bacteria co-transformed with expression and target site vectors, plasmids were subsequently restricted with either HhaI or HpaII restriction enzyme and the digest products were analysed on an agarose gel. In the absence of any methylation, restriction occurs at all sites and a standard fragment pattern is generated, which includes fragments of x and y bp in size derived from cleavage at a, b and the target site, indicated by a filled lollipop. Where targeted methylation has occurred, restriction at that site is blocked, thus modulating the normal restriction pattern by the generation of a unique band of predicted length (designated the target band), of size (x + y) bp as indicated in the figure. As further confirmation, this ‘target fragment’ should restrict at the EcoRV restriction sites within the target region. (B) Plasmid-based methylation analysis of targeted and non-targeted HpaII wild-type and FH mutant proteins. Linearly increasing amounts of purified protein (20–100 fmol, in 10 fmol increments) were incubated with either target site or non-target site plasmids (1–2 μg). After HpaII cleavage, the restriction products were analysed by gel electrophoresis. Arrows denote the appearance (or expected location for wild-type enzyme panels) of a DNA fragment corresponding to 560 bp in size [(x + y) fragment in Figure 1B], denoting targeted methylation. M is a DNA marker mix (NEB, 100 bp + 1 kb ladders). Lane C represents DNA digested in the absence of Mtase incubation. Plasmids were additionally restricted with EcoRV to confirm the identity of the 560 bp fragment. A typical result is shown for the 4aFH protein (centre panel).
Bacterial in vivo analysis of targeted HpaII enzymes
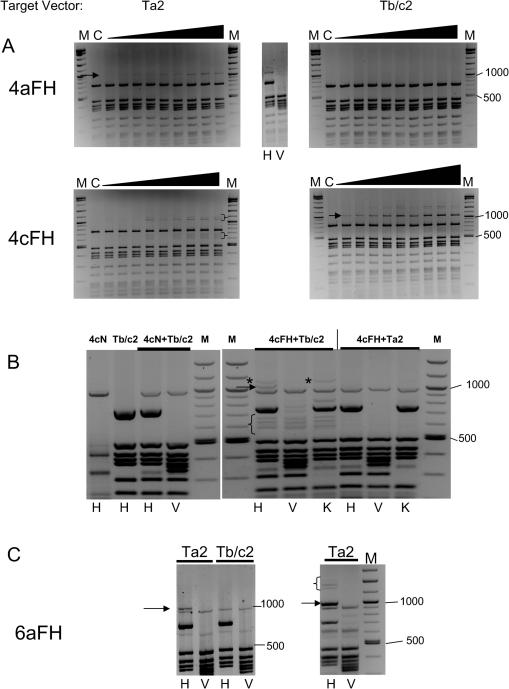
The ability of mutant enzymes to specifically methylate at a single targetable HpaII site on a plasmid in vitro, coupled with the observation that respective expression vectors were not methylated when isolated from E.coli, suggested to us that we could develop an in vivo bacterial-based assay for the detection of targeted methylation by this class of enzyme. The assay comprises co-transformation of E.coli with a compatible expression vector harbouring the targeted methylase protein and the relevant target site vector, followed by plasmid isolation and restriction with the relevant methylation-sensitive restriction enzyme. As in the case with in vitro assays, a unique band should similarly appear indicative of targeted methylation. In order to enable visualization of the targeted methylation-derived restriction fragment against the expected background of cleavage products from both expression and target site vectors, new target site vectors were constructed. These were designed to produce larger DNA fragments as a consequence of targeted methylation and subsequent restriction (designated Ta2 and Tb/c2). Preliminary in vitro methylation analysis using recombinant purified 4aFH and 4cFH proteins confirmed that both new target vectors were specifically recognized by the respective protein (see Figure 4A). A unique fragment of ∼1060 bp appears as expected (shown using arrows in Figure 4A, target vector panels), and is destroyed by combined HpaII and EcoRV restriction (shown for 4aFH, top-middle panel in Figure 4A). A low level of non-specific methylation is observed for the 4cFH protein with non-target site vector, which is shown in brackets (panel Ta2, Figure 4A).
Figure 4.
(A) Plasmid-based methylation analysis of targeted and non-targeted HpaII wild-type and FH mutant proteins for ‘large fragment’ target vector. Increasing amounts of purified 4aFH and 4cFH protein (as described in Figure 3) were incubated with target site or non-target site plasmids. After HpaII cleavage, the restriction products were analysed via gel electrophoresis. Arrows denote the appearance of a DNA fragment corresponding to 1060 bp in size, denoting targeted methylation. M is a DNA marker described previously. Bands derived from non-specific methylation are shown using brackets. (B) Bacterial-based in vivo analysis of targeted methylation by 4cFH protein. Details are described in Materials and Methods. The left-hand panel shows the HpaII restriction profile of the 4cNull and target vectors alone, and after co-isolation from bacteria, for comparative purposes. The right-hand panel shows the restriction profile of 4cFH after co-isolation with either target (Tb/c2) or non-target (Ta2) vector. The band indicative of targeted methylation is arrowed. Low molecular weight target vector associated products are shown using brackets. A higher molecular weight product, just above the target band, resistant to KpnI cleavage, is shown using asterisks. H, HpaII; V, HpaII + EcoRV; K, HpaII + KpnI restriction in all cases. (C) Bacterial-based in vivo analysis of targeted methylation by 6aFH protein. Experimental details are essentially as in (B) above. The 1060 bp fragment indicative of targeted methylation is arrowed in the figure. The right-hand panel shows the results of digestion of plasmids isolated from culture grown from 3-day-old bacterial colonies. The 1060 bp target band is arrowed. Higher molecular weight bands related to the targeted methylation event are shown using brackets.
Bacterial in vivo evaluation of this new vector system was performed, initially using the 4cFH protein, which consistently gave good levels of targeted methylation in in vitro assays. Following co-transformation with expression and target or non-target vectors, bacteria were grown overnight and plasmids were isolated and digested with rHpaII and in concert with EcoRV. Control experiments involved use of expression vectors coding for the null activity targeted Mtases which retain the same Mtase (and hence restriction) site distribution as functional enzyme coding vectors. When 4cFH was expressed in the presence of the wrong target site vector (Ta2), the same restriction pattern is seen as with the null activity mutant (Figure 4B, lanes 4cN+Ta2 versus 4cFH+Ta2). Expression with the correct target site vector (Tb/c2), however, produces the expected band (arrowed), which is further shown to be destroyed by EcoRV. An additional higher molecular weight band, only seen with the target site vector, is shown using asterisks in Figure 4B and fails to disappear upon additional cleavage with KpnI. This band is derived from methylation at the most proximal 5′ HpaII site to the target region (site ‘a’ in Figure 3A), but with no methylation occurring at the target site itself. A cluster of low molecular weight bands were also observed, but again only in the presence of the target site vector, and persisted after additional restriction (shown using brackets in Figure 4B).
As a further test of this system, we analysed a six zinc-finger FH Mtase fusion protein, 6aFH, the targeting component of which was a tandem repeat of the Zif268 protein, linked together by the canonical TGEKP linker sequence previously described to be effective in enlarging zinc-finger arrays (30). Such Zif268-based six zinc-finger proteins have been shown to bind their recognition sequence with high affinity [Kds in the pM/fM range (30,31)], whereas four zinc-finger proteins generally have binding constants in the nM range (19,21). The target site for this six zinc-finger protein was 5′-cgcccacgctcgcccacgc-3′, the underlined base separating the two original zif268 recognition sites. We had not been able to purify a sufficient amount of this protein for in vitro analysis, due largely to solubility issues, and thus this protein was an ideal candidate for evaluation by the ‘in vivo’ system. Figure 4C shows the results of this assay. In this instance, the unique band indicative of targeted methylation is strong and seen only with the target site vector (Ta2). This band is destroyed by additional EcoRV restriction. Interestingly, the intensity of the target band increases when plasmids derived from older bacterial cultures were examined (shown using arrow in Figure 4C, right panel). Additional higher molecular weight bands were also found, which also disappear upon cleavage with EcoRV and are indicative of possible processive action from the target site (shown using bracket in Figure 4C, right-hand panel). A similar effect was seen when freshly co-transformed cells were grown under conditions in which expression of targeted enzyme was induced (data not shown).
Bacterial in vivo analysis of targeted HhaI enzymes
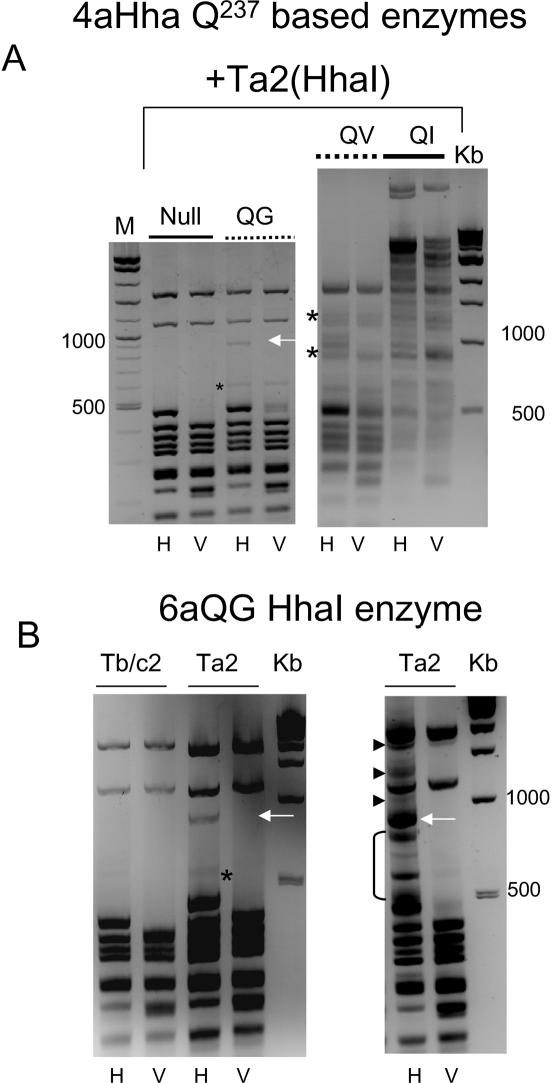
We further used the bacterial in vivo system to evaluate the 4aHhaI Q237 to G/V and I series of mutants for their relative abilities to methylate specifically at the target site. The results of this assay are shown in Figure 5A and suggest that only the QG mutant gives specific targeted methylation under these conditions. The unique band is shown using an arrow (left panel), and destroyed upon additional EcoRV restriction as expected. The QV, and QI mutants are less targeted in their specificity, giving rise to an increasing background of non-specific bands, respectively (non-specific methylation-derived bands are shown using asterisks for the QV mutant), although for the QV enzyme there is evidence of increased targeted methylation. The 6aQG mutant gave evidence of targeted methylation similar to that seen for 4aQG (see band arrowed in Figure 5B, left panel). Transiently increasing the expression vector copy number relative to the target vector (using DNA obtained directly from freshly transformed bacteria before liquid culture) resulted in an increase in intensity of the target band, as well as the generation of additional higher molecular weight bands (short arrows in Figure 5B, right panel) that disappeared upon further cleavage with EcoRV, again indicative of processive Mtase action.
Figure 5.
Methylation-sensitive restriction analysis of plasmids isolated from bacteria transformed with 4aHhaI or 6aHhaI expression vectors and target vector [Ta2(HhaI)]. Experimental details are essentially as in Figure 4B. (A) Restriction analysis of 4aQ237HhaI mutant or null vectors and 4aHhaI-specific target vector isolated from co-transformed bacteria. The 940 bp band denoting targeted methylation for 4aQG is shown using an arrow. Evidence of non-specific methylation by the 4QV mutant is shown using asterisks. (B) Left-hand panel shows the results of the bacterial assay described above, for the 6aQG protein. The band denoting targeted methylation is indicated (white arrow). The non-specific band is shown using an asterisk. The right-hand panel shows the effects of a transient increase in 6aQG expression vector copy number on the intensity of the 940 bp band and the appearance of higher molecular weight, targeted methylation related bands that disappear upon EcoRV cleavage (small arrows). Low molecular weight non-specific bands are shown using bracket. H, HhaI; V, HhaI + EcoRV restriction, throughout this figure. M is marker mix, Kb is 1 kb ladder (NEB).
Effects of targeted methyltransferase expression on genomic DNA methylation status
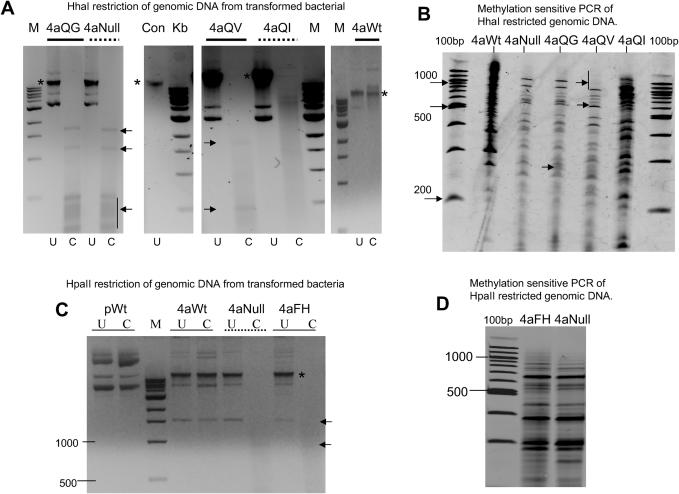
Genomic DNA isolated from bacteria expressing various targeted Mtase enzymes was digested with appropriate methylation-sensitive restriction enzymes (shown in Figure 6A and C). The results of arbitrary primed PCR (AP-PCR) analysis of the digest products are shown in Figure 6B and D. The methodology of combining methylation-sensitive restriction digest followed by non-specific PCR of the digest product is an approach often used for analysing subtle differences between methylation patterns of different tissue and cancer types (32,33). Targeted HhaI mutants demonstrated the same broad range of functional activity on genomic DNA as they did from plasmid restriction assays, although perhaps less marked (Figure 6A). The trend of increasing non-specific Mtase activity of these enzymes is mirrored in the AP-PCR profiles obtained, which results in differences in product levels and pattern generated from equivalent quantities of template. For example, the 4aNull and 4aQG PCR profiles are broadly similar in terms of band intensities and pattern, whereas the 4aQV and more noticeably the 4aQI profiles, which showed moderate and high levels of non-specific activity, respectively, in bacterial assays (see Figure 5A), are markedly different (compare 4aNull and 4aQG lanes with 4aQV and 4aQI and especially 4aWt lanes in Figure 6B). Genomic DNA isolated from bacteria expressing the 4aFH HpaII mutant appeared fully restricted (Figure 6C). The AP-PCR profile between the 4aNull (HpaII) and 4aFH enzyme were similar (Figure 6D), although the intensities of certain bands varied between the two samples. These small differences were of about the same levels as seen between the HhaI 4aNull and 4aQG experiments, rather than the larger differences seen for the QV or QI mutants. This again suggests that little non-specific genomic methylation had occurred for these targeted enzymes.
Figure 6.
Analysis of genomic DNA from bacteria expressing targeted methyltransferase enzymes. (A) Restriction analysis of genomic DNA recovered from bacteria expressing various 4aHhaI enzyme derivatives (and containing the target site vector), described above the relevant lanes. The lane denoted ‘Con’ represents genomic DNA isolated from untransformed bacteria. U, uncut; C, cut with HhaI restriction enzyme. (Fragments seen in some ‘cut’ lanes are derived from plasmid carryover during genomic DNA preparation, shown using arrows. Input genomic DNA is shown using asterisks.) (B) Arbitrary primed PCR of the HhaI restricted genomic DNA described in (A). (C) Restriction analysis of genomic DNA recovered from bacteria expressing various 4aHpaII enzyme derivatives, described above the relevant lanes. pWt is plasmid DNA coding for the wild-type targeted enzyme. U, uncut, C, cut with HpaII. (D) Arbitrary primed PCR of HpaII restricted genomic DNA described in (C). M is 1 kb ladder (NEB), 100bp is 100 bp ladder (NEB).
Targeted methylation of an integrated sequence in HeLa cells
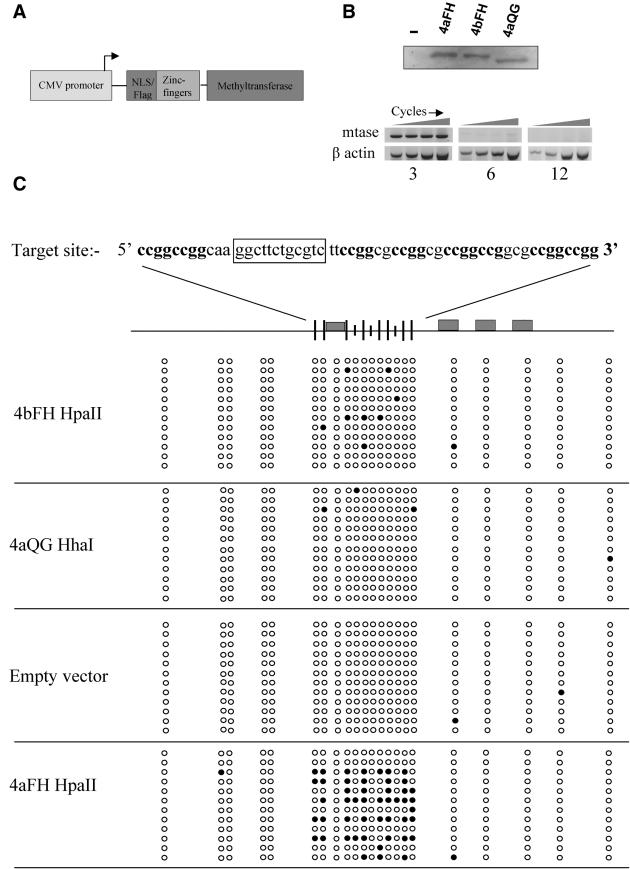
Having generated enzymes which produce only very low levels of non-specific methylation against a relatively complex bacterial genome, we next employed these enzymes to deliver de novo methylation to a specific locus in mammalian cells. We constructed a cell line harbouring a stably integrated plasmid which comprised 4aZf recognition sites adjacent to eight HpaII and three HhaI sites (see Figure 7C). We have previously shown that this four zinc-finger protein as a fusion with VP16 is able to bind chromatin in mammalian cells and upregulate transcription from promoters harbouring its recognition site (21). We elected to employ a short, defined target region containing a low number of methylatable sites so that the likelihood of any endogenous de novo Mtase action and subsequent cellular response would be reduced, thus allowing a ‘clean’ assessment of methylation by the transiently expressed targeted enzymes, but providing us with the opportunity to monitor maintenance and spread of methylation to flanking CG sequences by endogenous enzymes over time. The target cell line was transfected with either empty expression vector, 4aFH or 4aQG expression vectors, or a vector expressing the 4bFH protein, which does not recognize the target site (see Figure 7A for general vector schematic and 7B, upper panel, for protein expression). Western analysis indicated that the expression of targeted Mtase enzymes was undetectable at day 6 (data not shown). More sensitive RT–PCR analysis confirmed complete loss of Mtase expression at 12 days post-transfection. An approximate 95% reduction in targeted Mtase RNA levels was observed at only 6 days post-transfection, suggesting that cellular mechanisms were responsible for perpetuating the de novo methylation pattern at this stage (Figure 7B, lower panel). Bisulphite sequencing of the genomic DNA isolated from each experiment 14 days post-transfection is shown in Figure 7C. Cells transfected with empty vector did not become significantly methylated over the course of the experiment, confirming that the CpG sequences associated with the target site were not subject to methylation by the endogenous methyltransferases. We were therefore able to eliminate contributions from endogenous de novo activity in our assessment of targeted methylation.
Figure 7.
(A) Schematic diagram describing mammalian vectors used for the expression of targeted Mtases. (B) Upper panel showing western blot confirming expression levels of 4aFH, 4bFH and 4aQC targeted Mtase enzymes from vector described in (A). Lower panel showing the results of RT–PCR of 4aZfFH RNA at 3, 6 and 12 days post-transfection with 4aZfFH targeted Mtase expression vector. β-Actin was used an internal control. PCR was performed for 28, 30, 32 and 34 cycles. (C) Bisulphite sequencing results for genomic target site DNA, 14 days post-transient transfection with targeted Mtase or control vector. The target site sequence is given at the top of the figure. HpaII sites are in boldface and the 12 bp zinc-finger recognition site is boxed. Below this is a schematic diagram showing target site and flanking CpG site distribution (represented by circles in sequencing data) spanning ∼160 bp. Large ticks indicate HpaII sites, small ticks indicate HhaI sites. Zinc-finger recognition sites for the 4aZf protein are represented by filled rectangles. The expression vector transiently transfected in each experiment is indicated next to each sequencing results block. Filled circles indicate observed methylation.
Methylation was only observed to a significant extent when the 4aFH enzyme was expressed in these cells. Only 3 of the 12 clones sequenced for this experiment were devoid of methylation, 7 of the remaining 9 were methylated extensively. Significantly, the pattern of methylation found within the target site varies, confirming that our analysis was derived from a number of different PCR clones. Most methylation was observed 3′ to the zinc-finger recognition site, a result not unexpected due to the orientation of the zinc-finger motif when bound to DNA and to the Mtase being fused at the C-terminus. A few non-HpaII sites were observed to be methylated within the target site and at some CpG sites in flanking regions, suggesting a low level of de novo methylation by endogenous enzymes. A low density and sporadic pattern of methylation was observed with the 4bFH enzyme, predominantly at HpaII sites. However, the level of methylation observed was significantly lower than that seen for 4aFH, confirming zinc-finger interaction with DNA as the dominant targeting event for the 4aFH enzyme.
DISCUSSION
Initial analysis showed that wild-type methyltransferases are not targetable, producing unacceptably high levels of background methylation when expressed as fusions with the zinc-finger proteins used in this study. Binding analysis of a series of different zinc-finger proteins fused with the HpaII FH mutant showed the expected recognition specificity as well as variations in affinity for their respective sites. This represents a general confirmation of the accuracy of current zinc-finger DNA recognition code information.
Significantly, comparative analysis of the targeted 4aFH and non-targeted M.HpaII methyltransferase enzymes for methylating DNA fragments containing either single HpaII sites or with a flanking zinc-finger recognition site, revealed that the 4aFH protein was largely target-site specific. Its interaction with DNA was dominated by the zinc-finger component, whereas the M.HpaII enzyme methylated all DNA sequences equally. The degree of methylation achieved by the 4aFH enzyme for target site DNA was also roughly equivalent to that generally seen for the non-targeted wild-type enzyme, suggesting that the lower catalytic affinity of the 4aFH Mtase component is being compensated for by targeting of the Mtase at or near the targeted HpaII site, to the extent that non-targeted methylation is virtually zero for the 4aFH enzyme under conditions where targeted methylation is relatively strong. As we have observed previously, however, at excessive protein concentrations non-specific Mtase activity is observed to some degree for all enzymes studied, either as a consequence of increased non-specific zinc-finger–DNA interactions (21), or due to high local protein concentrations that compensate for the low activity/affinity attributes of the Mtase component. It should be noted however that targeted mutant enzymes can be used at much higher concentrations than targeted wild-type enzymes, before non-targeted methylation becomes an issue.
Methylation analyses of more complex sequences, where the target site represented only 4% of all HpaII sites present, revealed a trend similar to that seen in gel shift assays. The most significant aspects of this assay are the observation of specific targeted methylation in all cases, albeit variable, except for the M.HpaII and 4aHpaII WT enzymes. There was also a general lack of significant non-specific methylation when the non-target site vector was used, again except for M.HpaII and 4aWT enzymes. When the target site vector was used, however, all targeted enzymes displayed evidence of low methylation at a number of other sites to varying degrees, implying such methylation is derived from the initial targeted binding event in each case. Whether this is due to plasmid topology, for example, or due to variable effects of zinc-finger–DNA-binding affinity on methyltransferase procession and/or dissociation is unclear at this time.
Evaluation of the 4cFH enzyme using the bacterial in vivo assay system confirmed the targeted methylation also observed in in vitro plasmid-based assays. The increased intensity of non-specific methylation of target site vector relative to the ‘target band’ and to that seen in in vitro results again suggests that plasmid topology may play a role in bringing the targeted enzyme closer to non-targeted HpaII sites, similar to the effects of nucleosome formation in mammalian cells. Evidence for targeted methylation-derived events that did not include methylation at the target site itself was also seen for the 4cFH enzyme, as evidenced by the appearance of a slightly higher molecular weight band than the target band, which was cleaved by EcoRV, but not cut by KpnI. This effect was not seen in in vitro assays, again suggesting that plasmid topology may be a factor in the methylation of these vectors in vivo.
Similar in vivo analysis of the 6aFH protein in bacteria revealed a high degree of targeting with no real evidence of non-targeted methylation. Analysis of the longer term effects of protein expression revealed that targeted methylation had become more pronounced, with evidence of procession of Mtase activity from the target site to the nearest flanking HpaII sites. This suggests that even with the expected build up of protein within the cell over time, it still falls within the range that allows monofunctionality of the enzymes. Evidence of processive activity seemed less pronounced for the 6aFH than four finger derivatives which were also studied (data not shown), presumably due to tighter binding of the former protein to its recognition sequence. Therefore for targeted methylation purposes, it could be argued that four zinc-finger derivatives would be more suitable for targeted promoter methylation in mammalian cells, due to a relatively higher processivity away from the initial binding site.
The general attributes of all the targeted FH mutants examined in this study, correlate with studies of the F18H (homology and functionally related) mutation in M.HhaI, which demonstrated a reduced catalytic activity for this enzyme relative to the wild type, although no quantitative assessment was made in that instance (27).
Analysis of the 4aHhaI mutants in bacterial assays demonstrated variable levels of targeted methylation. Targeted methylation by the 6aQG mutant was also evident. Similarly to the time-dependent 6aFH HpaII action, evidence of processive methyltransferases activity from the initial binding site was observed for this enzyme under conditions of transiently increased protein concentration. While targeted methylation was also seen to increase significantly, background methylation rose proportionally. It therefore seems that as a general rule, duration and level of expression of targeted methyltransferases in the cell may be key factors in controlling methylation delivery, non-specific action and potential processivity.
Variability in the activity of HhaI Q237 mutants has been observed previously for the M.HhaI protein and our choice of mutants for this particular enzyme was largely based on such studies. Methylation analysis of Q237G/V and I mutants revealed that these proteins had catalytic activities of 2.6%, 1.3% and 2.5%, respectively, of that of the wild-type enzyme (28). Binding analysis in the same study revealed that the Q237G/V and I mutants bound to oligonucleotide substrates with 0.1%, 0.1% and 3% of wild-type enzyme binding affinity, respectively. It has also recently been shown that Q237 may be involved in active flipping of the target cytosine from the DNA helix into the catalytic binding pocket of the protein (34). In this study, substitutions of Q237 with G and A amino acids, all bound DNA, but no base flipping was observed. The QG mutant enzyme however does possess a low but demonstrable methyltransferase activity in our hands. This might suggest that base flipping is actually still occurring, but at a much slower rate than normal, and it is only the anchoring of the methylase at or near its recognition site due to zinc-finger tethering that allows the methyltransfer reaction to proceed at a moderate level, before the enzyme might ordinarily dissociate. This is corroborated to some extent by the observation of a dramatically reduced exchange of hydrogen atoms on the target cytosine base with protons from water when DNA is bound by the Q237G mutant (34).
Analysis of the effects of expression of various targeted Mtases on the bacterial genome mirrors the activities seen in in vivo bacterial-based plasmid assays, suggesting that the latter method is sufficient on its own to determine the efficacy of these enzymes.
Expression of the targeted 4aFH enzyme in mammalian cells resulted in specific methylation of an integrated 4aZf-specific target site at a significantly higher level than that seen for the control 4bFH enzyme, which was only a little above the background level in these experiments. Given that both 4aFH and 4bFH Mtases are broadly similar in their non-specific methyltransferase activity, differences between the two methylation patterns observed at the target site can be attributed directly to zinc-finger targeting. A strong methylation signal is maintained 14 days post-transient transfection with the 4aFH vector, strongly suggesting that the de novo methylation is being maintained by the cell, in the absence of a measurable targeted Mtase presence. It is perhaps surprising that no significant methylation spread was observed for this experiment, given the existence of flanking CpG sites around the target sequence and the action of the endogenous Dnmt1 maintenance Mtase in maintaining and possibly expanding methylation signals through replication. However, this may simply reflect a time issue; since, for example, in patients with myelodysplastic syndrome (MDS), where an initial low density of methylation associated with tumour suppressor gene promoters is observed, higher density methylation attributed to methylation spread is only seen with disease progression over relatively long time scales (35). This may be similar to the gradual process of gene silencing seen for the mouse Aprt gene (36).
The 4aQG enzyme's inability to deliver methylation to the target site in mammalian cells is not surprising, given that this enzyme was not very active in bacterial assays. The activity associated with this enzyme may represent a threshold level, only above which will these enzymes function to an observable extent in mammalian cells and as such may serve as a useful reference point for further studies.
In summary, we have developed enzymes in which the dominant interaction with DNA is clearly driven by the zinc-finger component. Moreover, the combination of strong zinc-finger binding and reduced methyltransferase activity gained from rational mutagenesis, has allowed us to target methylation predominantly to specific DNA sequences without incurring significant background methylation. These enzymes are fully functional in mammalian cells, where the targeted methylation pattern delivered is demonstrated to be inherited through successive cell divisions. The development of targeted Mtase enzymes and methodologies to analyse their function rapidly and within a relatively complex model system, together with the further characterization of their functional activity in vivo in mammalian cells, represents an important step towards the ultimate employment of this class of molecule in both research and therapeutic arenas.
Acknowledgments
Funding to pay the Open Access publication charges for this article was provided by King's College London, Department of Haematological and Molecular Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Nakao M. Epigenetics: interaction of DNA methylation and chromatin. Gene. 2001;278:25–31. doi: 10.1016/s0378-1119(01)00721-1. [DOI] [PubMed] [Google Scholar]
- 2.Jones P.L., Veenstra G.J., Wade P.A., Vermaak D., Kass S.U., Landsberger N., Strouboulis J., Wolffe A.P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 3.Wade P.A., Gegonne A., Jones P.L., Ballestar E., Aubry F., Wolffe A.P. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nature Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- 4.Robertson K.D., Ait-Si-Ali S., Yokochi T., Wade P.A., Jones P.L., Wolffe A.P. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nature Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 5.Rountree M.R., Bachman K.E., Baylin S.B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nature Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- 6.Lorincz M.C., Dickerson D.R., Schmitt M., Groudine M. Intragenic DNA methylation alters chromatin structure and elongation efficiency in mammalian cells. Nature Struct. Mol. Biol. 2004;11:1068–1075. doi: 10.1038/nsmb840. [DOI] [PubMed] [Google Scholar]
- 7.Kawasaki H., Taira K. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature. 2004;431:211–217. doi: 10.1038/nature02889. [DOI] [PubMed] [Google Scholar]
- 8.Morris K.V., Chan S.W., Jacobsen S.E., Looney D.J. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289–1292. doi: 10.1126/science.1101372. [DOI] [PubMed] [Google Scholar]
- 9.Murayama A., Sakura K., Nakama M., Yasuzawa-Tanaka K., Fujita E., Tateishi Y., Wang Y., Ushijima T., Baba T., Shibuya K., et al. A specific CpG site demethylation in the human interleukin 2 gene promoter is an epigenetic memory. EMBO J. 2006;25:1081–1092. doi: 10.1038/sj.emboj.7601012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park C.W., Chen Z., Kren B.T., Steer C.J. Double-stranded siRNA targeted to the huntingtin gene does not induce DNA methylation. Biochem. Biophys. Res. Commun. 2004;323:275–280. doi: 10.1016/j.bbrc.2004.08.096. [DOI] [PubMed] [Google Scholar]
- 11.Svoboda P., Stein P., Filipowicz W., Schultz R.M. Lack of homologous sequence-specific DNA methylation in response to stable dsRNA expression in mouse oocytes. Nucleic Acids Res. 2004;32:3601–3606. doi: 10.1093/nar/gkh697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ting A.H., Schuebel K.E., Herman J.G., Baylin S.B. Short double-stranded RNA induces transcriptional gene silencing in human cancer cells in the absence of DNA methylation. Nature Genet. 2005;37:906–910. doi: 10.1038/ng1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishii T., Fujishiro M., Masuda M., Teramoto S., Matsuse T. A methylated oligonucleotide induced methylation of GSTP1 promoter and suppressed its expression in A549 lung adenocarcinoma cells. Cancer Lett. 2004;212:211–223. doi: 10.1016/j.canlet.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Yao X., Hu J.F., Daniels M., Shiran H., Zhou X., Yan H., Lu H., Zeng Z., Wang Q., Li T., et al. A methylated oligonucleotide inhibits IGF2 expression and enhances survival in a model of hepatocellular carcinoma. J. Clin. Invest. 2003;111:265–273. doi: 10.1172/JCI15109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klug A. Towards therapeutic applications of engineered zinc-finger proteins. FEBS Lett. 2005;579:892–894. doi: 10.1016/j.febslet.2004.10.104. [DOI] [PubMed] [Google Scholar]
- 16.Xu G.L., Bestor T.H. Cytosine methylation targetted to pre-determined sequences. Nature Genet. 1997;17:376–378. doi: 10.1038/ng1297-376. [DOI] [PubMed] [Google Scholar]
- 17.Carvin C.D., Parr R.D., Kladde M.P. Site-selective in vivo targeting of cytosine-5 DNA methylation by zinc-finger proteins. Nucleic Acids Res. 2003;31:6493–6501. doi: 10.1093/nar/gkg853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNamara A.R., Hurd P.J., Smith A.E., Ford K.G. Characterisation of site-biased DNA methyltransferases: specificity, affinity and subsite relationships. Nucleic Acids Res. 2002;30:3818–3830. doi: 10.1093/nar/gkf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNamara A.R., Ford K.G. A novel four zinc-finger protein targeted against p190(BcrAbl) fusion oncogene cDNA: utilisation of zinc-finger recognition codes. Nucleic Acids Res. 2000;228:4865–4872. doi: 10.1093/nar/28.24.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choo Y., Sanchez-Garcia I., Klug A. In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence. Nature. 1994;372:642–645. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- 21.Smith A.E., Farzaneh F., Ford K.G. Single zinc-finger extension: enhancing transcriptional activity and specificity of three-zinc-finger proteins. Biol. Chem. 2005;2386:95–99. doi: 10.1515/BC.2005.012. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A laboratory Manual, 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1989. [Google Scholar]
- 23.Wislon G.G. Amino acid sequence arrangements of DNA-methyltransferases. Nucleic Acids Res. 1992;17:2421–2435. [Google Scholar]
- 24.Cheng X., Kumar S., Posfai J., Pflugrath J.W., Roberts R.J. Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-l-methionine. Cell. 1993;74:299–307. doi: 10.1016/0092-8674(93)90421-l. [DOI] [PubMed] [Google Scholar]
- 25.Klimasauskas S., Kumar S., Roberts R.J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994;76:357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- 26.O'Gara M., Horton J.R., Roberts R.J., Cheng X. Structures of HhaI methyltransferase complexed with substrates containing mismatches at the target base. Nature Struct. Biol. 1998;5:872–877. doi: 10.1038/2312. [DOI] [PubMed] [Google Scholar]
- 27.Sankpal U.T., Rao D.N. Mutational analysis of conserved residues in HhaI DNA methyltransferase. Nucleic Acids Res. 2002;30:2628–2638. doi: 10.1093/nar/gkf380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mi S., Alonso D., Roberts R.J. Functional analysis of Gln-237 mutants of HhaI methyltransferase. Nucleic Acids Res. 1995;23:620–627. doi: 10.1093/nar/23.4.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vilkaitis G., Dong A., Weinhold E., Cheng X., Klimasauskas S.R. Functional roles of the conserved threonine 250 in the target recognition domain of HhaI DNA methyltransferase. J. Biol. Chem. 2000;275:38722–38730. doi: 10.1074/jbc.m005278200. [DOI] [PubMed] [Google Scholar]
- 30.Liu Q., Segal D.J., Ghiara J.B., Barbas C.F., III Design of polydactyl zinc-finger proteins for unique addressing within complex genomes. Proc. Natl Acad. Sci. USA. 1997;94:5525–5530. doi: 10.1073/pnas.94.11.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J.S., Pabo C.O. Getting a handhold on DNA: design of poly-zinc-finger proteins with femtomolar dissociation constants. Proc. Natl Acad. Sci. USA. 1998;95:2812–2817. doi: 10.1073/pnas.95.6.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang G., Salem C.E., Yu M.C., Nguyen H.D., Gonzales F.A., Nguyen T.T., Nichols P.W., Jones P.A. DNA methylation differences associated with tumor tissues identified by genome scanning analysis. Genomics. 1998;53:260–268. doi: 10.1006/geno.1998.5502. [DOI] [PubMed] [Google Scholar]
- 33.Liang G., Gonzalgo M.L., Salem C., Jones P.A. Identification of DNA methylation differences during tumorigenesis by methylation-sensitive arbitrarily primed polymerase chain reaction. Methods. 2002;27:150–155. doi: 10.1016/s1046-2023(02)00068-3. [DOI] [PubMed] [Google Scholar]
- 34.Daujoyte D., Serva S., Vilaitis G., Merkiene E., Venclovas C., Klimasauskas S. HhaI DNA methyltransferase uses the protruding Gln237 for active flipping of its target cytosine. Structure. 2004;12:1047–1055. doi: 10.1016/j.str.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 35.Christiansen D.H., Andersen M.K., Pedersen-Bjergaard J. Methylation of p15INK4B is common, is associated with deletion of genes on chromosome arm 7q and predicts a poor prognosis in therapy-related myelodysplasia and acute myeloid leukemia. Leukemia. 2003;17:1813–1819. doi: 10.1038/sj.leu.2403054. [DOI] [PubMed] [Google Scholar]
- 36.Yates P.A., Burman R., Simpson J., Ponomoreva O.N., Thayer M.J., Turker M.S. Silencing of the mouse Aprt is a gradual process in differentiated cells. Mol. Cell. Biol. 2003;23:4461–4470. doi: 10.1128/MCB.23.13.4461-4470.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]