Abstract
We show that the extent of stable DNA wrapping by Escherichia coli RNA polymerase (RNAP) in the RNAP–promoter open complex depends on the sequence of the promoter and, in particular, on the sequence of the upstream region of the promoter. We further show that the extent of stable DNA wrapping depends on the presence of the RNAP α-subunit carboxy-terminal domain and on the presence and length of the RNAP α-subunit interdomain linker. Our results indicate that the extensive stable DNA wrapping observed previously in the RNAP–promoter open complex at the λ PR promoter is not a general feature of RNAP–promoter open complexes.
Introduction
Atomic force microscopy (AFM) allows straightforward detection and quantification of DNA compaction by DNA-binding proteins (Rivetti et al, 1999; Verhoeven et al, 2001; Heddle et al, 2004). DNA compaction is observed as a reduction of the DNA contour length in the presence of the DNA-binding protein of interest with respect to free DNA.
In previous work, using AFM, it has been shown that Escherichia coli RNA polymerase (RNAP) results in massive, ∼30 nm, apparent DNA compaction on formation of a catalytically competent RNAP–promoter open complex (RPo) at the λ PR promoter (Rivetti et al, 1999). On the basis of the dimensions of RNAP (∼10 nm × ∼10 nm × ∼15 nm; Zhang, 1999), the structure of RNAP (Zhang, 1999), and the modelled structure of RPo (Naryshkin et al, 2000), this massive apparent DNA compaction must arise from wrapping or spooling of upstream promoter DNA (promoter positions −100 to −40) around RNAP (Naryshkin et al, 2000). To account for the full extent of the apparent DNA compaction in RPo at the λ PR promoter, it is necessary to invoke wrapping of upstream promoter DNA by nearly 300° around RNAP (Coulombe & Burton, 1999; Rivetti et al, 1999). Interactions between upstream promoter DNA and RNAP, presumably dependent on at least partial wrapping of upstream promoter DNA around RNAP, have been shown to affect the rate of formation of RPo (Davis et al, 2005; Ross & Gourse, 2005) and have been proposed to affect the rate of entry of DNA into, and unwinding of DNA in, the RNAP active centre cleft (Davis et al, 2005).
In this work, using AFM, we assess the effects of the promoter sequence, the RNAP α-subunit carboxy-terminal domain (αCTD) and the RNAP α-subunit interdomain linker (α-linker) on DNA compaction by RNAP. We find that DNA compaction depends on the sequence of the upstream region of the promoter, the presence of αCTD and the presence and length of the α-linker. Our results indicate that the sequence of the upstream region of the promoter affects DNA compaction not only through effects on αCTD–DNA interaction but also through other effects—presumably effects on intrinsic DNA curvature. Our results further indicate that, in the absence of αCTD–DNA interaction with upstream promoter DNA and intrinsic DNA curvature in the upstream region of the promoter, DNA compaction by RNAP is only ∼2–4 nm, consistent with the expectation from the modelled structure of RPo (Naryshkin et al, 2000). Overall, our results indicate that the massive, ∼30 nm, DNA compaction observed previously in RPo at the λ PR promoter (Rivetti et al, 1999) is not a general feature of RPo at all promoters.
Results
AFM measurements of DNA compaction in RPo
In this work, RNAP and RNAP derivatives were used to prepare RPo at the λPR and lacUV5 promoters, or at substituted derivatives of these promoters (Fig 1A). Complexes were deposited onto freshly cleaved mica and imaged in air by AFM, and DNA contour lengths were measured (Fig 1B). Protein-induced apparent DNA compaction is defined as the difference between (i) the DNA contour length in the absence of protein (Fig 1C, top) and (ii) the DNA contour length of RNAP–DNA complexes (Fig 1C, bottom). This compaction was used as a quantitative indicator of protein-induced stable DNA wrapping, with a large compaction interpreted as a large protein-induced stable DNA wrapping (Fig 1D; Rivetti et al, 1999; Verhoeven et al, 2001; Heddle et al, 2004). Free contour length measurements were made by tracing the DNA path from end to end, as described previously (Rivetti & Codeluppi, 2001). The contour length of RNAP–DNA complexes was measured by tracing the DNA path from end to end and passing through the centre of the protein (Rivetti et al, 1999). In the case of protein–DNA complexes, the measurements are complicated because DNA in contact, or in close proximity, with the protein potentially can be hidden by tip-broadening and out-of-plane effects (Bustamante, 1993; Bustamante & Rivetti, 1996). This might result in underestimation of protein-free DNA contour lengths and overestimation of apparent DNA compaction. In the case of a large protein, such as RNAP, this effect can be up to ∼5 nm. Accordingly, in this work, an apparent DNA compaction is deemed significant as an indicator of stable DNA wrapping only if it is >5 nm.
Figure 1.
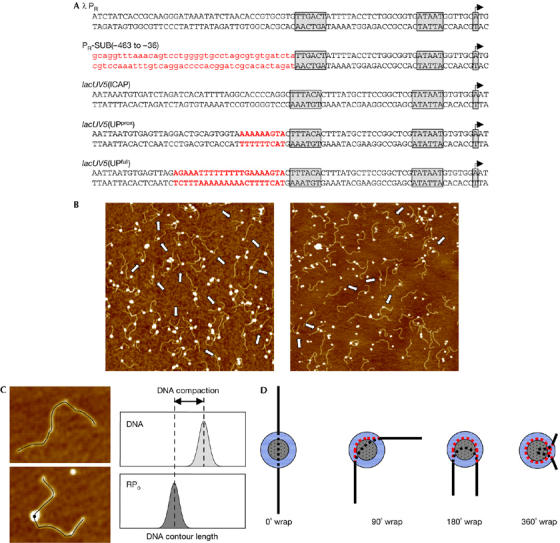
Atomic force microscopy measurements of DNA compaction. (A) Promoters analysed in this work. In each sequence, the transcription start site, the −10 and −35 hexamers are boxed. In PR-SUB(−463 to −36), heterologous DNA is shown in red, lower case; in lacUV5(UPprox), the consensus proximal UP-element subsite (Estrem et al, 1999) is shown in red; in lacUV5(UPfull), the consensus full UP element (Estrem et al, 1998) is shown in red. (B) Representative atomic force microscopy images of RPo formed with wild-type RNAP at the λ PR promoter (left) or the lacUV5(ICAP) promoter (right). Arrows point to individual complexes. The image scan size is 2 μm. (C) Measurement of apparent DNA contour lengths in the absence (top) and presence (bottom) of RNAP. (D) Relationship between apparent DNA compaction and stable DNA wrapping. Grey dotted circle, RNAP; blue circle, tip broadening effect; black line, protein-free DNA; red dashed line, hidden DNA path; black dashed line, inferred DNA path through the centre of the RNAP. Apparent DNA compaction equals the difference in length between the red dashed line and the black dashed line. RNAP, RNA polymerase; RPo, RNA polymerase–promoter open complex.
Surface interactions on deposition onto mica can also potentially cause an artefactual apparent DNA compaction (Rivetti et al, 1996). Accordingly, in this work, we used deposition conditions that allow complete configurational equilibration of DNA (Rivetti et al, 1996) as confirmed by the observation that mean-square end-to-end distances of DNA in the absence of RNAP were in the range predicted for an ideal worm-like chain polymer at equilibrium in two dimensions (data not shown; see Rivetti et al, 1996). It is thus believed that, under the conditions used, complexes were not significantly distorted by the deposition process (Bustamante & Rivetti, 1996).
DNA compaction depends on promoter sequence
Figure 2 and Table 1 present data for RNAP–promoter complexes at the λ PR and lacUV5(ICAP) promoters. For λ PR, an apparent DNA compaction of 30±0.4 nm was observed, consistent with previous results (Rivetti et al, 1999). By contrast, for lacUV5(ICAP), a strikingly smaller apparent DNA compaction, only 4±0.3 nm, was observed. Control experiments established that in the presence of nucleotides, 69% of the complexes formed at λ PR and 70% of the complexes formed at lacUV5(ICAP) can actively transcribe the downstream DNA. This indicates that most of the promoter-bound complexes were active RPo (supplementary Table S1 and Fig S1 online). We conclude that the extent of apparent DNA compaction in RPo is not constant from promoter to promoter, but, instead, depends on promoter sequence.
Figure 2.
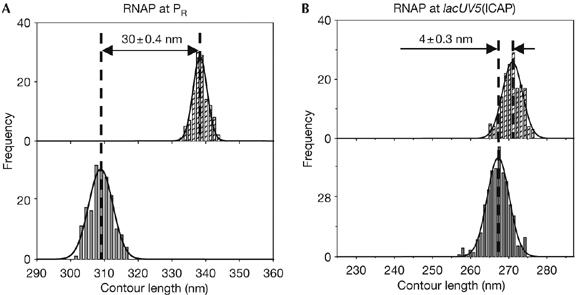
The extent of DNA compaction depends on the sequence of the promoter. (A,B) Contour length distributions of DNA in the absence (hatched bars; top) and presence (grey bars; bottom) of RNAP are shown. Apparent DNA compaction is presented as mean±s.e.m. RNAP, RNA polymerase.
Table 1.
DNA compaction in RPo
| RNAP | Promoter | DNA length (bp) | Bare DNA contour length (nm) | DNA contour length of RPo (nm) | DNA compaction (nm) |
|---|---|---|---|---|---|
| RNAP | λ PR | 1,054 | 338±0.2 (N=126) | 308±0.3 (N=177) | 30±0.4 |
| RNAP | PR-SUB(−463 to −36) | 963 | 317±0.6 (N=112) | 311±0.5 (N=168) | 6±0.8 |
| RNAP | lacUV5(ICAP) | 832 | 271±0.2 (N=166) | 267±0.2 (N=354) | 4±0.3 |
| RNAP | lacUV5(UPprox) | 1,050 | 347±0.3 (N=109) | 345±0.4 (N=130) | 2±0.5 |
| RNAP | lacUV5(UPfull) | 1,191 | 378±0.3 (N=101) | 357±0.6 (N=130) | 21±0.7 |
| ΔαCTDI /ΔαCTDII RNAP | λ PR | 1,054 | 344±0.3 (N=76) | 334±0.6 (*) (N=130) | 10±0.7 |
| ΔαCTDI /ΔαCTDII RNAP | lacUV5(ICAP) | 832 | 269±0.5 (N=83) | 265±0.7 (N=73) | 4±0.8 |
| ΔαCTDI /ΔαCTDII RNAP | lacUV5(UPfull) | 1,050 | 343±0.3 (N=91) | 330±0.5 (N=79) | 13±0.6 |
| ΔαCTDII RNAP | λ PR | 1,054 | 348±0.3 (N=141) | 335±0.2 (N=466) | 13±0.4 |
| ΔαCTDII RNAP | lacUV5(ICAP) | 832 | 266±0.7 (N=93) | 261±2.5 (§) (N=41) | 5±2.0 |
| ΔαCTDII RNAP | lacUV5(UPfull) | 1,050 | 345±0.3 (N=92) | 332±0.5 (N=55) | 13±0.5 |
| Δ6-αI/Δ6-αII RNAP | λ PR | 1,054 | 349±0.3 (N=203) | 334±0.4 (*) (N=182) | 15±0.4 |
| Δ6-αI/Δ6-αII RNAP | lacUV5(UPfull) | 1,050 | 345±0.3 (N=81) | 332±0.6 (N=51) | 13±0.6 |
| Δ12-αI/Δ12-αII RNAP | λ PR | 1,054 | 349±0.3 (N=121) | 335±0.6 (*) (N=122) | 14±0.6 |
| Δ12-αI/Δ12-αII RNAP | lacUV5(UPfull) | 1,050 | 341±0.5 (N=40) | 329±1.0 (N=35) | 12±1.1 |
Contour length values of DNA in the absence of RNAP and RPo represent the mean±standard error of the mean (s.e.m.) obtained from the fitting of the DNA contour length distributions shown in Figs 2, 3, 4 and 5. In the case of bimodal distributions (*), values refer to the leftmost peak. (§) Because of the paucity and scatter of the data, this value represents the arithmetical average of the distribution. The basis for the no-compaction peak observed in Figs 3D, 5A,B is not known, but might represent a subpopulation of RNAP–promoter closed complexes that exhibit no compaction (supplementary Fig S2 online; Rippe et al, 1997). N represents the number of molecules measured for each data set. The DNA compaction is given by the difference between the DNA contour length of DNA in the absence of RNAP and that of RPo.
DNA compaction depends on upstream elements
To determine whether DNA compaction depends on sequence determinants in the upstream region of the promoter, we assessed DNA compaction in RNAP–promoter complexes at PR-SUB(−463 to −36), a PR derivative having a substitution replacing all sequences between positions −463 and −36 (Fig 1A). A large DNA compaction of 30±0.4 nm was observed for PR, whereas a strikingly smaller DNA compaction, of only 6±0.8 nm, was observed for PR-SUB(−463 to −36; Fig 3A; Table 1). Control experiments established that, in the presence of nucleotides, 77% of the complexes formed at PR-SUB(−463 to −36) were active RPo (supplementary Table S1 and Fig S1 online). We conclude that DNA compaction in RPo depends on sequence determinants in the upstream region of the promoter.
Figure 3.
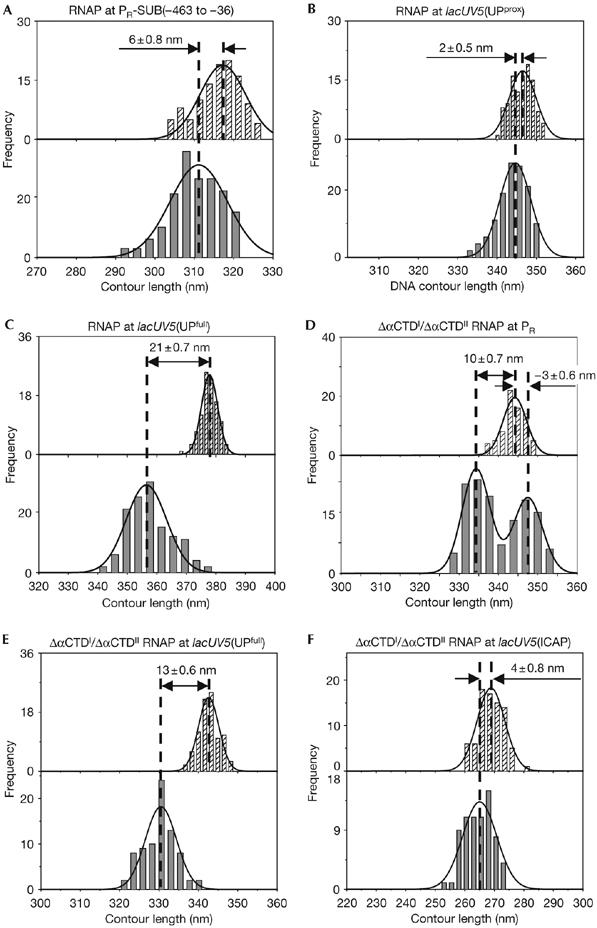
The extent of DNA compaction depends on sequence determinants in the upstream region of the promoter and on αCTD. Contour length distributions of DNA in the absence (hatched bars; top) and presence (grey bars; bottom) of RNAP, from experiments with promoter derivatives having substituted upstream regions (A–C) and with RNAP derivatives lacking both αCTDs (D–F). Apparent DNA compaction is presented as mean±s.e.m. αCTD, α-subunit carboxy-terminal domain; RNAP, RNA polymerase.
Two classes of upstream region promoter elements have been defined: the UP-element subsite and the full UP element (Estrem et al, 1998, 1999; Gourse et al, 2000). The UP-element subsite (a short A/T-rich sequence centred in the −93, −83, −73, −63, −53 or, optimally, −43 region) stimulates transcription up to ∼200-fold through interactions with one RNAP αCTD (Estrem et al, 1999; Gourse et al, 2000). The full UP element (two adjacent UP-element subsites, optimally one centred in the −53 region and the other centred in the −43 region) stimulates transcription up to ∼300-fold through interactions with two RNA αCTDs (Estrem et al, 1998; Gourse et al, 2000).
To determine whether DNA compaction can be affected by a UP-element subsite or a full UP element, we assessed DNA compaction in RNAP–promoter complexes at the lacUV5(UPprox) and lacUV5(UPfull) promoters, respectively, a lacUV5 derivative containing a consensus UP-element subsite centred in the −43 region and a lacUV5 derivative containing a consensus full UP element centred in the −53/−43 region (Fig 1A; Estrem et al, 1998, 1999). A DNA compaction of only 2±0.5 nm was observed for the lacUV5(UPprox) promoter (Fig 3B; Table 1), whereas a DNA compaction of 21±0.7 nm was observed for lacUV5(UPfull) promoter (Fig 3C; Table 1). Control experiments established that, in the presence of nucleotides, 76% of the complexes formed at lacUV5(UPfull) were active RPo (supplementary Table S1 and Fig S1 online). We again conclude that DNA compaction in RPo depends strongly on sequence determinants in the upstream region of the promoter. We further conclude that a full UP element, but not a single UP-element subsite (at least not a single proximal UP-element subsite), represents a sequence determinant for large DNA compaction in RPo.
DNA compaction depends on the presence of the αCTD
RNAP (subunit composition β′βαIαIIωσ70) contains two identical α-subunits, αI and αII. Each α-subunit consists of (i) an amino-terminal domain (αNTD) with determinants for dimerization and for interaction with β′ and β, (ii) an αCTD with determinants for protein–DNA interaction with UP-element subsites and for protein–protein interactions with transcriptional regulators and (iii) a long, flexible interdomain linker (α-linker; Blatter et al, 1994; Busby & Ebright, 1994). The long, flexible α-linker allows αCTD to occupy different positions relative to αNTD—and thus relative to the remainder of RNAP—in different RNAP–promoter complexes (i.e. to interact nonspecifically with upstream DNA at a simple promoter, to interact with a UP-element subsite or UP element at a UP-element-subsite- or UP-element-containing promoter or to interact with an activator at an activator-dependent promoter; Busby & Ebright, 1994).
To determine whether DNA compaction is dependent on the presence of αCTD, we assessed DNA compaction in RPo prepared using ΔαCTDI/ΔαCTDII RNAP, an RNAP derivative lacking both αCTDI and αCTDII, at the λ PR and lacUV5(UPfull) promoters. At PR, a DNA compaction of 30±0.4 nm was observed with wild-type (WT)-RNAP, but a significantly smaller DNA compaction, of only 10±0.7 nm, was observed with ΔαCTDI/ΔαCTDII RNAP (Fig 3D; Table 1). Similarly, at lacUV5(UPfull), a DNA compaction of 21±0.7 nm was observed with WT-RNAP, but a significantly smaller DNA compaction, of only 13±0.6 nm, was observed with ΔαCTDI/ΔαCTDII RNAP (Fig 3E; Table 1). As expected, a DNA compaction of 4±0.8 nm was observed with ΔαCTDI/ΔαCTDII RNAP at lacUV5(ICAP) (Fig 3F; Table 1). We conclude that the extent of DNA compaction depends on the presence of αCTD.
To determine whether DNA compaction is dependent on the presence of both αCTDI and αCTDII, we assessed DNA compaction in RPo assembled with ΔαCTDII RNAP, an RNAP derivative having αCTDI but lacking αCTDII (Estrem et al, 1999) at the λ PR and lacUV5(UPfull) promoters. At PR, only a small DNA compaction, of 13±0.4 nm, was observed with ΔαCTDII RNAP (a DNA compaction comparable with that observed with ΔαCTDI/ΔαCTDII RNAP; Fig 4A; Table 1). Similarly, at lacUV5(UPfull), only a small DNA compaction, of 13±0.5 nm, was observed with ΔαCTDII RNAP (Fig 4B; Table 1). As expected, a DNA compaction of 5±2.0 nm was observed with ΔαCTDII RNAP at lacUV5(ICAP) (Fig 4C; Table 1). We conclude that αCTDI alone is insufficient to mediate large-scale DNA compaction.
Figure 4.
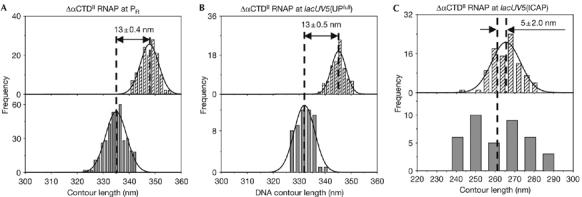
αCTDI alone is insufficient to mediate large-scale DNA compaction. (A–C) Contour length distributions of DNA in the absence (hatched bars; top) and presence (grey bars; bottom) of RNAP, from experiments with ΔαCTDII RNAP derivatives. Apparent DNA compaction is presented as mean±s.e.m. Because of the paucity and scatter of the data, the mean value in (C) represents the arithmetical average of the data. αCTD, α-subunit carboxy-terminal domain; RNAP, RNA polymerase.
DNA compaction depends on the α-linker
The α-linker is ∼15 amino acids in length and comprises α-residues from residue ∼236 to ∼251 (Blatter et al, 1994; Meng et al, 2000).
To determine whether DNA compaction is dependent on the presence and length of the α-linker, we assessed DNA compaction in RPo assembled with Δ6-αI/Δ6-αII RNAP, an RNAP derivative lacking six amino-acid residues of the α-linker (α-residues 235–241; Meng et al, 2000), and with Δ12-αI/Δ12-αII RNAP, an RNAP derivative lacking 12 amino-acid residues of the α-linker (α-residues 235–247; Meng et al, 2000).
At PR, only a small DNA compaction, of 15±0.4 or 14±0.6 nm, was observed with Δ6-αI/Δ6-αII RNAP or Δ12-αI/Δ12-αII RNAP (a DNA compaction comparable with that observed with ΔαCTDI/ΔαCTDII RNAP; Fig 5A,B; Table 1). Similarly, at lacUV5(UPfull), only a small DNA compaction, of 13±0.6 or 12±1.1 nm, was observed with Δ6-αI/Δ6-αII RNAP and Δ12-αI/Δ12-αII RNAP (a DNA compaction comparable with that observed with ΔαCTDI/ΔαCTDII RNAP; Fig 5C,D; Table 1). We conclude that the α-linker is required for large-scale DNA compaction.
Figure 5.
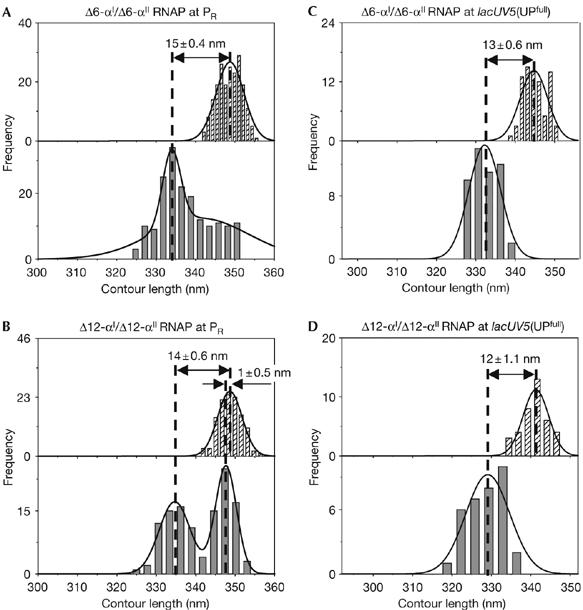
The extent of DNA compaction depends on the length of the α-linker. (A–D) Contour length distributions of DNA in the absence (hatched bars; top) and presence (grey bars; bottom) of RNAP derivatives, from experiments with RNAP derivatives lacking residues of the α-linker. Apparent DNA compaction is presented as mean±s.e.m. α-linker, α-subunit interdomain linker; RNAP, RNA polymerase.
The DNA bend angle is coupled to the DNA compaction
The DNA bend angle, defined as the deviation from linearity of the double helix, was measured for each complex by means of tangents drawn at the exit points of DNA from RNAP. The data are shown in supplementary Figs S3–S6 online. Overall, there is a good correlation between DNA compaction and bend angle. For example, the DNA bend angle of WT-RNAP at PR is 55±2.4°, which is consistent with an almost complete turn of the DNA around the RNAP at this promoter (see also figure 8 in Rivetti et al, 1999). Conversely, at lacUV5(ICAP), the measured DNA bend angle is 16±3.3°, which is consistent with the little DNA compaction observed at this promoter. At PR-SUB(−463 to −36) and lacUV5(UPprox) promoters, the DNA bend angles are 0±4.8° and 18±4.7°, respectively. These values correlate well with the little DNA compaction observed with these promoters. At lacUV5(UPfull), the measured DNA bend angle is 49±1.4°, which is again consistent with the 21 nm DNA compaction observed at this promoter. Thus, we conclude that a large DNA compaction associated with a high DNA bend angle is the result of DNA wrapping around the RNAP.
A statistically significant correlation between DNA compaction and DNA bend angle cannot be attributed for complexes assembled with αCTD-RNAP mutants. This might be due to the intermediate DNA compaction observed in some of these cases, the presence of different types of complex as shown by the bimodal DNA contour length distributions and difficulties of obtaining narrow bend angle distributions.
Discussion
Our results establish that the extent of stable DNA wrapping in RPo depends on the sequence of the promoter and, in particular, on sequence determinants in the upstream region of the promoter (UP elements). The presence of αCTD and an intact α-linker is required to maintain extensive stable DNA wrapping. Our results further indicate that the sequence of the upstream region of the promoter can affect DNA wrapping even in the absence of αCTD and thus even in the absence of αCTD–DNA interactions. For example, RPo prepared using ΔαCTDI/ΔαCTDII RNAP shows an apparent DNA compaction of 13±0.6 nm at lacUV5(UPfull) but only 4±0.8 nm at lacUV5(ICAP) (Fig 3E,F). We infer that the sequence of the upstream region of the promoter can affect compaction not only through effects on αCTD–DNA interaction but also through other effects. We suggest that these other effects involve intrinsic DNA curvature, noting that UP-element subsites and UP elements are A/T-rich sequences (Fig 1A; Ross et al, 1993) and that A/T-rich sequences are associated with intrinsic DNA curvature (Koo et al, 1986). In the absence of αCTD–DNA interaction with upstream promoter DNA and of intrinsic DNA curvature in upstream DNA, stable DNA wrapping in RPo is small.
Overall, we show that tight, stable DNA wrapping is not a general feature of RPo, but rather depends on the promoter sequence. The tight, stable DNA wrapping observed at λ PR implies the presence, in the upstream region of this promoter, of sequence determinants that promote stable DNA wrapping—sequence determinants that, similar to the UP element in lacUV5(UPfull), make specific protein–DNA interactions with RNAP, favour intrinsic DNA curvature or both. A detailed analysis of the sequence determinants responsible for stable DNA wrapping at λ PR will be presented elsewhere.
Interestingly, as described in the supplementary information online, inactive promoter complexes (complexes in which RNAP is bound to the promoter but is unable to initiate RNA synthesis), in particular those formed at PR and lacUV5(UPfull), have a mean DNA compaction that is significantly smaller than that of the corresponding active RPo (supplementary Fig S2 and Table S2 online).
The results of photocrosslinking studies indicate that the αCTDs can interact sequence-nonspecifically with the upstream region of the lacUV5 promoter, making sequence-nonspecific interactions with the DNA minor groove near positions −43, −53, −63, −73, −83 and −93 (Naryshkin et al, 2000). The results of promoter-truncation experiments indicate that the upstream region of the lacUV5 promoter can influence the association of RNAP (Ross & Gourse, 2005). How can this evidence for upstream interactions at lacUV5 be reconciled with the observed near-absence of apparent DNA compaction and thus the near-absence of stable DNA wrapping at lacUV5? We suggest that αCTD interacts only transiently with the upstream region of lacUV5, making interactions sufficient to allow crosslinking and to allow effects on the association of RNAP, but not sufficient to yield measurable DNA compaction at equilibrium (as in imaging by AFM under the conditions used in this work; see the discussion in the first section of Results; see also Bustamante & Rivetti, 1996). In this regard, we note that (i) the photogenerated reactive species used in photocrosslinking studies is relatively long-lived, allowing efficient sampling of transient interactions (Naryshkin et al, 2000; N.N. & R.H.E., unpublished data), and (ii) rapid quenching of the photogenerated reactive species eliminates crosslinking of αCTD with the upstream region of lacUV5 (N.N. & R.H.E., unpublished data). Thus, results from photocrosslinking expressly support the occurrence of transient, as opposed to stable, interactions of αCTD with the upstream region of lacUV5.
An extended RNAP–DNA interaction can affect transcription in several, not mutually exclusive ways. A larger number of contacts between the RNAP and promoter DNA can increase the overall affinity of RNAP for the promoter (Aiyar et al, 1998; Estrem et al, 1998, 1999; Ross et al, 1993, 1998). An extended RNAP–DNA interaction, and corresponding DNA wrapping and wrapping-dependent distortion, also potentially can mechanically influence the isomerization from RNAP–promoter closed to RNAP–promoter open complex (Coulombe & Burton, 1999; Davis et al, 2005; Ross & Gourse, 2005). However, it should be emphasized that, as shown in this work, tight, stable DNA wrapping is not a general feature of all RPo at all promoters, but rather is strongly dependent on the promoter sequence.
Methods
DNA templates and proteins. The 1,054-bp-long DNA fragment carrying λ PR contains λ-DNA from –438 to +34 with respect to the PR start site, which is positioned 616 bp from the downstream end. The 963-bp-long DNA fragment PR-SUB(−463 to −36) contains λ-DNA from −35 to +34 with respect to the PR start site, which is positioned 500 bp from the downstream end. The 832-bp-long DNA fragment carrying lacUV5(ICAP) has the start site positioned 360 bp from the downstream end. The 1,191 and 1,050-bp-long DNA fragments carrying lacUV5(UPfull) have the start site positioned 706 and 602 bp from the downstream end, respectively. The 1,050-bp-long DNA fragment carrying lacUV5(UPprox) has the start site positioned 602 bp from the downstream end. Details about the preparation of DNA fragments and proteins are provided in the supplementary information online.
Complex formation and AFM imaging. RPo complexes were prepared by mixing 20 nM DNA with 20 nM RNAP in transcription buffer (20 mM Tris–HCl pH 7.9, 50 mM KCl, 5 mM MgCl2). The 10 μl reaction was incubated at 37°C for 15 min. The reaction was diluted to 1–2 nM complexes in 20 μl of deposition buffer (4 mM HEPES pH 7.4, 10 mM NaCl, 4 mM MgCl2) and deposited onto freshly cleaved mica. The sample was incubated for about 2 min before the surface was rinsed with water and dried with nitrogen. AFM imaging was carried out in air with the tapping mode using a Nanoscope III microscope (Veeco Digital Instruments, Santa Barbara, CA, USA).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Information
Acknowledgments
We thank M. Thomas for plasmids and Centro Interfacoltà Misure for the AFM facility. This work was supported by the Italian Ministry of University and Scientific and Technological Research grant FIRB-RBNE01KMT9 to C.R. and by National Institutes of Health grant GM41376 and a Howard Hughes Medical Institute investigatorship to R.H.E.
References
- Aiyar SE, Gourse RL, Ross W (1998) Upstream A-tracts increase bacterial promoter activity through interactions with the RNA polymerase α subunit. Proc Natl Acad Sci USA 95: 14652–14657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatter EE, Ross W, Tang H, Gourse RL, Ebright RH (1994) Domain organization of RNA polymerase α subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell 78: 889–896 [DOI] [PubMed] [Google Scholar]
- Busby S, Ebright RH (1994) Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79: 743–746 [DOI] [PubMed] [Google Scholar]
- Bustamante C, Rivetti C (1996) Visualizing protein–nucleic acid interactions on a large scale with the scanning force microscope. Annu Rev Biophys Biomol Struct 25: 395–429 [DOI] [PubMed] [Google Scholar]
- Bustamante C, Keller D, Yang G (1993) Scanning force microscopy of nucleic acids and nucleoprotein assemblies. Curr Opin Struct Biol 3: 363–372 [Google Scholar]
- Coulombe B, Burton ZF (1999) DNA bending and wrapping around RNA polymerase: a ‘revolutionary' model describing transcriptional mechanisms. Microbiol Mol Biol Rev 63: 457–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Capp MW, Record MT Jr, Saecker RM (2005) The effects of upstream DNA on open complex formation by Escherichia coli RNA polymerase. Proc Natl Acad Sci USA 102: 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem ST, Gaal T, Ross W, Gourse RL (1998) Identification of an UP element consensus sequence for bacterial promoters. Proc Natl Acad Sci USA 95: 9761–9766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrem ST, Ross W, Gaal T, Chen ZW, Niu W, Ebright RH, Gourse RL (1999) Bacterial promoter architecture: subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase α subunit. Genes Dev 13: 2134–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse RL, Ross W, Gaal T (2000) UPs and downs in bacterial transcription initiation: the role of the α subunit of RNA polymerase in promoter recognition. Mol Microbiol 37: 687–695 [DOI] [PubMed] [Google Scholar]
- Heddle JG, Mitelheiser S, Maxwell A, Thomson NH (2004) Nucleotide binding to DNA gyrase causes loss of DNA wrap. J Mol Biol 337: 597–610 [DOI] [PubMed] [Google Scholar]
- Koo HS, Wu HM, Crothers DM (1986) DNA bending at adenine·thymine tracts. Nature 320: 501–506 [DOI] [PubMed] [Google Scholar]
- Meng W, Savery NJ, Busby SJ, Thomas MS (2000) The Escherichia coli RNA polymerase α subunit linker: length requirements for transcription activation at CRP-dependent promoters. EMBO J 19: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naryshkin N, Revyakin A, Kim Y, Mekler V, Ebright RH (2000) Structural organization of the RNA polymerase–promoter open complex. Cell 101: 601–611 [DOI] [PubMed] [Google Scholar]
- Rippe K, Guthold M, von Hippel PH, Bustamante C (1997) Transcriptional activation via DNA-looping: visualization of intermediates in the activation pathway of E. coli RNA polymerase-s54 holoenzyme by scanning force microscopy. J Mol Biol 270: 125–138 [DOI] [PubMed] [Google Scholar]
- Rivetti C, Codeluppi S (2001) Accurate length determination of DNA molecules visualized by atomic force microscopy: evidence for a partial B- to A-form transition on mica. Ultramicroscopy 87: 55–66 [DOI] [PubMed] [Google Scholar]
- Rivetti C, Guthold M, Bustamante C (1996) Scanning force microscopy of DNA deposited on mica: equilibration versus kinetic trapping studied by polymer chain analysis. J Mol Biol 264: 919–932 [DOI] [PubMed] [Google Scholar]
- Rivetti C, Guthold M, Bustamante C (1999) Wrapping of DNA around the E. coli RNA polymerase open promoter complex. EMBO J 18: 4464–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Gourse RL (2005) Sequence-independent upstream DNA-αCTD interactions strongly stimulate Escherichia coli RNA polymerase–lacUV5 promoter association. Proc Natl Acad Sci USA 102: 291–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL (1993) A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science 262: 1407–1413 [DOI] [PubMed] [Google Scholar]
- Ross W, Aiyar SE, Salomon J, Gourse RL (1998) Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J Bacteriol 180: 5375–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhoeven EE, Wyman C, Moolenaar GF, Hoeijmakers JH, Goosen N (2001) Architecture of nucleotide excision repair complexes: DNA is wrapped by UvrB before and after damage recognition. EMBO J 20: 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA (1999) Crystal structure of Thermus aquaticus core RNA polymerase at 3.3Å resolution. Cell 98: 811–824 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Information


