Abstract
The cullin CUL-2 is a crucial component of a subclass of multisubunit cullin-RING ubiquitin-ligases. The specificity of CUL-2-based complexes is provided by variable substrate-recognition subunits that bind to specific substrates. In Caenorhabditis elegans, CUL-2 regulates several key processes in cell division and embryonic development, including meiotic progression, anterior–posterior polarity and mitotic chromatin condensation. However, the substrate recognition subunits that work in these CUL-2-dependent processes were unknown. Here, we present evidence that ZYG-11 is the substrate-recognition subunit for a CUL-2-based complex that regulates these functions. We show that ZYG-11 interacts with CUL-2 in vivo and binds to the complex adaptor protein Elongin C using a nematode variant of the VHL-box motif. We show that the ZYG11 gene family encompasses two main branches in metazoa, and provide evidence that members of the extended ZYG11 family in nematodes and humans are conserved components of CUL2-based ubiquitin-ligases.
Introduction
Members of the cullin-RING family of ubiquitin-ligases (E3s) function in a wide range of dynamic cellular processes, including the cell cycle, signal transduction and transcription (Petroski & Deshaies, 2005). Ubiquitin ligases facilitate the transfer of ubiquitin from a ubiquitin-conjugating enzyme (E2) to the substrate. The cullin CUL2 is a core component of multisubunit E3 complexes (designated CBC) that include the adaptor protein Elongin C, a ubiquitin-like protein Elongin B, the RING finger protein Rbx1/Roc1 and a variable substrate recognition subunit (SRS; Petroski & Deshaies, 2005). In mammals, three CBC complex SRSs have been identified: the von Hippel–Lindau (VHL) tumor suppressor, which degrades hypoxia-inducible factor-α (HIF-α) under normoxic conditions; LRR-1, which suppresses 4-1-BB receptor signalling in CD4+ and CD8+ T cells; and FEM1b, which regulates glucose-stimulated insulin secretion (Jang et al, 2001; Kamura et al, 2004; Kim & Kaelin, 2004; Lu et al, 2005).
Genetic studies of CUL2 complexes in mammals have been limited to analysis of individual SRSs, whereas the loss of core components, which would inactivate all CBC complexes, has not been analysed. In Caenorhabditis elegans, analysis of cul-2 mutants has revealed roles for CUL-2 in important cell cycle and developmental processes. In germ cells, CUL-2 is required for the negative regulation of the CDK inhibitor CKI-1 to allow cell-cycle progression from G1 to S phase (Feng et al, 1999). CUL-2 is also required for several processes in the early embryo: the meiosis II metaphase-to-anaphase transition; establishment of anterior–posterior polarity; cyclin B degradation; chromosome condensation; mitotic progression; proper cytoplasmic organization; and the degradation of CCCH-finger polarity proteins (Feng et al, 1999; DeRenzo et al, 2003; Liu et al, 2004; Sonneville & Gonczy, 2004). Significantly, RNA interference (RNAi) and mutant analysis of the C. elegans homologues of the known mammalian SRSs indicates that they do not function in these processes (Epstein et al, 2001; data not shown). An additional SRS, ZIF-1, has been identified and is required for the degradation of CCCH polarity proteins, but not for the other known CUL-2 phenotypes (DeRenzo et al, 2003). Therefore, the SRSs for the CBC complexes that execute most of the known CUL-2 functions have not been identified.
Here, we identify ZYG-11 as the SRS for a CBCZYG-11 complex that is responsible for most of the known CUL-2 functions. We show that ZYG-11 binds to the CUL-2 complex in vivo through direct interaction with Elongin C (ELC-1). ZYG-11 binds to ELC-1 using a nematode variant of the VHL-box motif. Phylogenetic analysis identifies two branches of zyg-11 homologues in diverse metazoa: the ZYG11 and ZER1 subfamilies. We provide evidence that C. elegans ZER-1 and human ZYG11B and ZYG11BL associate with CUL-2 in vivo through direct interaction with Elongin C. Our work therefore suggests that members of the extended metazoan ZYG11 family function as conserved SRS components for CUL-2 ubiquitin-ligase complexes.
Results And Discussion
ZYG-11 is the SRS component of a CBC complex
In C. elegans, zyg-11 was identified as a mutant that disrupted zygote development (Kemphues et al, 1986). zyg-11 mutant phenotypes are similar to a subset of phenotypes observed in cul-2 mutants, including a prolonged delay in meiosis II, a failure to degrade cyclin B, defects in anterior–posterior polarity and defects in chromatin condensation (Table 1). Here, we extend the analysis of cul-2 and zyg-11 phenotypes. Both cul-2 and zyg-11 mutant zygotes enter an extended period of arrest in metaphase II (Liu et al, 2004; Sonneville & Gonczy, 2004). After the meiosis II delay, zyg-11(mn40)-null mutants complete anaphase (8 out of 8 embryos), although DNA bridges are observed between the separating DNA strands and the second polar body is generally not extruded (7 out of 8 embryos). By contrast, cul-2(ek1)-null mutants either have no discernible anaphase II (3 out of 5 embryos) or have aberrant splitting of DNA after decondensation has started (2 out of 5 embryos). In both zyg-11 and cul-2 mutant zygotes, maternal pronuclei subsequently enter mitosis with the paternal pronucleus.
Table 1.
Comparison of mutant phenotypes of cul-2 and substrate recognition subunit genes
| cul-2 phenotypes | zyg-11 | zif-1 | vhl-1 |
|---|---|---|---|
| Meiotic metaphase II delay* | Yes* | No# | No‡,§ |
| Failure to degrade maternal cyclins B1 and B3* | Yes* | ND | ND |
| Ectopic localization of PAR-2* | Yes* | No# | ND |
| Cytoplasmic extensions¶ | Yes∥ | No# | No‡ |
| Granule-free areas in zygote‡ | Yes∥ | No# | No‡ |
| Failure of chromosome condensation¶ | Yes∥ | No# | No‡,§ |
| Early embryonic developmental arrest¶ | Partial∥,‡ | No# | No‡,§ |
| Mitotic delay¶ | Partial‡ | No# | No‡ |
| Germ cell arrest in G1 phase, accumulation of CKI-1¶ | No‡ | No# | No‡,§ |
| Failure to degrade CCCH polarity proteins# | No‡ | Yes# | ND |
*Liu et al (2004) and Sonneville & Gonczy (2004). ‡Our observations. §Epstein et al (2001). ∥Kemphues et al (1986). ¶Feng et al (1999). #DeRenzo et al (2003). ND: not done.
During meiosis II, cul-2 mutants have one or more large cytoplasmic regions devoid of granules, similar to zyg-11 mutants (Fig 1A; Kemphues et al, 1986). In both mutants, these granule-free regions are temporally restricted to the extended meiosis II period (Fig 1B). The underlying cause of the cleared regions is unknown, but they do not arise from an accumulation of tubulin or actin-based cytoskeleton (supplementary Fig 1 online). We observe that the length of mitosis is extended in both cul-2 and zyg-11 mutant embryos; however, mitotic timing is significantly longer for cul-2 than for zyg-11 (Fig 1C). cul-2 mutants also show some phenotypes that are not observed in zyg-11 mutants. cul-2 mutant embryos undergo an early embryonic arrest with fewer cells (approximately 24) and more uneven DNA distribution than is generally observed for arrested zyg-11 mutant embryos (Fig 1D). Germ cells in cul-2 mutants undergo a G1-phase arrest, so that there are fewer, larger cells, whereas zyg-11 germ cells show normal proliferation (Fig 1E).
Figure 1.
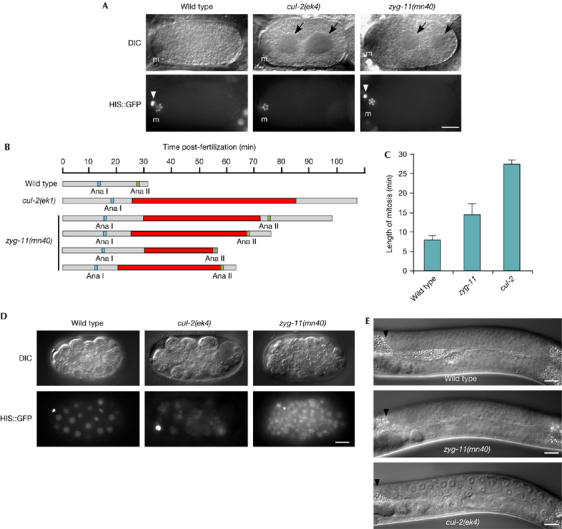
cul-2 and zyg-11 phenotypes. (A) Differential interference contrast (DIC) and histone H2B∷GFP epifluorescence images of in utero metaphase II wild-type, zyg-11(mn40) and cul-2(ek4) zygotes. ‘m', meiotic spindle region. White arrowheads indicate the first polar body (out of focus in the cul-2 image) and black arrows the granule-free areas. (B) Graphical representation of the timing of meiotic divisions and the appearance of granule-free areas (red boxes) for wild-type, cul-2(ek1) and zyg-11(mn40) embryos. Anaphase I, blue box; anaphase II, green box. (C) The length of the first mitosis (in min) in wild-type, zyg-11(mn40) and cul-2(ek4) embryos. Mitotic timing was calculated from nuclear envelope breakdown to nuclear envelope reformation. Error bars, s.e.m. Statistical significance: wild type versus cul-2, P<1 × 10−6; wild type versus zyg-11, P<0.05; cul-2 versus zyg-11, P<0.005. (D) DIC and histone H2B∷GFP images of an early-stage wild-type embryo, and cul-2 and zyg-11 arrested embryos collected 24 h after being laid. Note that the cul-2 mutant embryo has a more unequal DNA distribution between cells than wild-type or zyg-11 mutant embryos. (E) DIC micrograph of wild-type, zyg-11(mn40) and cul-2(ek4) L4-stage gonad arms. The black arrowhead indicates the distal tip cell. Scale bars, 10 μm. GFP, green fluorescent protein.
The substantial overlap of cul-2 and zyg-11 mutant phenotypes suggests that the two proteins might function together. We affinity-purified transgenic Flag-tagged CUL-2 from gravid adults to identify the associated proteins. We observed that ZYG-11 co-purified with CUL-2–Flag, by using both western blot and mass spectrometry, indicating physical association in vivo (Fig 2A; data not shown).
Figure 2.
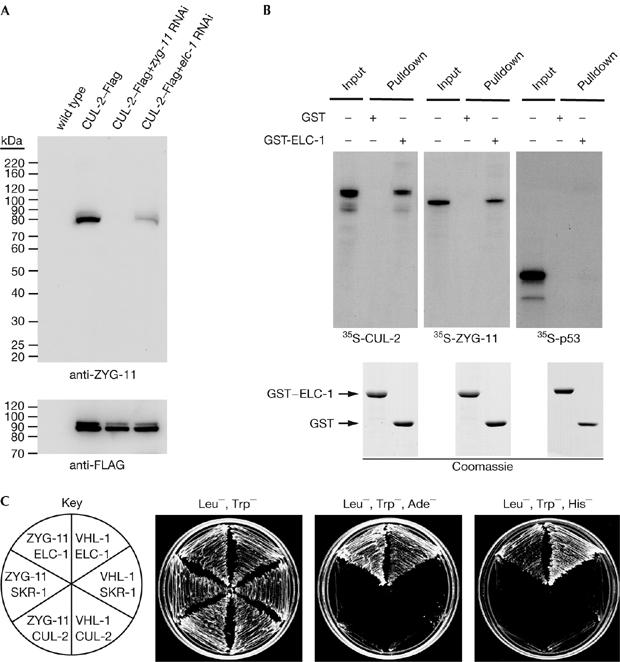
ZYG-11 interacts with CBC components in vivo and in vitro. (A) ZYG-11 interacts with CUL-2 in vivo. Anti-Flag affinity purification from animals expressing no transgenes (lane 1) or a CUL-2-Flag transgene without RNAi treatment (lane 2), with zyg-11 RNAi (lane 3) or with elc-1 RNAi (lane 4) was analysed by western blot with ZYG-11 (top) and Flag (bottom) antibodies. (B) ZYG-11 interacts with glutathione-S-transferase (GST)–ELC-1 in vitro. 35S-labelled in vitro-translated CUL-2 (left panel), ZYG-11 (middle panel) or human p53 (right panel) were incubated with GST or GST-ELC-1. GST-associated proteins were precipitated, separated by SDS–PAGE and analysed by autoradiography (top) or Coomassie blue staining (bottom). Input denotes 100% of the in vitro-translated protein used for the binding reactions. (C) ZYG-11 interacts with ELC-1 in the two-hybrid system. Yeast expressing the indicated two-hybrid fusion proteins were streaked on nonselective media (Leu−, Trp−) or selective media (Leu−, Trp−, Ade−; and Leu−, Trp−, His−, with 2 mM 3-AT, 3-amino-1,2,4-triazole). VHL-1, a predicted SRS, is included as a positive control. CBC, CUL2-Elongin BC ubiquitin-ligase; ELC, Elongin C; SDS–PAGE, sodium dodecylsulphate–PAGE; RNAi, RNA interference; VHL, von Hippel–Lindau.
The observation that zyg-11 mutants show a subset of cul-2 mutant phenotypes led to the proposal that ZYG-11 might function as an SRS (Liu et al, 2004; Sonneville & Gonczy, 2004), as individual SRSs govern only a subset of CUL-2 functions. SRSs are expected to interact physically with the adaptor ELC-1 (Stebbins et al, 1999). We investigated whether ZYG-11 interacts with ELC-1 by using three systems. First, in the yeast two-hybrid system, we observed that ZYG-11 interacted specifically with ELC-1 but not with the related protein SKR-1, which acts as an adaptor for CUL-1 complexes (Fig 2C; Nayak et al, 2002; Yamanaka et al, 2002). SRSs do not interact independently with the cullin and, consistently, we did not observe any interaction between ZYG-11 and CUL-2 (Fig 2C). Second, in vitro-translated 35S-labelled ZYG-11 interacted with recombinant GST–ELC-1 (GST for glutathione-S-transferase; Fig 2B). Finally, RNAi depletion of elc-1 in C. elegans reduced the amount of ZYG-11 that co-precipitated with CUL-2–Flag, suggesting that ELC-1 is required for the association of ZYG-11 with CUL-2 in vivo (Fig 2A). The elc-1 RNAi appeared to be impenetrant because approximately 15% of elc-1(RNAi) embryos hatched, whereas 100% of cul-2(RNAi) and zyg-11(RNAi) embryos arrested.
The ZYG-11 protein contains four variant leucine-rich repeat (vLRR) motifs and an armadillo-like helical domain (ARM-like) (supplementary Fig 2 online). The regions of ZYG-11 that interact with ELC-1 were mapped by using deletion constructs in the two-hybrid system (Fig 3A). The amino-terminal region of ZYG-11, which does not contain the vLRR or ARM-like domains, is sufficient for interaction with ELC-1. Mammalian SRSs bind to Elongin C by a three α-helix structure called the VHL box (Kamura et al, 2004), and we identified a divergent VHL-box motif in the N terminus of ZYG-11 (Fig 3B).
Figure 3.
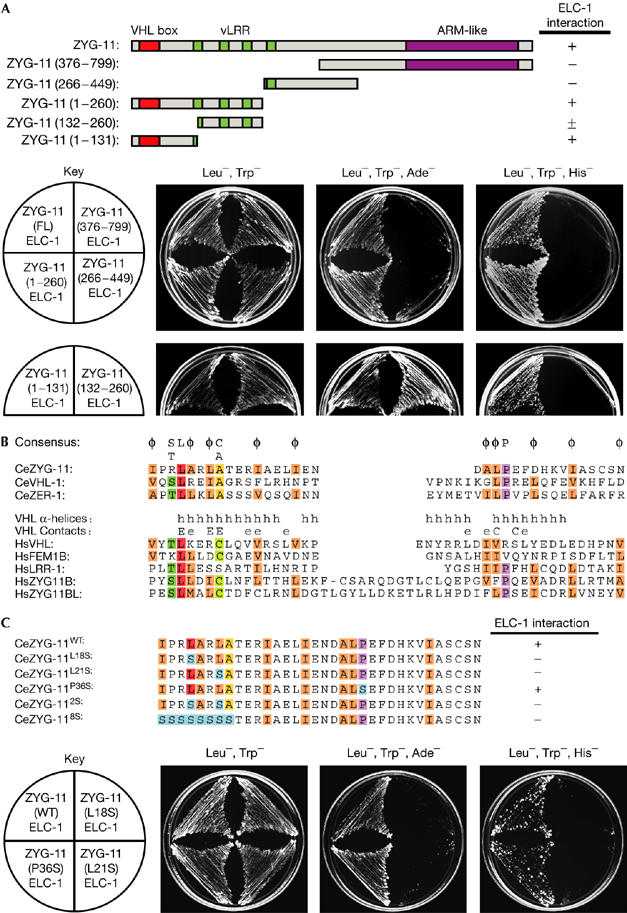
ZYG-11 binds to Elongin C through a von Hippel–Lindau-box sequence. (A) Schematic representation of ZYG-11 deletion mutants (top) and two-hybrid interactions with ELC-1 (bottom). The ZYG-11 132–260 amino-acid region interacted with ELC-1 under adenine-deficient selection, but not under the more stringent histidine-deficient+2 mM 3-AT selection. (B) Alignment of VHL-box sequences of C. elegans (Ce) ZYG-11, VHL-1 and ZER-1, and human (Hs) VHL, FEM1B, LRR-1, ZYG11B and ZYG11BL. Amino acids that are conserved in more than half of the aligned sequences are highlighted: dark green (Serine or Threonine), red (Leucine), purple (Proline), yellow (Alanine), light green (Cysteine) and orange (aliphatic residues). A consensus is provided above the alignment; φ=aliphatic amino acid. Human VHL-box features: α-helices (h), residues that make contact with CUL2 (C) and major (E) or minor (e) contacts with Elongin C (Stebbins et al, 1999; Kamura et al, 2004). (C) The ZYG-11 VHL-box sequence and residues mutated to Serine (blue; top). Interactions of ELC-1 with the ZYG-11 VHL-box mutant constructs in the two-hybrid system are shown (bottom). ARM-like, armadillo-like; ELC, Elongin C; VHL, von Hippel–Lindau; vLRR, variant leucine-rich repeat.
The C. elegans VHL-box motif is notably different from that in humans. In particular, there is an aliphatic amino acid rather than a cysteine in the conserved eighth position, and the residues LP in the distal region. The LP corresponds to the LPXP sequence of the mammalian SOCS-box motif, which is related to the VHL-box motif but binds to CUL5 complexes rather than to CUL2 complexes (Kamura et al, 2004). In humans, the LPXP sequence is in the CUL5-box region of the SOCS box, and the terminal proline is required for interaction with CUL5 (Kamura et al, 2004). However, the LPXP sequence does not seem to be specific for CUL-5 binding in C. elegans, as we observed that a protein containing the LPXP sequence, ZER-1, physically associated with CUL-2 in vivo (Fig 4A).
Figure 4.
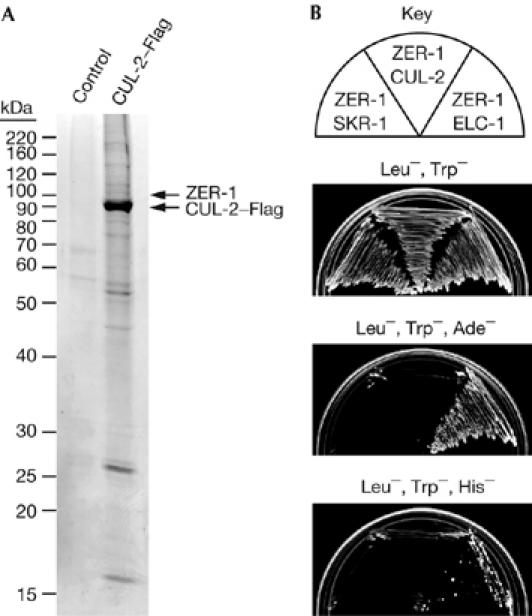
ZER-1 interacts with CBC components in vivo and in vitro. (A) Coomassie-stained gel of CUL-2–Flag affinity purification. ZER-1 and CUL-2 were identified by mass spectrometry in the marked protein bands of the CUL-2–Flag affinity purification, but not from gel slices taken from the equivalent positions of the control purification lane. (B) Two-hybrid analysis of interaction of ZER-1 with ELC-1, SKR-1 and CUL-2; ELC-1, Elongin C.
To determine whether the interaction of ZYG-11 with ELC-1 is dependent on the VHL-box motif, we generated full-length ZYG-11 with missense mutations in the VHL box. When either leucine 18 or leucine 21 of the VHL box were replaced with serine, the interaction of ZYG-11 with ELC-1 was abolished (Fig 3C). The second half of the VHL-box motif is the CUL2-box region, which is required for binding to CUL2 but not Elongin C (Kamura et al, 2004). Consistently, mutation of the proline of the LP motif located in the CUL2 box did not disrupt ZYG-11 binding to ELC-1. Thus, our results indicate that ZYG-11 interaction with ELC-1 uses the VHL box.
In total, several lines of evidence support ZYG-11 as an SRS of a new CBC complex. ZYG-11 physically associates with CUL-2 in vivo. ZYG-11 binds to the CBC complex adaptor ELC-1, and this association is required for the interaction of ZYG-11 with CUL-2 in vivo. ZYG-11 and ELC-1 interaction requires the VHL-box motif of ZYG-11, as expected for a CBC-complex SRS. The phenotypic overlap suggests that a CBCZYG-11 complex is required for promoting meiosis II metaphase-to-anaphase transition, the degradation of maternal cyclin B, mitotic chromosome condensation, proper localization of the PAR-2 polarity protein and cytoplasmic organization (Table 1).
Five genes have been identified as interactors of ZYG-11 in the two-hybrid assay: pal-1, mut-2/rde-3, ftt-2, him-3 and W03D8.9 (Li et al, 2004). We investigated whether RNAi depletion of any of these genes could specifically enhance or suppress the viability of the temperature-sensitive zyg-11(b2ts) mutant. pal-1 RNAi produces 100% arrested embryos (Gonczy et al, 2000), and therefore could not be analysed for its effect on viability. RNAi depletions of mut-2/rde-3, ftt-2 and W03D8.9 did not affect zyg-11(b2ts) viability (supplementary Table 1 online). By contrast, him-3 RNAi increased the percentage of zyg-11(b2ts) mutant progeny that hatched at the non-permissive temperature of 25°C (5.6% viability for zyg-11(b2ts) alone compared with 11.8% viability for zyg-11(b2ts), him-3(RNAi); P<0.01; supplementary Table 1 online). HIM-3 is required for synapsis and chiasma formation, and also for chromosome segregation during meiosis (Zetka et al, 1999). RNAi depletion of him-3 does not significantly affect the timing of meiosis II in zyg-11(mn40) mutant zygotes (data not shown). However, him-3 RNAi promotes the anaphase II extrusion of polar bodies in zyg-11(b2ts) mutants; a higher percentage of zyg-11(b2ts) embryos grown at 25°C have two polar bodies after him-3 RNAi treatment (57%; 12 out of 21) than with no RNAi treatment (30%; 9 out of 30; P<0.01). We analysed an integrated GFP∷him-3 strain and found that GFP∷HIM-3 localized to the region between meiosis I chromosomes during metaphase I, but was not present on chromosomes during anaphase I or meiosis II; this is similar to that reported for anti-HIM-3 staining (Zetka et al, 1999). zyg-11 RNAi treatment did not change the meiotic localization pattern of GFP∷HIM-3, suggesting that ZYG-11 does not regulate this event (supplementary Fig 3 online).
ZYG-11 homologues
C. elegans has two ZYG11 family members, ZYG-11 and ZER-1, Drosophila has a single homologue, sea urchin has two, and mice and humans have three each. All of the ZYG11 family members contain an ARM-like helical domain and at least three vLRR (variant leucine-rich repeat) motifs (Fig 5A; supplementary Fig 2 online). Phylogenetic analysis shows two major clades in the ZYG11 family that are supported by a 100% bootstrap score: (i) a ZYG11 clade that contains sea urchin ZYG11B, and mammalian ZYG11A and ZYG11B; and (ii) a ZER1 clade that contains sea urchin ZYG11BL, Drosophila ZYG11BL and mammalian ZYG11BL (Fig 5B). We have been unable to detect homologues in prokaryotes, protozoa, plants or fungi, suggesting that ZYG-11 homologues are restricted to metazoa.
Figure 5.
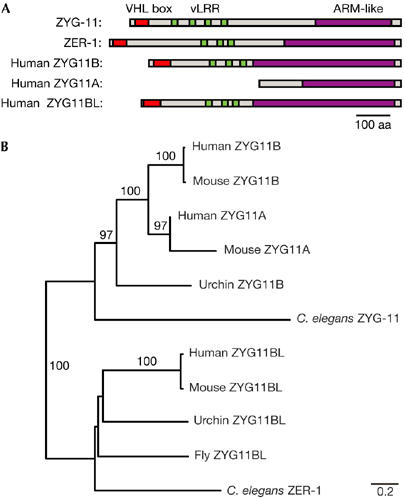
Evolution of the ZYG11 family. (A) Schematic representation of C. elegans and human ZYG-11 homologues showing the locations of the VHL box (red), vLRR (green) and Armadillo-like (ARM-like) domains (purple). (B) Neighbour-joining phylogeny of C. elegans, Strongylocentrotus purpuratus (Urchin), Drosophila melanogaster (Fly), Mus musculus (Mouse) and Homo sapiens (Human) ZYG11 family members. Bootstrap support values above 50% are listed. Scale bar, amino-acid substitutions per site. VHL, von Hippel–Lindau; vLRR, variant leucine-rich repeat.
The phylogeny suggests that the last common ancestor of both Ecdysozoa (which includes insects and nematodes) and Deuterostomes (which includes vertebrates and sea urchin) contained both ZYG11 and ZER1 ancestral genes. The absence of a ZYG11 family member in flies and mosquitoes suggests that the ancestral ZYG11 gene has been lost in the insect lineage. In mammals, ZYG11A and ZYG11B seem to have arisen by a recent duplication, and are tandemly adjacent to each other in the mouse and human genomes.
ZYG-11 homologues are CBC complex components
ZER-1 was identified by using mass spectrometry among proteins that co-purified with CUL-2–Flag from C. elegans lysate, indicating that ZER-1 associates with CUL-2 in vivo (Fig 4A). ZER-1 contains a putative VHL box (Fig 3B) and interacts with ELC-1 in the two-hybrid system, as would be expected for an SRS (Fig 4B). The function of ZER-1 is unknown at present; zer-1 RNAi depletion does not produce overt phenotypes in either wild-type or zyg-11(mn40) genetic backgrounds (Gonczy et al, 2000; data not shown). However, zer-1 RNAi modestly increases the lethality of the temperature-sensitive zyg-11(b2ts) allele (65% lethality, 96 out of 146, at 20°C with zer-1 RNAi versus 45% lethality, 53 out of 118, without RNAi, P=0.001 χ2 test).
Human ZYG11B and ZYG11BL have predicted VHL-box motifs in their N termini (Fig 3B). The largest open reading frame in the human ZYG11A cDNA lacks the N-terminal region containing the VHL box. The 5′ region of the human ZYG11A cDNA could encode an N-terminal ZYG11 polypeptide, but frameshift mutations prevent its inclusion with the larger open reading frame (data not shown). By contrast, the murine ZYG11A cDNA has a full, intact coding region. Therefore, it is possible that human ZYG11A is an expressed pseudogene.
In the two-hybrid system, human ZYG11B interacts with Elongin C alone, whereas interaction of human ZYG11BL with Elongin C requires coexpression of Elongin B (Fig 6A), which is similar to that observed with human VHL (Pause et al, 1999). Mutation of the VHL box of ZYG11B and ZYG11BL by substituting a conserved leucine residue with a serine significantly reduced or abolished interaction with Elongin C, indicating that the interactions are VHL-box-dependent (Fig 6A). To determine whether ZYG11B and ZYG11BL interact with the endogenous CUL2 protein in vivo, we transfected human 293T cells with vectors expressing VSV-G-tagged ZYG11B or ZYG11BL. Immunoprecipitation of ZYG11B-VSV-G and ZYG11BL-VSV-G co-precipitated endogenous CUL2, indicating that the human ZYG-11 homologues physically associate with CUL2 in vivo (Fig 6B). Interestingly, ZYG11B-VSV-G protein accumulated only after treatment with the proteasome inhibitor LLnL, indicating that the expressed protein is subjected to proteasome-mediated degradation.
Figure 6.
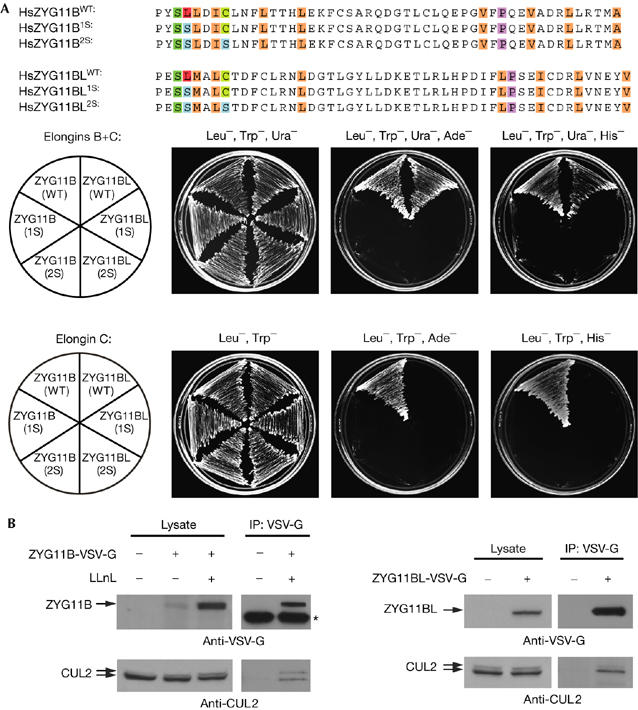
Human ZYG11B and ZYG11BL associate with Elongin C and CUL2. (A) The VHL-box sequences of human ZYG11B and ZYG11BL, with residues mutated to Serine shown highlighted in blue. Two-hybrid analysis of interaction of ZYG11B and ZYG11BL wild-type (WT) and serine mutants with Elongin C in the presence or absence of Elongin B (upper and lower panels, respectively). (B) ZYG11B and ZYG11BL associate with endogenous human CUL2 in vivo. Expression constructs that were transfected into 293T cells and LLnL pretreatment are noted above the lanes. Immunoprecipitation (IP) was performed with VSV-G antibody. Lysate and IP were analysed by western blotting with VSV-G (top) and human CUL2 (bottom) antibodies. The asterisk marks antibody heavy chain. LLnL, N-acetyl-L-leucyl-L-leucyl-L-norleucinal; VSV-G, vesicular stomatitis virus glycoprotein epitope tag.
In humans, the ZER-1 family member ZYG11BL has high levels of expression in skeletal muscle and the testes, in which it is expressed in late pachytene spermatocytes and spermatids (Feral et al, 2001). However, the functions of mammalian ZYG-11 homologues have not been explored. We propose that the ZYG11 family arose in the metazoan lineage specifically to function as SRSs in CBC E3 complexes. Given the many essential functions carried out by ZYG-11 in C. elegans, other ZYG11 homologues are predicted to function in important CUL2-dependent cellular processes.
Methods
ZYG-11 antibody. The rabbit polyclonal ZYG-11 antibody was raised against the carboxy-terminal peptide CHSLSSSPVRLVRRV conjugated to KLH (Imject Conjugation kit, Pierce Biotechnology, Rockford, IL, USA). The antiserum was affinity-purified by binding to a His-tagged ZYG-11 protein (residues 55–799) and precleared by incubation with PVDF membrane containing zyg-11(RNAi) lysate.
CUL-2–Flag affinity purification. Frozen gravid adults expressing CUL-2–Flag or control vector were ground in liquid nitrogen and then suspended in lysis buffer (50 mM HEPES, pH 7.8, 300 mM NaCl, 10% glycerol, 0.2% Triton X-100, 2 mM dithiothreitol (DTT), 1 mM EDTA and protease inhibitor cocktail (Roche, Basel, Switzerland)). Lysate was clarified by high-speed (100,000 g 40 min) centrifugation. Extracts were incubated with Flag M2 antibody–agarose beads (Sigma-Aldrich, St Louis, MO, USA) in lysis buffer with 200 mM NaCl. After washing in the same buffer, CUL–2-Flag was eluted by incubation with 0.4 mg/ml Flag peptide; Sigma). CUL-2–Flag and associated proteins were separated by sodium dodecyl sulphate–PAGE. Gels were stained with Coomassie R-250 (Sigma-Aldrich). Protein bands were in-gel digested with trypsin and analysed by matrix-assisted laser desorption/ionization-time of flight mass spectrometry at the University of Georgia Proteomics Center (Athens, GA, USA).
See the supplementary information online for other details on Methods.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Fig 1
supplementary Fig 2
supplementary Fig 3
Supplementary Table 1
Supplementary Material
Acknowledgments
We thank C. Dowd for technical assistance, H. Feng for creating the CUL-2–Flag strain, Y. Kohara for complementary DNA clones, the Caenorhabditis Genetics Center for strains and members of the Kipreos laboratory for comments on the manuscript. This work was supported by American Cancer Society grant RSG-01-251-01-DDC.
References
- DeRenzo C, Reese KJ, Seydoux G (2003) Exclusion of germ plasm proteins from somatic lineages by cullin-dependent degradation. Nature 424: 685–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein AC et al. (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107: 43–54 [DOI] [PubMed] [Google Scholar]
- Feng H, Zhong W, Punkosdy G, Gu S, Zhou L, Seabolt EK, Kipreos ET (1999) CUL-2 is required for the G1-to-S-phase transition and mitotic chromosome condensation in Caenorhabditis elegans. Nat Cell Biol 1: 486–492 [DOI] [PubMed] [Google Scholar]
- Feral C, Wu YQ, Pawlak A, Guellaen G (2001) Meiotic human sperm cells express a leucine-rich homologue of Caenorhabditis elegans early embryogenesis gene, Zyg-11. Mol Hum Reprod 7: 1115–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy P et al. (2000) Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408: 331–336 [DOI] [PubMed] [Google Scholar]
- Jang LK, Lee ZH, Kim HH, Hill JM, Kim JD, Kwon BS (2001) A novel leucine-rich repeat protein (LRR-1): potential involvement in 4-1BB-mediated signal transduction. Mol Cells 12: 304–312 [PubMed] [Google Scholar]
- Kamura T, Maenaka K, Kotoshiba S, Matsumoto M, Kohda D, Conaway RC, Conaway JW, Nakayama KI (2004) VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev 18: 3055–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues KJ, Wolf N, Wood WB, Hirsh D (1986) Two loci required for cytoplasmic organization in early embryos of Caenorhabditis elegans. Dev Biol 113: 449–460 [DOI] [PubMed] [Google Scholar]
- Kim WY, Kaelin WG (2004) Role of VHL gene mutation in human cancer. J Clin Oncol 22: 4991–5004 [DOI] [PubMed] [Google Scholar]
- Li S et al. (2004) A map of the interactome network of the metazoan C. elegans. Science 303: 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Vasudevan S, Kipreos ET (2004) CUL-2 and ZYG-11 promote meiotic anaphase II and the proper placement of the anterior–posterior axis in C. elegans. Development 131: 3513–3525 [DOI] [PubMed] [Google Scholar]
- Lu D, Ventura-Holman T, Li J, McMurray RW, Subauste JS, Maher JF (2005) Abnormal glucose homeostasis and pancreatic islet function in mice with inactivation of the Fem1b gene. Mol Cell Biol 25: 6570–6577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S, Santiago FE, Jin H, Lin D, Schedl T, Kipreos ET (2002) The Caenorhabditis elegans Skp1-related gene family: diverse functions in cell proliferation, morphogenesis, and meiosis. Curr Biol 12: 277–287 [DOI] [PubMed] [Google Scholar]
- Pause A, Peterson B, Schaffar G, Stearman R, Klausner RD (1999) Studying interactions of four proteins in the yeast two-hybrid system: structural resemblance of the pVHL/elongin BC/hCUL-2 complex with the ubiquitin ligase complex SKP1/cullin/F-box protein. Proc Natl Acad Sci USA 96: 9533–9538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20 [DOI] [PubMed] [Google Scholar]
- Sonneville R, Gonczy P (2004) zyg-11 and cul-2 regulate progression throughmeiosis II and polarity establishment in C. elegans. Development 131: 3527–3543 [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Kaelin WG Jr, Pavletich NP (1999) Structure of the VHL–ElonginC–ElonginB complex: implications for VHL tumorsuppressor function. Science 284: 455–461 [DOI] [PubMed] [Google Scholar]
- Yamanaka A, Yada M, Imaki H, Koga M, Ohshima Y, Nakayama K (2002) Skp1-related proteins in Caenorhabditis elegans: diverse patterns of interaction with Cullins and F-box proteins. Curr Biol 12: 267–275 [DOI] [PubMed] [Google Scholar]
- Zetka MC, Kawasaki I, Strome S, Muller F (1999) Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev 13: 2258–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Fig 1
supplementary Fig 2
supplementary Fig 3
Supplementary Table 1
Supplementary Material


