Abstract
The ubiquitin-binding zinc finger (UBZ) domain of human DNA Y-family polymerase (pol) η is important in the recruitment of the polymerase to the stalled replication machinery in translesion synthesis. Here, we report the solution structure of the pol η UBZ domain and its interaction with ubiquitin. We show that the UBZ domain adopts a classical C2H2 zinc-finger structure characterized by a ββα fold. Nuclear magnetic resonance titration maps the binding interfaces between UBZ and ubiquitin to the α-helix of the UBZ domain and the canonical hydrophobic surface of ubiquitin defined by residues L8, I44 and V70. Although the UBZ domain binds ubiquitin through a single α-helix, in a manner similar to the inverted ubiquitin-interacting motif, its structure is distinct from previously characterized ubiquitin-binding domains. The pol η UBZ domain represents a novel member of the C2H2 zinc finger family that interacts with ubiquitin to regulate translesion synthesis.
Introduction
The genetic material of human cells is under constant attack by endogenous oxidation and hydrolysis reactions and exogenous carcinogens such as UV light and alkylation agents. Although the majority of DNA damage is removed by nucleotide- and base-excision repair machineries, the remaining DNA lesions block the activity of the high-fidelity replicase at the replication fork during genome duplication (Hoeijmakers, 2001). To maintain genetic stability and synthesize DNA past the lesion sites (translesion DNA synthesis (TLS)), specialized DNA polymerases are used to accommodate various damaged DNA bases during replication (Prakash et al, 2005). Most of these TLS polymerases, including polymerase (pol) η, belong to the recently classified Y-family of DNA polymerases (Ohmori et al, 2001). The functional importance of TLS with pol η is highlighted by the association of defective pol η with the variant form of xeroderma pigmentosum (XP-V), a genetic disorder characterized by UV hypermutability of the cells and increased susceptibility to skin cancer (Johnson et al, 1999).
Accumulating evidence suggests that the switch between the replicase and TLS polymerase is regulated by post-translational modifications of the proliferating cell nuclear antigen (PCNA), a homotrimeric protein encircling DNA (Hoege et al, 2002; Kannouche et al, 2004). PCNA has a crucial role in the replication factory, not only contributing to the processivity of polymerases, but also acting as a scaffold to recruit proteins involved in cell-cycle control and DNA repair. Recently, two independent studies identified a novel C2H2 ubiquitin-binding zinc finger (UBZ) domain located between the catalytic domain and the PCNA-interacting peptide (PIP box) in pol η (supplementary Fig S1A online; Bienko et al, 2005; Plosky et al, 2006). Pol η can bind to unmodified PCNA, but it binds with much higher affinity to monoubiquitinated PCNA (Kannouche et al, 2004; Watanabe et al, 2004). Although mutations within this region do not alter the catalytic activity of the polymerase or its interaction with unmodified PCNA, they impair the ability of pol η to form UV-induced replication foci, presumably owing to a decreased affinity for binding to monoubiquitinated PCNA (Bienko et al, 2005; Plosky et al, 2006).
To understand the molecular basis for ubiquitination-mediated recruitment of pol η, we determined the solution structure of the UBZ domain of human DNA pol η. The binding between the pol η UBZ domain and ubiquitin was probed by nuclear magnetic resonance (NMR) titration, isothermal titration calorimetry (ITC) and site-directed spin labelling. On the basis of these data, we propose a model for the pol η UBZ domain–ubiquitin interaction.
Results And Discussion
The UBZ domain of human DNA pol η was purified under non-denaturing conditions. Mass spectrometry was used to verify the presence of a single, covalently bound zinc ion in the UBZ domain (supplementary Fig S1B online).
The solution structure of the pol η UBZ domain (Fig 1) was determined by NMR using 767 NOE, 74 dihedral angle and 45 residual dipolar coupling constraints. Except for residues at both termini, the UBZ domain is well structured, with mean pairwise r.m.s. deviations of 0.37 and 0.92 Å for the backbone and heavy atoms of residues 631–659, respectively. Additional statistics on the structures are given in the supplementary Table S1 online.
Figure 1.
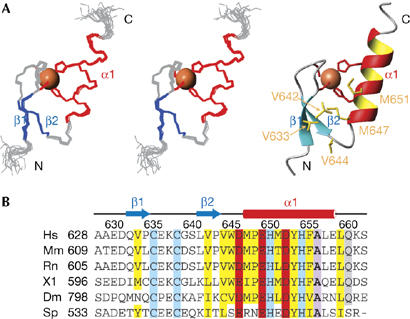
Solution structure of the polymerase η UBZ domain. (A) Backbone traces of the nuclear magnetic resonance ensemble (15 structures) in stereo view and the ribbon diagram of the UBZ domain. The zinc ion and side chains of the zinc ligands are shown in sphere and stick models, respectively. Side chains of the conserved hydrophobic core residues are also shown in stick model and are labelled in the ribbon diagram. (B) Sequence alignment of UBZ domains of pol η from different species, with zinc ligands highlighted in blue, conserved hydrophobic residues in yellow, acidic residues in red, an absolutely conserved alanine residue in purple and a highly conserved glutamine in grey. Secondary structures and residue numbers of the human pol η UBZ domain are denoted above the sequences. Abbreviations, boundaries and accession numbers for the pol η UBZ domains are as follows: Homo sapiens (Hs): residues 628–662, NP_006493; Mus musculus (Mm): residues 609–643, NP_109640; Rattus norvegicus (Rn): residues 605–639, XP_236934; Xenopus laevis (Xl): residues 596–630, AAX35543; Drosophila melanogaster (Dm): residues 798–832, BAB20905; Schizosaccharomyces pombe (Sp): residues 533–566, BAA95122. pol, polymerase; UBZ, ubiquitin-binding zinc finger.
The pol η UBZ domain consists of two short antiparallel β-strands formed by residues 632–634 and 641–643, respectively, and a carboxy-terminal α-helix spanning residues 647–658. As with other classical C2H2 zinc fingers, the α-helix packs against the β-strands with a zinc ion sandwiched between the α-helix and the β-strands. The zinc ion is coordinated through the completely conserved residues C635, C638, H650 and H654 (Fig 1). Both cysteines are located on the fingertip formed by the two β-strands, whereas the histidines are found on the α-helix and are separated by one helical turn. Mutation of H654A, which disrupts the formation of the zinc finger, abolished the UBZ domain–ubiquitin interaction (Plosky et al, 2006). The packing of the α-helix against the β-sheet is further buttressed by hydrophobic interactions mediated by the conserved residues V633, V642, V644, M647 and M651 (Fig 1).
The binding surface of the UBZ domain to ubiquitin was probed by using NMR titration. Resonances from residues D652, F655, A656, L657 and K661 of the UBZ domain were significantly perturbed (>2 s.d., 2σ) or attenuated. In addition, residues E649, H650, Y653, H654, E658, L659 and Q660 were perturbed, but to lesser extents (>1σ). All these perturbed residues are localized to the α-helix and C terminus of the UBZ domain, with the significantly perturbed residues found on the outward face of the helix (Fig 2A–C). A reciprocal NMR titration experiment was used to map the binding surface of ubiquitin to the pol η UBZ domain. Several residues of ubiquitin, including T7, L8, I13, T14, R42, F45, A46, G47, K48, Q49, H68 and L71, were either attenuated or significantly perturbed (>2σ); residues L43, L67 and L69 were also perturbed (>1σ). Most of these perturbed residues are located on the highly conserved, hydrophobic, concave surface of ubiquitin defined by residues L8, I44 and V70 (Fig 2D–F). The binding affinity (Kd) of the UBZ domain–ubiquitin complex was determined to be 73±15 μM on the basis of NMR titration and 81 μM on the basis of ITC measurement. Consistent with the binding interfaces shown by NMR titration, mutations of A656FUBZ or I44AUb completely abolished the UBZ domain–ubiquitin interaction, and mutations of Q660UBZ or V70AUb weakened the affinity of the complex approximately 1.7-fold (supplementary Fig S2 and supplementary Table S2 online).
Figure 2.
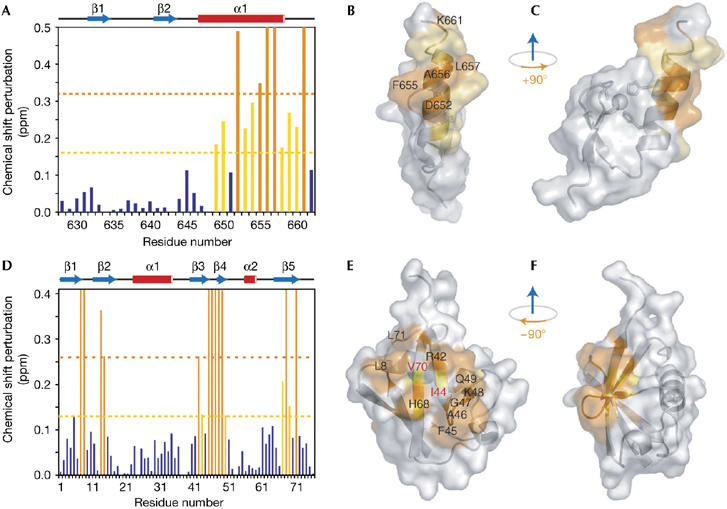
Nuclear magnetic resonance titration shows the binding interface between the polymerase η UBZ domain and ubiquitin. (A) Chemical shift perturbations of the UBZ domain plotted against residue number. Residues with chemical shift changes greater than 2σ and 1σ are shown in orange (significantly perturbed) and yellow (perturbed), respectively. (B) A surface representation of the UBZ domain with significantly perturbed residues (labelled in black) shown in orange and perturbed residues in yellow. (C) 90° right-handed rotation of (B). (D) Chemical shift perturbations of residues in ubiquitin calculated and coloured as in (A). (E) A surface representation of ubiquitin. Significantly perturbed residues (shown in orange and labelled in black) and perturbed residues (shown in yellow) are distributed along the canonical hydrophobic surface defined by residues V70, I44 (both labelled in red) and L8. (F) 90° left-handed rotation of (E). Surface representations were generated by PyMol (DeLano, 2002). pol, polymerase; UBZ, ubiquitin-binding zinc finger.
Our NMR titration and site-directed mutagenesis studies suggest that the UBZ domain binds ubiquitin through the exposed surface of its C-terminal α-helix; therefore we investigated whether the UBZ domain resembles other ubiquitin-binding single helix motifs, such as the UIM (ubiquitin-interacting motif; Hofmann & Falquet, 2001) or the recently described MIU (motif interacting with ubiquitin)/IUIM (inverted UIM) of Rabex-5 (Lee et al, 2006; Penengo et al, 2006).
The UIM forms a single helix that binds to the β-sheet of ubiquitin (Swanson et al, 2003; Wang et al, 2005; Hirano et al, 2006). It contains a signature motif, ##xxφxxAφxφSxx, in which φ and # represent large hydrophobic and acidic residues, respectively, A denotes a central invariant alanine residue and x denotes any amino acid. Structural studies of UIM–ubiquitin complexes show that the methyl group of the invariant alanine is buried in the hydrophobic cleft of ubiquitin formed by residues L8Ub, I44Ub and V70Ub (Fig 3A). This interaction is reinforced by van der Waals contacts mediated by the hydrophobic residues on the same face of the α-helix as the central alanine in the UIM and the concave hydrophobic surface of ubiquitin.
Figure 3.
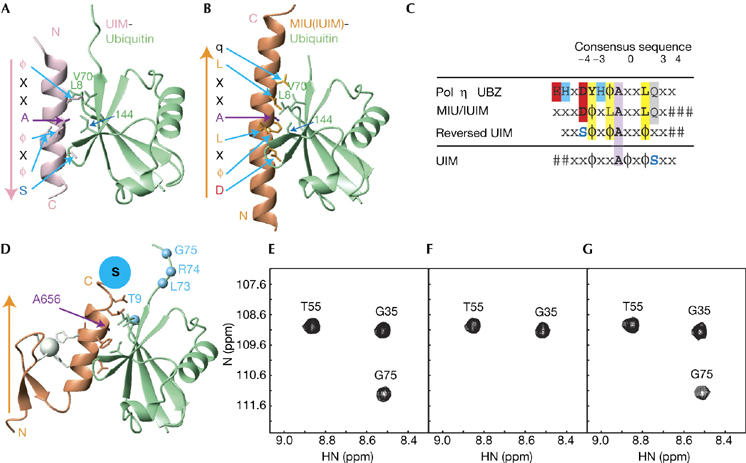
Proposed model of the polymerase η UBZ domain–ubiquitin complex. (A) A ribbon diagram of the UIM–ubiquitin complex (PDB entry 1Q0W), with conserved residues shown in stick model. The orientation of the UIM α-helix (pink) bound to ubiquitin (green) is indicated by an arrow, directed from the N to C terminus. (B) A ribbon diagram of the MIU/IUIM motif (brown) bound to ubiquitin (PDB entry 2FIF), with the orientation of the α-helix and conserved residues indicated as in (A). (C) Alignment of the consensus sequences of the UBZ domain with MIU/IUIM and reversed UIM. The central invariant alanine is highlighted in purple, conserved hydrophobic residues in yellow, acidic residues in red, zinc ligands in blue and a highly conserved glutamine residue in grey. A serine residue at the +4 position in the UIM, which is replaced by an aspartate in the MIU/IUIM and the UBZ domains, is shown in blue. (D) A model of the UBZ domain–ubiquitin complex. The C-terminal cysteine of the UBZ domain, which was modified by the spin-label reagent MTSL (denoted as S), and ubiquitin residues, the resonances of which were severely attenuated during the spin-label titration, are coloured in blue. (E) Sections of 1H-15N HSQC spectra of ubiquitin, (F) in the presence of the spin-labelled UBZ domain and (G) after addition of 5 mM dithiothreitol. Note that the amide resonance of G75Ub disappears in the presence of the spin-labelled UBZ domain (F). IUIM; inverted UIM; MIU, motif interacting with ubiquitin; PDB, Protein Data Bank; UBZ, ubiquitin-binding zinc finger; UIM, ubiquitin-interacting motif.
The UIM–ubiquitin binding is further stabilized by polar interactions at both termini of the α-helix. The hydroxyl group of the invariant C-terminal serine forms a hydrogen bond with the amide of either A46Ub or G47Ub. In addition, amino-terminal acidic residues of UIM form electrostatic interactions with basic residues (R42Ub and R72Ub) located peripherally to the hydrophobic I44Ub pocket of ubiquitin on the third and fifth β-strands.
By contrast, the single α-helical MIU/IUIM motif of Rabex-5 contains a signature motif, xxxDφxLAxxLqx###, which is similar to a UIM motif in the reverse orientation (xxSφxφAxxφxx##), with the central invariant alanine as the ‘reflection' point (Fig 3B,C; Lee et al, 2006; Penengo et al, 2006). Also similar to the UIM, the methyl group of the central alanine in the MIU/IUIM motif wedges into the hydrophobic pocket formed by L8Ub, I44Ub and V70Ub, with additional hydrophobic residues, located at positions −3, −1 and +3 with respect to the alanine, generating a contiguous hydrophobic interface with ubiquitin.
The most notable change in the MIU/IUIM consensus sequence is the replacement of a serine residue in the UIM at the +4 position with respect to the central alanine by an aspartate residue at the −4 position. In this inverted orientation, the aspartate substitution allows the formation of simultaneous hydrogen bonds with the amides of A46Ub and G47Ub. In addition, a highly conserved glutamine at the +4 position contributes to the binding affinity by forming favourable van der Waals interactions with the V70Ub side chain.
Alignment of the UBZ domain of pol η from several organisms shows a conserved sequence of EHxDYHφAxxLqxx along the α-helix (Fig 3C). The UBZ consensus is similar to the signature MIU/IUIM motif, although it contains a pair of conserved histidines for the coordination of the zinc ion and a glutamate residue at the N terminus. In the centre of the consensus sequences of the pol η UBZ domain is an invariant alanine (A656). Its resonance was severely attenuated during NMR titration with ubiquitin, and ubiquitin binding was abolished by the mutation of A656FUBZ. These observations suggest that A656 forms crucial contacts with ubiquitin, most likely through interactions with the hydrophobic pocket of L8Ub, I44Ub and V70Ub. With respect to the central alanine, the UBZ domain contains a strictly conserved leucine (L659) at the +3 position, which is also highly conserved in the MIU/IUIM motif. Similar to the MIU/IUIM consensus sequences, large hydrophobic residues are located at the −1 and the −3 positions of the UBZ domain motif.
At the −4 position, there is an invariant aspartate in the UBZ consensus, which is located at the same position as the corresponding aspartate in the MIU/IUIM consensus, and presumably forms the same hydrogen bonds with the amide groups of A46Ub and G47Ub as the latter. The functional significance of this aspartate residue is supported by mutagenesis studies showing that a D652A mutation in the pol η UBZ domain abolished ubiquitin binding (Bienko et al, 2005). In addition, the MIU/IUIM and UBZ consensus sequences share a highly conserved glutamine (Q660) at the +4 position, which is likely to provide peripheral van der Waals contacts at the binding interface. Consistently, mutation of Q660AUBZ weakened the affinity of the UBZ domain–ubiquitin complex by approximately 1.7 fold.
On basis of these analyses, we propose a model in which the UBZ domain interacts through the exposed face of the α-helix with the canonical hydrophobic patch of ubiquitin in a manner analogous to the MIU/IUIM domain (Fig 3D). To confirm our model, we used site-directed spin labelling to determine the orientation of the UBZ domain in the ubiquitin-bound complex. The introduction of paramagnetic electrons was expected to result in severe line-broadening of resonances close (<14 Å) to the spin label (Battiste & Wagner, 2000). A spin label was introduced to the UBZ domain by reacting MTSL ((1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl)methanethiosulphonate) with a modified UBZ domain containing a free cysteine (through an SSC sequence) at the C terminus. This site, which on the basis of NMR titration data, was not expected to be directly involved in the binding of the UBZ domain to ubiquitin, was chosen to provide information on the orientation of the UBZ α-helix relative to its binding partner.
Four residues, T9Ub, L73Ub, R74Ub and G75Ub, were identified in which amide resonances were not perturbed by the wild-type UBZ domain, but were severely broadened on binding to the spin-labelled UBZ domain. Furthermore, these resonances reappeared after removal of the spin label by treatment with 5 mM dithiothreitol (DTT), suggesting that these residues are located close to the spin label at the C terminus of the UBZ domain. Most of these broadened resonances belong to the residues localized to the C-terminal tail of ubiquitin, suggesting that the orientation of the bound UBZ α-helix is similar to the MIU/IUIM α-helix (Fig 3D–G).
The UBZ domain of pol η is the first C2H2 zinc finger implicated in ubiquitin binding. The structure and mode of interaction of the UBZ domain of pol η are distinctly different from the other ubiquitin-associating zinc fingers, such as the NZF, ZnF-UBP, and RUZ domains (Alam et al, 2004; Lee et al, 2006; Penengo et al, 2006; Reyes-Turcu et al, 2006), but more similar to the recognition motif used by classical zinc fingers to interact with nucleic acids (Wolfe et al, 2000). On the basis of sequence alignment, NMR titration and mutagenesis studies, we propose that the UBZ helix binds to ubiquitin in a manner similar to the binding by a MIU/IUIM helix, a model further supported by the site-directed spin-labelling experiment. The existence of a single α-helix in different structural folds that recognizes the same canonical hydrophobic surface of ubiquitin highlights the conserved architecture in the ubiquitin-signalling pathways. The UBZ domain represents a new class of ubiquitin-binding domains and its interaction with ubiquitin adds a novel function to the already diverse roles of the C2H2 zinc fingers.
The solution structure of the UBZ domain and the proposed model for its interaction with ubiquitin provide insights into the recruitment of pol η to monoubiquitinated PCNA at the stalled replication fork. The moderate binding affinity between ubiquitin and the UBZ domain of pol η is important for the rapid and reversible assembly of transient protein–protein complexes crucial for regulating translesion synthesis. Together with the C-terminal PIP box, the UBZ domain enhances the specificity and binding of the polymerase to monoubiquitinated PCNA, allowing the timely recruitment of pol η to the stalled replication fork. The moderate affinity of this interaction and the reversibility of PCNA monoubiquitination are crucial for subsequent dissociation of this highly specialized polymerase from the replication machinery beyond the DNA lesion site to restore high-fidelity genomic replication.
Methods
Molecular cloning and protein purification. The UBZ domain (residues 628–662) of human DNA pol η, a modified UBZ domain containing three additional residues SSC at the C terminus, human ubiquitin, and point mutants of the UBZ domain and ubiquitin were cloned into the pET15b vector as N-terminal His6-tagged fusion proteins and were overexpressed in and purified from bacterial cells (see the supplementary information online). The His6 tags of the proteins were removed by thrombin cleavage. The fragment of the wild-type UBZ domain, containing GSHM at the N terminus, was used for NMR studies. The isotopically enriched UBZ domain was overexpressed in M9 minimal media using [15N]-NH4Cl and [13C]-glucose as the sole nitrogen and carbon sources (Cambridge Isotope Laboratories, Andover, MA, USA). NMR samples were prepared with U-15N, U-13C/15N or 10% 13C labelling. All NMR samples were exchanged into a buffer containing 25 mM sodium phosphate, 100 mM KCl, 2 mM DTT and 5% or 100% D2O (pH 7.0) before experiments.
Mass spectrometry, isothermal titration calorimetry and binding affinity measurement. See the supplementary information online.
Structure determination and NMR titration. NMR experiments were conducted at 25°C using Varian INOVA 600 or 800 MHz spectrometers. Following standard protocols (see supplementary information online), we obtained a final structural ensemble of 15 structures (Protein Data Bank (PDB) entry 2I5O) with no NOE violations more than 0.4 Å and no dihedral angle violations more than 4°. The quality of these structures can be evaluated in supplementary Table S1 online. To map the binding interface between the pol η UBZ domain and ubiquitin, we collected a series of 1H-15N HSQC spectra of the 15N-labelled UBZ domain in the presence of increasing molar ratios of unlabelled ubiquitin; a reciprocal titration was carried out with the unlabelled UBZ domain added to 15N-labelled ubiquitin. Normalized chemical shift changes were calculated as: 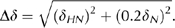
Site-directed spin labelling. Site-directed spin labelling was achieved by reacting the modified pol η UBZ domain containing a free C-terminal cysteine with a 5-fold molar excess of the spin-label reagent MTSL at 25°C for 4 h (Toronto Research Chemicals, Toronto, Canada). Excess reagent was removed by buffer exchange, and the presence of the spin label was verified by the shift of the corresponding protein band on SDS–polyacrylamide gel electrophoresis gels. A series of 1H-15N HSQC spectra were obtained to monitor the disappearance of a subset of resonances of 15N-labelled ubiquitin in the presence of increasing molar ratios of the unlabelled, MTSL-coupled UBZ domain. A control spectrum was collected at the end of titration after addition of 5 mM DTT to remove the spin label MTSL.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary Information
Acknowledgments
This work was supported by the Whitehead Foundation (to P.Z.) and by a Canadian Institute of Health Research grant (to S.S.-C.L.).
References
- Alam SL, Sun J, Payne M, Welch BD, Blake BK, Davis DR, Meyer HH, Emr SD, Sundquist WI (2004) Ubiquitin interactions of NZF zinc fingers. EMBO J 23: 1411–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battiste JL, Wagner G (2000) Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry 39: 5355–5365 [DOI] [PubMed] [Google Scholar]
- Bienko M et al. (2005) Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310: 1821–1824 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMol Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific [Google Scholar]
- Hirano S, Kawasaki M, Ura H, Kato R, Raiborg C, Stenmark H, Wakatsuki S (2006) Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nat Struct Mol Biol 13: 272–277 [DOI] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Falquet L (2001) A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci 26: 347–350 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L (1999) hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285: 263–265 [DOI] [PubMed] [Google Scholar]
- Kannouche PL, Wing J, Lehmann AR (2004) Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell 14: 491–500 [DOI] [PubMed] [Google Scholar]
- Lee S, Tsai YC, Mattera R, Smith WJ, Kostelansky MS, Weissman AM, Bonifacino JS, Hurley JH (2006) Structural basis for ubiquitin recognition and autoubiquitination by Rabex-5. Nat Struct Mol Biol 13: 264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmori H et al. (2001) The Y-family of DNA polymerases. Mol Cell 8: 7–8 [DOI] [PubMed] [Google Scholar]
- Penengo L, Mapelli M, Murachelli AG, Confalonieri S, Magri L, Musacchio A, Di Fiore PP, Polo S, Schneider TR (2006) Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell 124: 1183–1195 [DOI] [PubMed] [Google Scholar]
- Plosky BS, Vidal AE, Fernandez de Henestrosa AR, McLenigan MP, McDonald JP, Mead S, Woodgate R (2006) Controlling the subcellular localization of DNA polymerases ι and η via interactions with ubiquitin. EMBO J 25: 2847–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Johnson RE, Prakash L (2005) Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem 74: 317–353 [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Horton JR, Mullally JE, Heroux A, Cheng X, Wilkinson KD (2006) The ubiquitin binding domain ZnF UBP recognizes the C-terminal diglycine motif of unanchored ubiquitin. Cell 124: 1197–1208 [DOI] [PubMed] [Google Scholar]
- Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I (2003) Solution structure of Vps27 UIM–ubiquitin complex important for endosomal sorting and receptor downregulation. EMBO J 22: 4597–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Young P, Walters KJ (2005) Structure of S5a bound to monoubiquitin provides a model for polyubiquitin recognition. J Mol Biol 348: 727–739 [DOI] [PubMed] [Google Scholar]
- Watanabe K, Tateishi S, Kawasuji M, Tsurimoto T, Inoue H, Yamaizumi M (2004) Rad18 guides pol η to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J 23: 3886–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO (2000) DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct 29: 183–212 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary Information


