Abstract
In plants, chloroplast division is an integral part of development, and these vital organelles arise by binary fission from pre-existing cytosolic plastids. Chloroplasts arose by endosymbiosis and although they have retained elements of the bacterial cell division machinery to execute plastid division, they have evolved to require two functionally distinct forms of the FtsZ protein and have lost elements of the Min machinery required for Z-ring placement. Here, we analyse the plastid division component accumulation and replication of chloroplasts 3 (ARC3) and show that ARC3 forms part of the stromal plastid division machinery. ARC3 interacts specifically with AtFtsZ1, acting as a Z-ring accessory protein and defining a unique function for this family of FtsZ proteins. ARC3 is involved in division site placement, suggesting that it might functionally replace MinC, representing an important advance in our understanding of the mechanism of chloroplast division and the evolution of the chloroplast division machinery.
Introduction
Chloroplast division is a complex process involving components of both prokaryotic and eukaryotic origin (cf. Aldridge et al, 2005). Because of their prokaryotic origin, bacterial cell division has been used as a model for plastid division, resulting in the identification of the key plastid division components AtFtsZ1, AtFtsZ2 (Osteryoung & Vierling, 1995; Osteryoung et al, 1998), AtMinD1 (Colletti et al, 2000), AtMinE1 (Itoh et al, 2001; Maple et al, 2002) and GC1 (Maple et al, 2004; Raynaud et al, 2004). Cloning of the disrupted loci in several accumulation and replication of chloroplasts (arc) mutants (Pyke & Leech, 1991) has further shown that plastid division is controlled by prokaryote- and host-eukaryote-derived proteins residing in the plastid stroma and the cytosol.
Division of Escherichia coli is initiated by FtsZ polymerization into a contractile Z-ring and correct Z-ring placement is mediated by the Min system (cf. Rothfield et al, 2005). MinC is an FtsZ polymerization antagonist, and topological specificity is conferred on MinC by the coordinated action of MinD and MinE (cf. Rothfield et al, 2005). As in E. coli, AtFtsZ1 and AtFtsZ2 form a Z-ring at the chloroplast division site in Arabidopsis (McAndrew et al, 2001); they can interact in planta (Maple et al, 2005) and form polymers in vitro (El-Kafafi et al, 2005). Correct Z-ring placement in Arabidopsis is regulated by the higher plant Min homologues: disequilibrated AtMinD1 or AtMinE1 levels lead to chloroplast division site misplacement (Colletti et al, 2000; Maple et al, 2002). Similar to their bacterial homologues the Arabidopsis Min proteins form a complex (Maple et al, 2005) and AtMinD1 shows ATPase activity (Aldridge & Møller, 2005). However, higher plants do not harbour a MinC homologue, indicating that a non-typical MinC-like protein must have evolved to compensate for this loss.
ARC3 contains an amino-terminal domain with similarities to FtsZ proteins and a carboxy-terminal domain containing MORN (membrane occupation and recognition) motifs, linked by a novel middle domain, and was reported as a cytosolic chloroplast division component (Shimada et al, 2004). We have readdressed this and shown that ARC3 is a bona fide stromal plastid division component that interacts with several stromal division proteins and that altered ARC3 levels result in FtsZ-ring misplacement. Our data indicate that ARC3 might represent a MinC-like protein of the plastid division machinery.
Results And Discussion
ARC3 is a stromal plastid division component
ARC3 has been predicted to localize to a ring-like structure on the cytosolic face of chloroplasts (Shimada et al, 2004); however, in silico analysis predicts that the first 41 amino acids of ARC3 harbour a chloroplast-targeting transit peptide. We reinvestigated ARC3 localization using in vitro chloroplast import and protease protection assays. A full-length radiolabelled ARC3 translation product was synthesized (pARC3) in vitro, and import assays show that ARC3 is imported into pea chloroplasts, processed to the mature protein and protected from proteolytic degradation (Fig 1A, upper panel). This behaviour was identical to the stromal marker protein Rubisco small subunit precursor (Fig 1A, lower panel).
Figure 1.
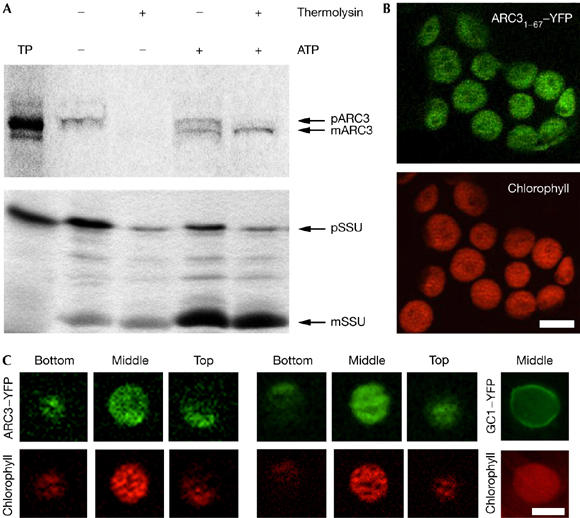
ARC3 is a stromal chloroplast protein. (A) Radiolabelled pARC3 (top panel) and pSSU (lower panel), the precursors of mature (m)ARC3 and mSSU, respectively, were incubated with isolated pea chloroplasts in the absence (−) or presence (+) of 3 mM ATP. Chloroplasts were incubated with (+) or without (−) thermolysin. Lane 1 shows 1/10 of the translation product (TP) added to each import reaction. (B) CaMV35S-driven ARC31–67–YFP transiently expressed in tobacco leaf cells. Extended focus images of YFP and autofluorescence are shown. (C) ARC3–YFP and GC1–YFP transiently expressed in tobacco leaf cells. Single plane images of the top, middle and bottom of individual chloroplasts are shown indicating that the ARC3–YFP fusion protein is distributed throughout the stroma. By contrast, GC1–YFP is associated with the chloroplast envelope. Scale bars, 5 μm. ARC, accumulation and replication of chloroplasts; CaMV, Cauliflower mosaic virus promoter; GC1, GIANT CHLOROPLAST 1; YFP, yellow fluorescent protein.
To confirm these findings in planta the N-terminal 201 bp of ARC3 were fused to YFP (yellow fluorescent protein) in pWEN18 (Kost et al, 1998). The ARC31–67–YFP fusion protein was transiently expressed in tobacco leaf cells by biolistic microprojectile transformation and samples analysed after 24 h. ARC31–67–YFP localizes exclusively to chloroplasts (Fig 1B). To define further the localization of ARC3, the full-length complementary DNA was fused to YFP in pWEN18. The 35S.ARC3.YFP cassette was cloned into the binary vector pBA002 and ARC3–YFP transiently expressed in tobacco leaf cells by infiltration. Samples were analysed after 72 h. Serial optical sections were taken through the chloroplasts, and the fusion protein was found to localize throughout the stroma (Fig 1C). As a control, a GC1–YFP (GC1 for GIANT CHLOROPLAST 1) fusion was included. Unlike ARC3.YFP, the GC1.YFP fusion protein was not distributed throughout the stroma but, as previously shown, was associated with the envelope (Fig 1C; Maple et al, 2004). These data show that ARC3 is a stromal plastid protein.
ARC3 interacts with stromal plastid division components
Stromal plastid division components do not act alone but form complexes (Maple et al, 2005). Because of the ARC3 stromal localization, we tested whether ARC3 could form part of a complex with known stromal division proteins. For all yeast two-hybrid interactions, the plastid division proteins were expressed as fusions to the Gal4-activation domain (AD) or the Gal4 DNA-binding domain (BD) from pGADT7 and pGBKT7 (Clontech, Mountain View, CA, USA). As a marker for protein–protein interactions, the relative growth of yeast on synthetic drop-out media lacking tryptophan and leucine, and synthetic drop-out media lacking tryptophan, leucine and histidine was calculated as described by Maple et al (2005). No interactions were detected between the full-length ARC3 protein fused to the BD and any of the other full-length stromal division proteins (Fig 2A; supplementary Fig 1 online). This lack of interaction might be due to membrane binding by the MORN domain in the full-length protein. Subsequently, we generated two truncations of ARC3 fused to BD to generate BD-ARC31–598 (FtsZ-like and middle domain) and BD-ARC3393–741 (middle and MORN domain). Quantitative analysis showed that BD-ARC31–598 interacts with ARC3, AtMinE1, AtMinD1 and AtFtsZ1 but not AtFtsZ2, whereas BD-ARC3393–741 interacts with AtMinE1 and AtMinD1 (Fig 2A; supplementary Fig 1 online).
Figure 2.
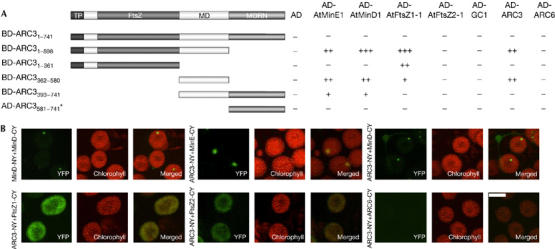
ARC3 forms part of a complex with stromal chloroplast division proteins. (A) Interactions of ARC3 with stromal plastid division components were assayed in yeast HF7c cells (*ARC3581–741 was fused to the AD and assayed for interaction with plastid division proteins fused to the BD). Domains are represented as follows: transit peptide (TP), FtsZ-like domain (FtsZ), middle domain (MD) and MORN repeat motifs. Interaction strengths are represented as three classes on the basis of the ratio of growth on −TL media to that on −HTL media: +++, ratio of >1.0; ++, ratio of 0.6–1.0; ++, ratio of 0.2–0.6; −, ratio equal to the relevant control (<0.2 in all cases). (B) BiFC assays were performed by coexpressing ARC3-NY with AtMinE1-CY, AtMinD1-CY, AtFtsZ1-CY, AtFtsZ2-CY or ARC6-CY in tobacco leaf cells. AtMinD1-NY and AtMinD1-CY were used as positive controls. Reconstituted YFP fluorescence and autofluorescence were captured by epifluorescence microscopy and the images were merged. Negative controls confirmed that these interactions were specific (see Methods; supplementary Fig 4 online). Scale bar, 5 μm. AD, Gal4 activation domain; ARC, accumulation and replication of chloroplasts; BD, Gal4 DNA-binding domain; BiFc, biomolecular fluorescence complementation; CY, YFP155–238; –HTL, media lacking histidine, tryptophan and leucine; MORN, membrane occupation and recognition; NY, YFP1–154; –TL, media lacking tryptophan and leucine; YFP, yellow fluoresent protein.
To define the domains of ARC3 that mediate these interactions, ARC3 was further divided into an FtsZ-like domain (ARC31–361), a middle domain (ARC3362–580) and a MORN domain (ARC3581–741). ARC3362–580, which has no homology with other proteins, is sufficient for the interaction of ARC3 with AtMinE1, AtMinD1 and ARC3. Further, AtFtsZ1 interacts with ARC31–361 and less strongly with ARC3362–580, indicating that determinants in both these domains mediate the interaction of ARC3 with AtFtsZ1 (Fig 2A; supplementary Fig 1 online). ARC31–361 has homology to FtsZ proteins of bacteria and higher plants; however, the tubulin signature motif and residues shown to be crucial for GTPase activity are not conserved. The FtsZ-like domain of ARC3 is therefore not a true FtsZ protein, but might have evolved to fulfil a role in the localization and interactions of ARC3. The MORN repeats (ARC3581–741) were not required for any of the ARC3 interactions, indicating that the C-terminal region of ARC3 might have a different function (Fig 2A; supplementary Fig 1 online).
To validate that the full-length ARC3 protein showed the same interaction patterns in living chloroplasts, bimolecular fluorescence complementation (BiFC) assays were preformed in planta. Tobacco leaf cells were co-infiltrated with binary vectors expressing ARC3-NY and AtMinE1-CY, AtMinD1-CY, AtFtsZ1-CY, AtFtsZ2-CY or ARC6-CY (supplementary Methods online). AtMinD1-NY and AtMinD1-CY BiFC interactions (Maple et al, 2005) were used as a positive control (Fig 2B). In agreement with the yeast interaction data, ARC3 was found to interact with AtMinD1 and AtMinE1, the reconstituted YFP fusion protein localizing to one or two spots at the poles of chloroplasts (Fig 2B). When the interaction of ARC3 with AtFtsZ1 was analysed, a strong signal of reconstituted YFP was detected, although as found for the interaction of ARC6 with AtCTD1 (Raynaud et al, 2005), YFP was detected throughout the chloroplast stroma (Fig 2B). A weak signal was also detected in cells expressing ARC3-NY and AtFtsZ2-CY, because AtFtsZ1 might act as a bridging protein in the Z-ring. In agreement with the yeast two-hybrid interaction studies, no detectable fluorescence was observed in cells expressing ARC3-NY and ARC6-CY (Fig 2B). BiFC assays were also used to confirm the domain interactions. ARC31–598.NY, ARC31–361.NY, TP.ARC3362–580.NY, TP.ARC3362–741.NY and TP.ARC3581–741.NY (TP, transit peptide; supplementary Methods online) were each coexpressed in tobacco leaf cells with AtMinE1-CY, AtMinD1-CY or AtFtsZ1-CY. The assays were in agreement with the yeast two-hybrid interaction data with the exception that no reconstituted fluorescence was detected when AtMinE1-CY or AtFtsZ1-CY was coexpressed with TP.ARC3362–580.NY. This is possibly because the interaction properties of the ARC3362–741 domain in isolation might not be sufficient to generate a detectable signal (Table 1; supplementary Fig 2 online).
Table 1.
Summary of the bimolecular fluorescence complementation assays between the domains of ARC3 and other stromal plastid division components
| CY fusion | ||||
|---|---|---|---|---|
| Empty | AtMinE1 | AtMinD1 | AtFtsZ1–1 | |
| NY fusion | ||||
| ARC31–598 | − | + | + | + |
| ARC31–361 | − | − | − | + |
| TP.ARC3362–580 | − | − | + | − |
| TP.ARC3362–741 | − | + | + | − |
| TP.ARC3581–741 | − | − | − | − |
+, reconstituted YFP fluorescence detected; −, no YFP detected. ARC, accumulation and replication of chloroplasts; TP, transit peptide.
AtFtsZ1 and AtFtsZ2 have essential, nonredundant roles during chloroplast division (Osteryoung et al, 1998; McAndrew et al, 2001), leading to speculation as to how they are functionally distinct. AtFtsZ2 interacts specifically with ARC6 (Maple et al, 2005), and the finding that ARC3 specifically interacts with AtFtsZ1 confirms a functional distinction between the two Arabidopsis FtsZ proteins. Additionally, C-terminal truncations of AtFtsZ1 previously generated in the laboratory (Maple et al, 2005) were found to be unable to interact with AD-ARC31–598 (supplementary Fig 3 online), indicating that this region of AtFtsZ1 is required for the interaction with ARC3 and that the FtsZ proteins diverged to enable different accessory components to interact with the Z-ring.
ARC3 localization
The FtsZ and Min proteins of Arabidopsis have distinct localization patterns, forming ring-like structures and discrete foci (McAndrew et al, 2001; Maple et al, 2002). Because ARC3 interacts with AtFtsZ1, AtMinE1 and AtMinD1, we investigated the detailed localization of ARC3. Full-length ARC3 was fused to YFP and transiently expressed in tobacco leaf cells. As previously reported, ARC3–YFP forms ring-like structures (Fig 3A; Shimada et al, 2004), however, ARC3–YFP also formed short filaments and discrete foci (Fig 3A).
Figure 3.
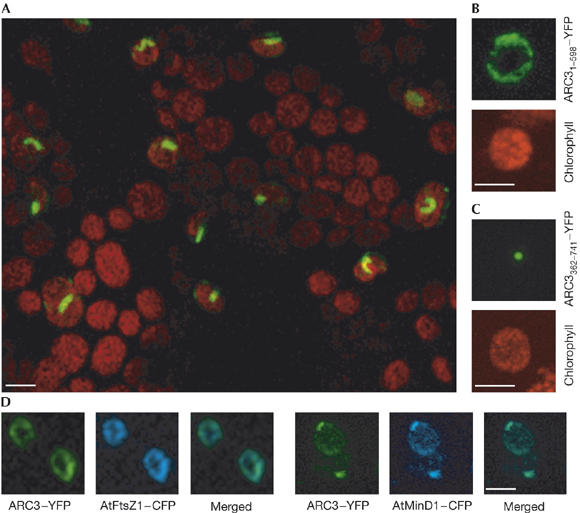
ARC3 localization is reminiscent of both the FtsZ and Min proteins. (A) ARC3–YFP visualized in the epidermal cells of tobacco leaf cells localizes to ring-like structures, short filaments and discrete foci. The merged YFP and autofluorescence image is shown. ARC31–598–YFP (B) localizes to ring-like structures and TP.ARC3362–741–YFP (C) localizes to discrete foci in chloroplasts. (D) ARC3–YFP colocalizes with both AtFtsZ1–CFP and AtMinD1–CFP. The green colour in the merged image of the CFP and YFP signals demonstrates colocalization. Scale bars, 5 μm. ARC, accumulation and replication of chloroplasts; CFP, cyan fluorescent protein; TP, transit peptide; YFP; yellow fluorescent protein.
The localization of ARC31–598 and of ARC3362–741 fused to the transit peptide of AtABC1 (Møller et al, 2001; TP.ARC3362–741) as fusions to YFP were analysed in tobacco leaf cells. ARC31–598–YFP formed ring-like structures (80%) and discrete foci (20%) inside chloroplasts (Fig 3B; data not shown) whereas TP.ARC3362–741–YFP localized exclusively to discrete spots in close proximity to the chloroplast envelope (Fig 3C). These localization patterns are in agreement with the protein–protein interactions observed (Fig 2). Subsequently, ARC3–YFP was coexpressed with either AtFtsZ1–CFP or AtMinD1–CFP. In cells expressing ARC3–YFP and AtFtsZ1–CFP, fluorescence was detected as colocalized ring-like structures (55%), whereas ARC3–YFP and AtMinD1–CFP were observed to colocalize tightly as one or two spots (80%; Fig 3D). In all other cases, only aggregates of the two proteins were observed, suggesting excessive levels of expression (data not shown). The ability of ARC3 to colocalize and interact with both AtFtsZ1 and AtMinD1 suggests that during the division process ARC3 is a dynamic protein. This behaviour might be associated with the MORN repeats of ARC3, as has recently been suggested for MORN1 of Toxoplasma (Gubbels et al, 2006).
Overexpression of ARC3 disrupts plastid division
Overexpression of stromal plastid division components leads to aberrant chloroplast division (cf. Aldridge et al, 2005). To determine whether ARC3 overexpression leads to altered chloroplast division, a CaMV35S-ARC3 (CaMV for Cauliflower mosaic virus) transgene was transformed into wild-type Arabidopsis. Analysis of 24 lines showed that mesophyll cells contain greatly enlarged chloroplasts (Fig 4A): 18 lines contained 1–5 giant chloroplasts/cell (Fig 4A; 35S.ARC3 4 and 10), whereas a small number of lines showed heterogeneity in chloroplast size and number (Fig 4A; 35S.ARC3 1). Division profiles were examined in petioles and showed that lines with greatly enlarged chloroplasts had few or no division events. However, in lines with greater heterogeneity many chloroplasts were arrested late in division, showing that ARC3 overexpression leads to chloroplast division arrest.
Figure 4.
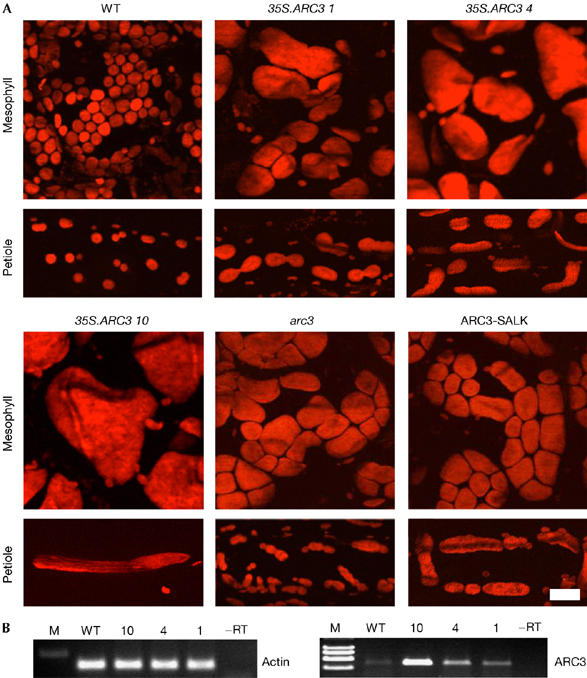
ARC3 shows MinC-like properties. (A) Chloroplast phenotypes in mesophyll and petiole cells of wild-type (WT), 35S.ARC3 primary transformants (lines 1, 4 and 10), arc3 and SALK_057144 Arabidopsis plants. Chlorophyll autofluorescence was detected, and extended focus images were generated. Scale bar, 25 μm. (B) 21-day-old wild type and 35S.ARC3 Arabidopsis primary transformants analysed for ARC3 and actin expression by RT–PCR. ARC, accumulation and replication of chloroplasts; RT–PCR, reverse transcription–PCR.
The interaction of ARC3 with AtMinE1, AtMinD1 and AtFtsZ1 suggests that ARC3 might be part of the machinery responsible for correct Z-ring placement. To investigate whether decreased levels of ARC3 lead to a chloroplast division site, misplacement arc3 was analysed (Pyke & Leech, 1992). Although arc3 mesophyll cells mostly contain 13 chloroplasts per cell, we observed cells with as few as three chloroplasts, in addition to cells frequently containing a heterogeneous chloroplast population (Fig 4A) indicative of division site misplacement. To characterize this further, we analysed chloroplast division profiles in arc3 petioles and found frequent division site misplacement, resulting in long chloroplasts with multiple division sites (Fig 4A) as in arc11 (Fujiwara et al, 2004) and in plants with elevated AtMinE1 (Maple et al, 2002). An identical phenotype was observed in an ARC3 T-DNA insertional line (Fig 4A; SALK_057144; Shimada et al, 2004). These findings show that ARC3 is involved in the correct division site placement in Arabidopsis.
Speculation
Because of its stromal localization, ARC3 can be included as part of the stromal division machinery. The ability of ARC3 to interact with both AtFtsZ1 and the Min proteins, in addition to its dual localization pattern, indicates that ARC3 is a key component of the machinery required for correct chloroplast division site placement. This hypothesis is strengthened by the finding that overexpression of ARC3 inhibits chloroplast division, whereas reduced levels of ARC3 result in aberrant placement of the chloroplast division site, analogous to the phenotypes caused by altered levels of the MinD and MinC bacterial cell division proteins (cf. Rothfield et al, 2005). Taken together, these data indicate that ARC3 fulfils a MinC-like function during chloroplast division. Further investigation of the function of ARC3—for example its novel interactions with both AtMinE1 and AtMinD1—will shed light on our understanding of the division machinery and evolution.
Methods
Plant material. Arabidopsis cv. Colombia and Nicotiana tabacum cv. Samsun were used for all experiments, unless otherwise stated. Identification of the arc3 mutant and the T-DNA insertion line (SALK_057144) has been previously described (Pyke & Leech 1992; Shimada et al, 2004).
Cloning of ARC3. All vectors used in the study and their construction are shown in the supplementary information online. Subscripts indicate the protein products (e.g., pGBKT7/ARC31–598: ARC3 fragment coding for amino acids 1–598).
A full-length ARC3 cDNA (At1g75010) was amplified from total RNA (GenElute™ Mammalian Total RNA Miniprep Kit, Sigma, Vienna, Austria) using Moloney murine leukaemia virus (M-MLV) reverse transcriptase (Promega, Mannheim, Germany), ACCUZYME™ DNA polymerase (Bioline, London, UK) and primers ARC3/6 5′-ATCATATGCCGATTTCTATGGAAC-3′ and ARC3/2 5′-ATGAGCTCTCA ATCTCCGGCGTCCACTTG-3′ (NdeI and SacI sites are underlined). The PCR product was cloned into pPCR-Script (Stratagene, La Jolla, CA, USA) to generate pPCR-Script/ARC3. The AtABC1 chloroplast transit peptide (Møller et al, 2001) was fused to ARC31084–2226 and ARC31741–2226 by splicing using overlap extension as described in the supplementary Methods online.
Image capture and analysis. Fluorescence image acquisition was carried out on a Nikon TE-2000U inverted fluorescence microscope and filters for YFP (exciter HQ500/20, emitter S535/30) and chlorophyll autofluorescence (exciter HQ630/30, emitter HQ680/40; Chroma Technologies, Rockingham, VT, USA) equipped with a Hamamatsu Orca ER 1394 cooled CCD camera. For localization studies, constructs were transfected into tobacco leaves by particle bombardment and analysed after 48 h. Volocity II software (Improvision, Coventry, UK) was used to capture 0.5 μm Z-sections to generate extended focus images.
In vitro import and protease protection assays. Transcription and translation were carried out as described by Waegemann & Soll (1995). The translation mixture was centrifuged (80,000 g, 10 min, 4°C), and the post-ribosomal supernatant was used for import studies.
Chloroplasts were isolated from the leaves of 10-day-old pea plants (Pisum sativum, var. Golf) (Waegemann & Soll, 1991). Import reactions contained chloroplasts equivalent to 20 μg chlorophyll in 100 μl import buffer (10 mM methionine, 10 mM cysteine, 20 mM potassium gluconate, 10 mM NaHCO3, 330 mM sorbitol, 50 mM HEPES–KOH, pH 7.6, 5 mM MgCl2) and 5% in vitro translation product and carried out for 20 min at 25°C. After chloroplast separation, chloroplasts were treated with thermolysin (0.5 μg/1 μg of estimated chlorophyll). Import products were separated by SDS–polyacrylamide gel electrophoresis and analysed by a phosphorimager.
Yeast two-hybrid analysis. Yeast HF7c cells were co-transformed with combinations of pGADT7 and pGBKT7 vectors according to the manufacturer's instructions (Clontech) followed by quantitative protein–protein interaction analysis (Maple et al, 2005).
Bimolecular fluorescence complementation. Fusions of full-length cDNAs to the N- and C-terminal halves of YFP (NY and CY, respectively) were constructed in pWEN-NY and pWEN-CY (Maple et al, 2005) and transferred to the binary vector pBA002 (supplementary information online). Assays were carried out by Agrobacterium co-infiltration (Yang et al, 2000) analysed after 48–72 h and repeated in triplicate. As negative controls, the expression of the NY and CY fragments alone or in combination with the stromal plastid division component cDNAs fused to NY or CY, was analysed and did not produce detectable fluorescence (supplementary Fig 4 online).
ARC3 overexpression. Full-length ARC3 cDNA, under the control of the CaMV35S promoter in pBA002 (pBA002/ARC3), was transformed into Arabidopsis by Agrobacterium-mediated floral dipping (supplementary information online) and primary transformants selected on 15 μg/ml DL-phosphinotricin (Melford Laboratories, Suffolk, UK). Reverse transcription (RT)–PCR of ARC3 and actin expression was conducted using gene-specific primers (supplementary Table 2 online). First-strand cDNA was synthesized using 2 μg total RNA, primer 5′-(T)17(A/G/C)N-3′ and M-MLV RT (Promega). One-twelfth of the RT reactions were used for each 20 cycle PCR reaction.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
supplementary methods
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft to J.S. and The Leverhulme Trust (F/00 212/M), The Functional Genomics (FUGE) Program/The Research Council of Norway and The EMBO Young Investigator Programme to S.G.M.
References
- Aldridge CP, Møller SG (2005) The plastid division protein AtMinD1 is a Ca2+-ATPase stimulated by AtMinE1. J Biol Chem 280: 31673–31678 [DOI] [PubMed] [Google Scholar]
- Aldridge CP, Maple J, Møller SG (2005) The molecular biology of plastid division in higher plants. J Exp Bot 56: 1061–1077 [DOI] [PubMed] [Google Scholar]
- Colletti KS, Tattersall EA, Pyke KA, Froelich JE, Stokes KD, Osteryoung KW (2000) A homologue of the bacterial cell division site-determining factor MinD mediates placement of the chloroplast division apparatus. Curr Biol 10: 507–516 [DOI] [PubMed] [Google Scholar]
- El-Kafafi ES, Mukherjee S, El-Shami M, Putaux JL, Block MA, Pignot-Paintrand I, Lerbs-Mache S, Falconet D (2005) The plastid division proteins, FtsZ1 and FtsZ2, differ in their biochemical properties and sub-plastidial localisation. Biochem J 387: 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara MT, Nakamura A, Itoh R, Shimada Y, Yoshida S, Moller SG (2004) Chloroplast division site placement requires dimerization of the ARC11/AtMinD1 protein in Arabidopsis. J Cell Sci 117: 2399–2410 [DOI] [PubMed] [Google Scholar]
- Gubbels MJ, Vaishnava S, Boot N, Dubremetz JF, Striepen B (2006) A MORN-repeat protein is a dynamic component of the Toxoplasma gondii cell division apparatus. J Cell Sci 119: 2236–2245 [DOI] [PubMed] [Google Scholar]
- Itoh R, Fujiwara M, Nagata N, Yoshida S (2001) A chloroplast protein homologous to the eubacterial topological specificity factor MinE plays a role in chloroplast division. Plant Phys 27: 1644–1655 [PMC free article] [PubMed] [Google Scholar]
- Kost B, Spielhofer P, Chua NH (1998) A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J 16: 393–401 [DOI] [PubMed] [Google Scholar]
- McAndrew RS, Froehlich JE, Vitha S, Stokes KD, Osteryoung KW (2001) Colocalization of plastid division proteins in the chloroplast stromal compartment establishes a new functional relationship between FtsZ1 and FtsZ2 in higher plants. Plant Physiol 127: 1656–1666 [PMC free article] [PubMed] [Google Scholar]
- Maple J, Chua NH, Møller SG (2002) The topological specificity factor AtMinE1 is essential for correct plastid division site placement in Arabidopsis. Plant J 31: 269–277 [DOI] [PubMed] [Google Scholar]
- Maple J, Fujiwara MT, Kitahata N, Lawson T, Baker NR, Yoshida S, Moller SG (2004) GIANT CHLOROPLAST 1 is essential for correct plastid division in Arabidopsis. Curr Biol 14: 776–781 [DOI] [PubMed] [Google Scholar]
- Maple J, Aldridge C, Møller SG (2005) Plastid division is mediated by combinatorial assembly of plastid division proteins. Plant J 43: 811–823 [DOI] [PubMed] [Google Scholar]
- Moller SG, Kunkel T, Chua NH (2001) A plastidic ABC protein involved in intercompartmental communication of light signaling. Genes Dev 15: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osteryoung KW, Vierling E (1995) Conserved cell and organelle division. Nature 376: 473–474 [DOI] [PubMed] [Google Scholar]
- Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY (1998) Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM (1991) Rapid image analysis screening procedure for identifying chloroplast number mutants in mesophyll cells of Arabidopsis thaliana (L.) Heynh. Plant Phys 96: 1193–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke KA, Leech RM (1992) Chloroplast division and expansion is radically altered by nuclear mutations in Arabidopsis thaliana. Plant Phys 99: 1005–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C, Cassier-Chauvat C, Perennes C, Bergounioux C (2004) An Arabidopsis homolog of the bacterial cell division inhibitor SulA is involved in plastid division. Plant Cell 16: 1801–1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud C, Perennes C, Reuzeau C, Catrice O, Brown S, Bergounioux C (2005) Cell and plastid division are coordinated through the prereplication factor AtCDT1. Proc Natl Acad Sci USA 102: 8216–8221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothfield L, Taghbalout A, Shih YL (2005) Spatial control of bacterial division-site placement. Nat Rev Microbiol 3: 959–968 [DOI] [PubMed] [Google Scholar]
- Shimada H, Koizumi M, Kuroki K, Mochizuki M, Fujimoto H, Ohta H, Masuda T, Takamiya K (2004) ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Phys 45: 960–967 [DOI] [PubMed] [Google Scholar]
- Waegemann K, Soll J (1991) Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J 1: 149–159 [Google Scholar]
- Waegemann K, Soll J (1995) Characterization and isolation of the chloroplast protein import machinery. Meth Cell Biol 50: 255–267 [DOI] [PubMed] [Google Scholar]
- Yang Y, Li R, Qi M (2000) In vivo analysis of plant promoters and transcription factors by agroinfiltration of tobacco leaves. Plant J 22: 543–551 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary methods


