Abstract
Recent population expansion and increased migration linked to urbanisation are assumed to be eroding the genetic structure of human populations. We investigated change in population structure over three generations by analysing both demographic and mitochondrial DNA (mtDNA) data from a random sample of 2351 men from twenty-two Iranian populations. Potential changes in genetic diversity (θ) and genetic distance (FST) over the last three generations were analysed by assigning mtDNA sequences to populations based on the individual's place of birth or that of their mother or grandmother. Despite the fact that several areas included cities of over one million inhabitants, we detected no change in genetic diversity, and only a small decrease in population structure, except in the capital city (Tehran), which was characterised by massive immigration, increased θ and a large decrease in FST over time. Our results suggest that recent erosion of human population structure might not be as important as previously thought, except in some large conurbations, and this clearly has important implications for future sampling strategies.
Keywords: population structure, human populations, mtDNA, Iran, demography
Introduction
The human population has grown dramatically over the last one hundred years and there has been a corresponding increase in migration to urban areas due to better healthcare, education and work opportunities. It has been suggested that demographic expansion and migration are scrambling the genetic structure of human populations (Cavalli-Sforza et al, 1991) and as a response, projects have been initiated to sample the genetic diversity of the world's populations before their genetic identity is lost for good (Cann et al, 2002, The Human Genome Diversity Project, HGDP, and more recently the Genographic Project). It remains unclear though whether these recent phenomena have already produced changes in the genetic structure of human populations and whether the suggestion that human populations are homogenising into a “global melting pot” is truly justified.
This is an important question from an intrinsic or historical perspective (e.g. for understanding the complexity of human migration and demographic history) but is also relevant to applied research in human genetics and medicine. In targeted drug administration, association studies and studies of gene function for example, even small amounts of admixture can lead to false positive results and failure to detect genuine associations (e.g. Pritchard et al, 1999, Deng et al, 2002, Hirschhorn et al, 2002, Freedman et al, 2004, Marchini et al, 2004, Helgason et al, 2005).
Although obtaining ancestral information from sampled individuals is quite a widespread practice (particularly for association studies e.g. Ardlie et al, 2002), this information has rarely been used to investigate changes in population structure over time (see Helgason et al, 2005 for an exception). Knowing the place of birth of a sampled individual, their parents and grandparents, provides a means to test whether increased migration during recent decades really has eroded human population structure. An important related question is whether increased migration (and the assumed erosion of population structure) is a general phenomenon or whether it is confined to major conurbations such as capital cities. Immigration to major cities, which seems a fair expectation, will have important consequences for sampling because limitations due to cost and accessibility mean that samples are often obtained from major cities and assumed to be representative.
Here, we explore the hypothesis that in recent generations, population expansion and increased migration (linked to urbanisation) have eroded signals of human population structure. We performed a case study of the Iranian population, using an intensive, unbiased sampling procedure and collecting information on the birthplace of sampled individuals, parents and grandparents. In the last three generations, the Iranian population has more than tripled in size to its present tally of >68 million. Iran is a large country (>1.6 million km2), and its populations, which are highly diverse in terms of culture and language, were relatively isolated geographically until the second half of twentieth century. Since then, major highways and railroads have been constructed to connect population centres, and the proportion of the population living in urban areas has more than doubled (from 27% in 1950 to 64% in 2000, United Nations Population Division). Migration, particularly from rural areas towards major cities, is therefore expected to have increased substantially in recent generations.
Methods
We sampled 2351 unrelated men from twenty-two populations, which comprehensively represent the diversity of Iran in terms of culture, language and geography (Fig. 1, Table 1). Samples were randomly collected from consenting volunteers at transfusion centers to ensure an unbiased sampling strategy. Information was obtained on the place of birth of the sampled individual (generation t) and on the place of birth of their parents (generation t−1) and grandparents (generation t−2). As mtDNA does not recombine, we could simply assign sequences to populations in each generation using place of birth of the sampled individual, their mother or grandmother. Such a strategy is equivalent to random sampling in the two previous generations as long as the sample does not comprise a large proportion of siblings and cousins, a situation we have been careful to avoid.
Fig. 1.
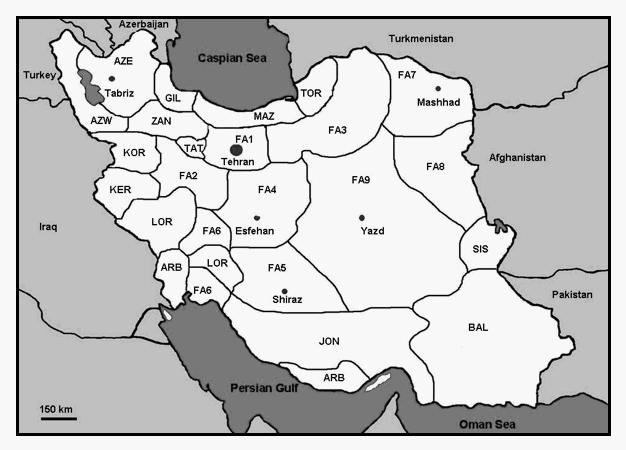
Sampling information.
Population codes and sample sizes (n=number of individual samples collected in each locality) are as in Table 1. The large dot represents the capital, Tehran (>10 million inhabitants), smaller dots are large conurbations ≥one million inhabitants. Borders in the figure roughly correspond to cultural regions based on ethnicity and language (see Ethnologue homepage for more information).
Table 1.
Sampling information
| Population code | Population name | Locality | Sample size |
|---|---|---|---|
| ARB | Arab | Khuzestan and Boushehr Provinces | 96 |
| AZE | Azeri | East Azerbaijan and Ardabil Provinces | 110 |
| AZW | Azeri | West Azerbaijan Province | 102 |
| BAL | Balochi | Sistan and Balochestan Province (Balochi area) | 135 |
| FA1 | Fars | Tehran and Qazvin Provinces | 390 |
| FA2 | Fars | Qom, Markazi and Hamedan Provinces | 75 |
| FA3 | Fars | Semnan Province | 55 |
| FA4 | Fars | Esfehan Province | 105 |
| FA5 | Fars | Shiraz Province | 45 |
| FA6 | Fars | North Khorasan Province | 70 |
| FA7 | Fars | South Khorasan Province | 50 |
| FA8 | Fars | Khuzestan Province | 52 |
| FA9 | Fars | Yazd and Kerman Provinces | 91 |
| GIL | Gilaki | Gilan Province | 125 |
| JON | Jonobi | Hormozghan Province | 100 |
| KER | Kerman | Kermanshah Province | 120 |
| KOR | Kords | Kordestan Province | 0* |
| LOR | Lorestani | Lorestan Province | 125 |
| MAZ | Mazandrani | Mazandaran Province | 120 |
| SIS | Sistani | Sistan and Balochestan Province (Sistani area) | 145 |
| TAT | Takestan | Qazvin Province | 70 |
| TOR | Torkaman | Golestan Province | 90 |
| ZAN | Zanjani | Zanjan Province | 80 |
| Total | 2351 | ||
DNA was extracted using the PAXgene blood DNA kit (Preanalytix). MtDNA control region sequences (both HVSI and HVSII) were obtained following PCR and sequencing with standard primers and protocols (Mogentale-Profizi et al, 2001, Torroni et al, 1996).
Genetic diversity was computed within populations (theta from the number of segregating sites, θs= 2Neμ, where Ne is the effective population size and μ the mutation rate (Watterson, 1975) and genetic structure estimated among populations (population specific FST, Wright, 1969) using DnaSP v4 (Rozas et al, 2003 and ARLEQUIN v3 (Excoffier et al, 2005) respectively. Both θs and FST were obtained per population for each of the three generations and then averaged over populations. The mtDNA sequence data generated in this article are freely available from the authors on request.
Results
Fig. 2 illustrates the percentage of randomly sampled individuals from each locality whose grandparents and/or parents were born in the same area, compared to those individuals that were born in the sampling locality, but whose parents or grandparents were born outside the area (“immigrants”). For simplicity, if the sampled individual and their grandparents were born in the same locality, it was assumed that the parents were also born in that locality (this was the case for all but three individuals in the whole sample set). Patterns of immigration in the last three generations are similar for females and males (Fig. 2 a and b respectively), although there is a very slight tendency for males to migrate more than females (over all populations χ2=4.77, 1 d.f., p<0.05). Immigration has been low in the majority of populations (less than 10% for all but three populations). In striking contrast, the majority of individuals sampled in Tehran (FA1) have parents or grandparents from outside the area (62% have immigrant mothers or grandmothers and 69% fathers or grandfathers). In the majority of cases, immigrants come from adjacent populations, and have therefore traveled very short distances (less than 300 km, data not shown). Although immigrants to Tehran have traveled further, immigration declines with distance and effectively ceases after 1000 km. In Tehran there is less than a 50% chance of sampling an mtDNA sequence that was present two generations ago, and approximately a one in six chance of sampling an Azeri East mtDNA (data not shown), which illustrates that caution is needed when sampling in the capital city.
Fig. 2.
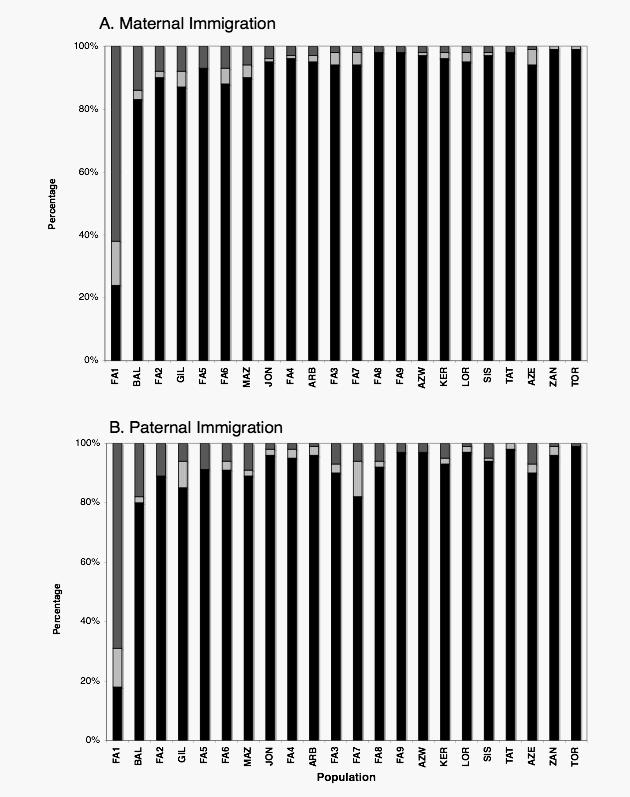
Demographic data.
Percentage of a) female and b) male immigrants per population. Dark grey: individuals with immigrant parents, Light grey: individuals with immigrant grandparents, Black: individuals with both parents and grandparents born locally. Population order corresponds to the decreasing percentage of individuals with immigrant mothers (a). Order of populations in b) is the same as for a) to allow direct comparison.
Mean within-population genetic diversity is very similar over the three generations (θ̅s(t-2) = 0.369, θ̅s(t-1) = 0.369, θ̅s(t) = 0.361, one-way repeated measures ANOVA F=1.926, d.f.=2, p=0.178, not significant, Fig. 3a). However three populations do show deviation from this pattern. A slight increase in θs over time is observed in the Balochi population (BAL θs(t−2)= 0.027, θs(t−1)= 0.029, θs(t)=0.035) and in the area of Masshad (FA6 θs(t−2)= 0.030, θs(t−1)= 0.032, θs(t) 0.036). By contrast a marked increase in genetic diversity over time is seen in Tehran (FA1 θs(t−2)= 0.043, θs(t−1)= 0.047, θs(t)= 0.057, Fig. 3a).
Fig. 3.
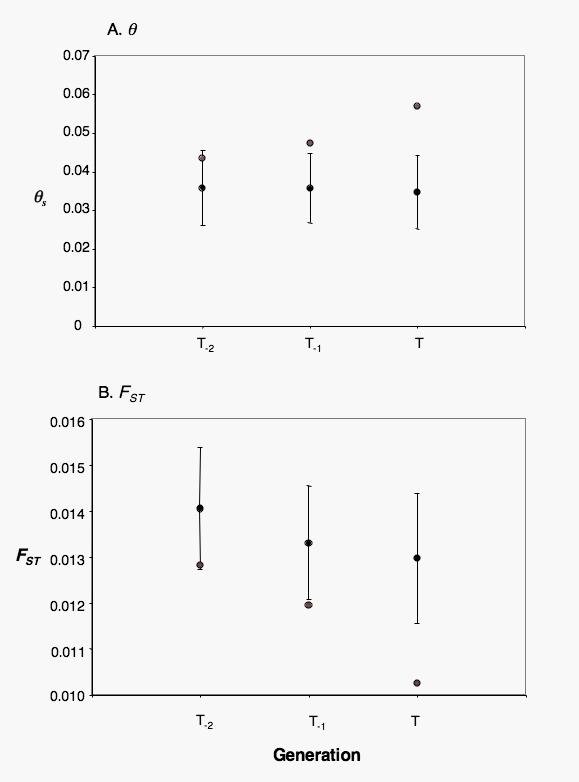
Genetic data: a) Mean within-population genetic diversity (θs) per generation and b) Mean population specific F̅ST per generation calculated from mtDNA. Black circles: mean (± SD error bars) for all populations except FA1 (Tehran); grey circles: FA1.
There is a slight decrease in mean pairwise population specific F̅ST per generation (Fig. 3b: F̅ST(t-2) = 0.0140, F̅ST(t-1) = 0.0133, F̅ST(t) = 0.0130 when Tehran is excluded from the calculations). Although this change is small, the effect is highly significant (one-way repeated measures ANOVA F=35.77, df=2, p<0.001). Note that this effect is not detected if FST is computed for each population against all other populations pooled together. A quantitatively more substantial decrease in FST over time is seen in Tehran (Fig. 3b: Tehran (FA1) FST(t−2)= 0.0128, FST(t−1) = 0.0120, FST(t)= 0.010).
Discussion
During the 20th century the average annual population growth rate in Iran was among the highest in the world (peaking at ∼3.6% between 1976 and 1986 in relation to the slackening of family planning laws following the Islamic Revolution in 1979). The increase in population size, as well as development of transport, industry and education and health services, is expected to have had a significant impact on recent migrations. Long distance migrations to major cities are assumed to have increased but more local scale migrations are also expected. We therefore predicted increased migration to be apparent in our demographic data and to translate to increased genetic diversity within populations and lowering of genetic distance among populations over time (i.e. decreased population structure). In contrast we found no difference in within-population diversity over time for most populations, and a quantitatively small, but significant decrease in population structure (Fig. 3). Only the capital, Tehran, showed considerable demographic and genetic evidence of immigration (Figs 2 and 3). In fact, sampling at random in Tehran would produce only a one in two chance of selecting an individual with native parents and grandparents. This result has important consequences for other sampling strategies, which often concentrate on capitals or other large cities, and reiterates the need to avoid major cities as sources of samples for population genetic analyses (e.g. Bowcock and Cavalli-Sforza 1991).
Patterns of migration between men and women generally suggest greater patrilocality and female-biased dispersal on local and regional scales (e.g. Salem et al, 1996, Seilestad et al, 1998, Mesa et al, 2000, Thomas et al, 2000, Oota et al, 2001, Wilson et al, 2001, Wen et al, 2004). By contrast, the demographic data presented here indicates that, while patterns of male and female migration to Tehran and overall are similar (Fig. 2) there is a slight tendency for men to migrate more often and further than women (over all populations χ2=4.77, 1 d.f., p<0.05). That this difference in migration patters is only slight suggests that our main mtDNA results should also hold for Y chromosomes.
Gathering data on an individual's place of birth clearly provides a unique insight into human population structure over time, and we highlight the importance of collecting this type of information during sampling. We expected recent migration within Iran to be considerable due to rapid population growth and increased urbanisation. However, despite extensive sampling, and inclusion of several major conurbations (six cities with over one million inhabitants), both our genetic and demographic data are consistent with only slight erosion of population structure in the last three generations, except in the capital city. These results do not therefore support the idea that human population structure has been extensively scrambled in recent decades, but do illustrate that sampling in capital cities (and preferably other very large cities) should be avoided unless ancestral information is obtained.
The dramatic population expansion and increased urbanisation seen in Iran during the twentieth century, as well as the size and location of the country, make it a particularly suitable case study for investigating recent changes in human population structure. The demographic processes that have shaped the diversity of the Iranian population are similar to those that have shaped the whole of the Middle East region. Among-population variation in this region is low compared to the worldwide average (1.3% compared to 5.4%, Rosenberg et al, 2002), which suggests that populations are subject to more migration (or have a recent common ancestry, which seems unlikely) than in more remote parts of the world. Finally, the rate of urbanisation in Iran is similar to that of other developing countries (e.g. China, Brazil, Philippines, Argentina, United Nations Population Division). Therefore, while the implications of this case study may not be appropriate for the human population as a whole, we expect them to be applicable on a broad scale, to other developing countries with similar demographic histories.
Acknowledgements
We would like to thank Laurent Lehmann, Hua Liu, Andrea Manica, Towfique Raj, Steve Russell, Camille Szmaragd, Chris Tyler-Smith, for discussion and suggestions, two anonymous reviewers for their helpful comments and the BBSRC of Great Britain for funding.
Footnotes
Web Resources
Ethnologue http://www.ethnologue.com/
Genographic Project, https://www3.nationalgeographic.com/genographic/
Human Genome Diversity Project, http://www.stanford.edu/group/morrinst/hgdp.html
United Nations Populations Division, http://www.un.org/esa/population/unpop.htm
References
- Ardlie KG, Lunetta KL, Seielstad M. Testing for population subdivision and association in four case-control studies. Am J Hum Genet. 2002;71:304–311. doi: 10.1086/341719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowcock A, Cavalli-Sforza L. The study of variation in the human genome. Genomics. 1991;11:491–498. doi: 10.1016/0888-7543(91)90170-j. [DOI] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand M-F, Morel V, Piouffre L, et al. A Human Genome Diversity Cell Line Panel. Science. 2002;296:261b–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Wilson AC, Cantor CR, Cook-Deegan RM, King M-C. Call for a worldwide survey of human genetic diversity: A vanishing opportunity for the Human Genome Project. Genomics. 1991;11:490–491. doi: 10.1016/0888-7543(91)90169-f. [DOI] [PubMed] [Google Scholar]
- Deng H-W, Gao G, Li J-L. Estimation of deleterious genomic mutation parameters in natural populations by accounting for variable mutation effects across loci. Genetics. 2002;162:1487–1500. doi: 10.1093/genetics/162.3.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, et al. Assessing the impact of population stratification on genetic association 215 studies. Nat Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- Helgason A, Yngvadottir B, Hrafnkelsson B, Gulcher J, Stefansson K. An Icelandic example of the impact of population structure on association studies. Nat Genet. 2005;37:90–95. doi: 10.1038/ng1492. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nat Genet. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- Mesa NR, Mondragon MC, Soto ID, Parra MV, Duque C, Ortiz-Barrientos D, et al. Autosomal, mtDNA, and Y-chromosome diversity in Amerinds: Pre- and post-Columbian patterns of gene flow in South America. Am J Hum Genet. 2000;67:1277–1286. doi: 10.1016/s0002-9297(07)62955-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogentale-Profizi N, Chollet L, Stevanovitch A, Dubut V, Poggi C, Pradie MP, et al. Mitochondrial DNA sequence diversity in two groups of Italian Veneto speakers from Veneto. Ann Hum Genet. 2001;65:153–166. doi: 10.1017/S0003480001008545. [DOI] [PubMed] [Google Scholar]
- Najmabadi H, Neishabury M, Sahebjam F, Kahrizi K, Shafaghati Y, Nikzat N, et al. The Iranian Human Mutation Gene Bank: A data and sample resource for worldwide collaborative genetics research. Hum Mutat. 2003;21:146–150. doi: 10.1002/humu.10164. [DOI] [PubMed] [Google Scholar]
- Oota H, Settheethamishida W, Tiwawech D, Ishida T, Stoneking M. Human mtDNA and Y-chromosomal variation is correlated with matrilocal versus patrilocal residence. Nature. 2001;29:20–21. doi: 10.1038/ng711. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Salem AH, Badr FM, Gaballah MF, Pääbo S. The genetics of traditional living: Y-chromosomal and mitochondrial lineages in the Sinai Peninsula. Am J Hum Genet. 1996;59:741–743. [PMC free article] [PubMed] [Google Scholar]
- Seielstad MT, Minch E, Cavalli-Sforza LL. Genetic evidence for a higher female migration rate in humans. Nat Genet. 1998;20:278–280. doi: 10.1038/3088. [DOI] [PubMed] [Google Scholar]
- Torroni A, Huoponen K, Francalacci P, Petrozzi M, Morelli L, Scozzari R, et al. Classification of European mtDNAs from an analysis three European populations. Genetics. 1996;144:1835–1850. doi: 10.1093/genetics/144.4.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas MG, Parfitt T, Weiss DA, Skorecki K, Wilson JF, le Roux M, Bradman N, Goldstein DB. Y chromosomes traveling south: The Cohen modal haplotype and the origins of the Lemba - the “black Jews of Southern Africa”. Am J Hum Genet. 2000;66:674–686. doi: 10.1086/302749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Wen B, Li H, Lu DR, Song XF, Zhang F, He YG, et al. Genetic evidence supports demic diffusion of Han culture. Nature. 2004;431:302–305. doi: 10.1038/nature02878. [DOI] [PubMed] [Google Scholar]
- Wilson JF, Weiss DA, Richards DA, Thomas MG, Bradman N, Goldstein DB. Genetic evidence for different male and female roles during cultural transitions in the British Isles. Proc Natl Acad Sci USA. 2001;98:5978–5983. doi: 10.1073/pnas.071036898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. Evolution and the Genetics of Populations II. Chicago: The Theory of Gene Frequencies University of Chicago Press; 1969. [Google Scholar]


