Abstract
OBJECTIVES
Most patients with Barrett’s esophagus do not progress to cancer, but those who do seem to have markedly increased survival when cancers are detected at an early stage. Most surveillance programs are based on histological assessment of dysplasia, but dysplasia is subject to observer variation and transient diagnoses of dysplasia increase the cost of medical care. We have previously validated flow cytometric increased 4N fractions and aneuploidy as predictors of progression to cancer in Barrett’s esophagus. However, multiple somatic genetic lesions develop during neoplastic progression in Barrett’s esophagus, and it is likely that a panel of objective biomarkers will be required to manage the cancer risk optimally.
METHODS
We prospectively evaluated endoscopic biopsies from 325 patients with Barrett’s esophagus, 269 of whom had one or more follow-up endoscopies, by a robust platform for loss of heterozygosity (LOH) analysis, using baseline 17p (p53) LOH as a predictor and increased 4N, aneuploidy, high-grade dysplasia, and esophageal adenocarcinoma as outcomes.
RESULTS
The prevalence of 17p (p53) LOH at baseline increased from 6% in negative for dysplasia to 57% in high-grade dysplasia (p < 0.001). Patients with 17p (p53) LOH had increased rates of progression to cancer (relative risk [RR] = 16, p < 0.001), high-grade dysplasia (RR = 3.6, p = 0.02), increased 4N (RR = 6.1, p < 0.001), and aneuploidy (RR = 7.5, p < 0.001).
CONCLUSIONS
Patients with 17p (p53) LOH are at increased risk for progression to esophageal adenocarcinoma as well as high-grade dysplasia, increased 4N, and aneuploidy. 17p (p53) LOH is a predictor of progression in Barrett’s esophagus that can be combined with a panel of other validated biomarkers for risk assessment as well as intermediate endpoints in prevention trials.
INTRODUCTION
The incidence of esophageal adenocarcinoma has been rising rapidly in the United States and several regions of Western Europe since the 1970s (1–4). Unfortunately, most esophageal adenocarcinomas present at an advanced stage in which the mortality is >90% (5). Barrett’s esophagus is the only established precursor of esophageal adenocarcinoma, and a systematic protocol of endoscopic biopsies can detect cancers arising in Barrett’s epithelium at an early, curable stage (6–9). However, multiple studies have shown that most patients with Barrett’s esophagus do not progress to cancer (10–13). These observations have led to dissociation between endoscopic practice patterns, in which most patients with Barrett’s esophagus are followed annually or biennially (14), and the results of cost-effectiveness analyses, which suggest that 5-yr surveillance intervals would be more appropriate (15).
One approach to management of the cancer risk in Barrett’s esophagus is to develop a panel of biomarkers that identify patient subsets with low and high risks for neoplastic progression (16, 17). We have recently shown by prospective evaluation of >300 patients with Barrett’s esophagus that flow cytometric abnormalities (increased 4N fractions and aneuploidy) are predictors of progression to esophageal adenocarcinoma (18). These results are consistent with other prospective studies indicating that DNA content flow cytometry identifies patients at increased risk for progression to intermediate endpoints (19–21). However, multiple somatic genetic lesions develop during the multistep progression to cancer in Barrett’s esophagus, and it is likely that a single biomarker will assess only one or a limited number of stages of progression. Therefore, a panel of biomarkers will probably be needed for risk stratification for endoscopic surveillance and intervention strategies tailored to stage of progression.
Much has been learned in recent years about the molecular pathogenesis of esophageal adenocarcinoma (17, 22–25). It is clear that genetic errors in Barrett’s epithelial cells lead to selection of abnormal clones that can spread by a process of clonal expansion to occupy large regions of esophageal mucosa (26–28). Some cells develop progressive instability, leading to the evolution of new clones with additional genetic errors and the emergence of cancer (20, 25–27, 29, 30). Biomarkers accurately identifying patients at risk for progressive instability and clonal evolution should therefore be useful in detecting the subset of patients with Barrett’s esophagus at increased risk for cancer. Once these biomarkers are validated as predictors of progression to esophageal adenocarcinoma, they could also be used as surrogate endpoints in prevention trials.
The p53 gene is a cell cycle control gene that prevents cells with DNA breaks from entering DNA synthesis where the breaks could be replicated, cause chromosome damage, and lead to progressive genetic instability and cancer (31, 32). The p53 gene is located at chromosome 17p13, and lesions in p53 are among the most common genetic abnormalities in human malignancies (33). Most people inherit two normal alleles of p53, one from the mother and one from the father, and most Barrett’s segments have two normal p53 alleles. However, during progression to cancer in Barrett’s esophagus, one allele of p53 is inactivated by mutation and the second is lost by a mechanism termed loss of heterozygosity (LOH). Loss of heterozygosity develops when there is loss of genetic information on chromosome 17p as a result of deletion of a portion of the chromosome or other events such as mitotic recombination or gene conversion (34). 17p (p53) LOH is the most common genetic lesion in esophageal adenocarcinomas and is found to occur in up to 95–100% of cases when highly purified neoplastic cell populations are evaluated (25, 35–39). The p53 gene has been strongly implicated as the target for 17p LOH because p53 mutations have been reported in up to 92% of esophageal adenocarcinomas (35, 37, 40–42). Several studies have reported 17p LOH in premalignant epithelium surrounding esophageal adenocarcinomas in esophagectomy specimens, indicating that it develops before cancer (29, 37, 42). Finally, inactivation of p53 by 17p (p53) LOH and mutation seems to be a relatively early event in neoplastic progression in Barrett’s esophagus because it develops in diploid cells before aneuploidy and other LOH events involving chromosomes 5, 13, and 18 (25, 26, 29, 43). Thus, 17p (p53) LOH is a promising biomarker for assessing risk of neoplastic progression in Barrett’s esophagus (24).
Traditional means of LOH analysis are labor intensive and not readily adaptable to the large sample numbers required for clinical or population science studies for several reasons. First, endoscopic biopsies are heterogeneous, containing genetically normal stromal cells that can obscure the detection of LOH in neoplastic epithelial cells (44). Second, endoscopic biopsies are small, making it difficult to obtain sufficient DNA to perform comprehensive genotyping (45). Third, manual autoradiographic LOH assessment cannot achieve the throughput required for clinical and population-based studies. Thus, it has been difficult previously to assess LOH as a predictor of progression in large numbers of biopsy samples to determine its potential utility in identifying risk subsets for progression to cancer in Barrett’s esophagus.
We have recently reported a method for LOH analysis for clinical studies that uses flow sorting to purify neoplastic cells, primer-extension preamplification to increase the quantity of DNA, and fluorescent genotyping to increase throughput (46). This method permits high-throughput analysis for large-scale clinical studies while simultaneously assessing 17p (p53) LOH, increased 4N, and aneuploidy in a single endoscopic biopsy (26). Here we report baseline characterization of a cohort of 325 patients with Barrett’s esophagus for the presence or absence of 17p (p53) LOH as well as determining its prognostic significance in identifying low and high risk patient subsets for progression to cancer and to validated intermediate markers of neoplastic progression, including increased 4N fractions, aneuploidy, and high-grade dysplasia.
MATERIALS AND METHODS
Patients
Eligible patients had metaplastic columnar epithelium in esophageal biopsies, no history of esophageal malignancy, and an endoscopic biopsy evaluation in the Seattle Barrett’s Esophagus Study between January 5, 1995, and December 2, 1999. A total of 325 patients, including 256 men and 69 women, met entry criteria for baseline evaluation; 269 of these have had one or more follow-up endoscopies in the Seattle Barrett’s Esophagus Study. The baseline endoscopy for this study was defined as the first after January 1, 1995. The mean and median follow-up times among the 243 patients without cancer at last endoscopy were 34 and 36 months, respectively. Patients were counseled concerning risks and benefits of surveillance and informed of alternatives, including esophagectomy for high-grade dysplasia. The Seattle Barrett’s Esophagus Study was approved by the Human Subjects Division of the University of Washington in 1983 and renewed annually thereafter, with reciprocity from the Fred Hutchinson Cancer Research Center since 1993.
Endoscopy and Biopsy
Endoscopy and biopsy were performed as described previously (47). Patients with a history of high-grade dysplasia had four-quadrant biopsies at 1-cm intervals in the Barrett’s segment, whereas those without a history of high-grade dysplasia underwent four-quadrant surveillance at 2-cm intervals. Multiple biopsies were taken of any endoscopically visible abnormalities.
Histology
Biopsies were fixed, processed, and interpreted as described previously (47). Patients were classified according to the maximum abnormality (negative, indefinite, low-grade dysplasia, high-grade dysplasia, and cancer) in any biopsy at a given endoscopy. Histological interpretations were made without knowledge of 17p or flow cytometric status. Histological criteria for diagnosing esophageal adenocarcinoma were described previously (48).
Flow Cytometry
Biopsy specimens were frozen in dimethyl sulfoxide and stored at −80°C. Each frozen specimen was minced and processed by DNA content or Ki67/DNA content flow cytometry as described previously (47). Ki67 is a proliferation-associated antibody that identifies an antigen expressed in G1, S, and G2, but not G0 phases of the cell cycle (49, 50). Using Ki67/DNA content flow cytometry, Ki67-positive G1 (2N) and, when present, increased 4N or aneuploid fractions were sorted. A DNA content 4N fraction of >6% was classified as abnormal as described previously (47). Flow cytometric interpretations were made without knowledge of 17p or histological status.
LOH Analysis
LOH analysis was typically performed on flow-purified biopsy samples taken at 2-cm intervals in the Barrett’s segment, as described previously (26). We evaluated 1824 flow cytometrically purified DNA samples. An average of 5.7 (range = 1–29) flow-purified DNA samples, depending on Barrett’s segment length, and a constitutive control were evaluated for each patient. DNA was extracted from the flow-purified neoplastic populations of one biopsy specimen per level of the Barrett’s segment using either standard phenol/chloroform or the Puregene DNA Isolation Kit as recommended by the manufacturer (Gentra Systems, Minneapolis, MN). Whole genome amplification using primer extension preamplification (PEP) was performed as described previously (45, 46). Locus-specific primers labeled with either FAM, TET, or HEX from Research Genetics (Huntsville, AL) included D17S1298 (17p13.3), TP53 (17p13.1) pentanucleotide repeat, TP53 (17p13.1) dinucleotide, D17S1537 (17p13.2), D17S786 (17p13.1), D17S974 (17p12), D17S1303 (17p12), and D17S1288 (17p11). Locus-specific PCR reagents and conditions were described previously (26). PCR products were run on an ABI 373 or 377 DNA sequencer using the internal size standard Genescan-500. After each gel run, lanes were manually tracked and data were visually inspected using Genescan and processed using Genotyper software (ABI). Allele ratios were determined by measuring the fluorescence intensity (peak height) of the smaller (basepair) allele “A” relative to the fluorescent unit intensity of the larger allele “B”(A/B). LOH was determined by assessing the ratio of peak heights in tumor or neoplastic tissue samples relative to the ratio in the corresponding normal control.
Depending on whether the smaller or larger allele was lost, QLOH could have any value between zero and infinity, with 0.0 being 100% loss of allele A, and infinity being 100% loss of allele B. QLOH values of <0.4 or >2.5 were considered to be clearly indicative of LOH, as described previously (26, 46). 17p (p53) LOH was defined as LOH at or spanning the p53 locus, as described previously (26).
Statistical Analysis
The relationship between grade of dysplasia and prevalence of 17p LOH was evaluated by testing the contribution of an ordered covariate for histological grade to a logistic regression model of 17p LOH. Relative risk (RR) estimates for the effect of 17p loss on the incidence of cancer, high grade dysplasia, aneuploidy, and 4N abnormality were obtained from univariable Cox proportional hazards regression models. Outcome and censoring times were defined relative to baseline endoscopy. Wald test ps for association of 17p loss with progression to these outcomes were based on coefficient estimates for this predictor in the corresponding models. Analysis of progression from HGD to cancer was restricted to patients with HGD detected at baseline endoscopy.
The Kaplan-Meier estimator was used for cumulative incidence curves and incidence estimates for cancer and HGD. Observations were censored at time of last surveillance endoscopy for patients without the outcome of interest. Corresponding 95% CIs were based on Greenwood SE estimates.
RESULTS
Prevalence of 17p LOH, Increased 4N, and Aneuploidy at the Baseline Endoscopy
We determined the prevalence of 17p (p53) LOH, increased 4N fractions, and aneuploidy in different grades of dysplasia at the baseline endoscopy for the 325 patients (Fig. 1). Only 312 patients are represented for 17p (p53) LOH because the remaining 13 were not informative for polymorphic markers at or spanning the p53 locus. The prevalence of 17p (p53) LOH increased from 6% in patients whose biopsy specimens were negative for dysplasia to 57% in patients who had high-grade dysplasia (p < 0.001). Similarly, the prevalence of increased 4N and/or aneuploidy increased from 11% in patients who were negative for dysplasia to 54% in those with high-grade dysplasia. One interpretation of this pattern of cross-sectional results would be that 17p (p53) LOH arises as a late event during neoplastic progression in Barrett’s esophagus, at about the stage of high-grade dysplasia. An alternative interpretation is that it arises early and that those patients with 17p (p53) LOH have an increased risk for progressing to high-grade dysplasia and esophageal adenocarcinoma. To distinguish between these two hypotheses, we evaluated baseline 17p (p53) LOH as a predictor of neoplastic progression during prospective endoscopic biopsy evaluation of the cohort of patients.
Figure 1.
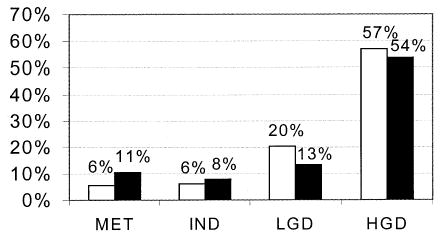
Percentage of patients with 17p (p53) LOH and flow cytometric abnormalities (increased 4N, aneuploidy) as a function of histological grade of dysplasia in Barrett’s esophagus. The y axis represents percent of patients, and the x axis represents histological grade. HGD = high-grade dysplasia; IND = indefinite for dysplasia; LGD = low-grade dysplasia; MET = metaplasia negative for dysplasia; □ = 17p (p53) LOH; ▪ = increased 4N or aneuploidy.
17p (p53) LOH and Progression to Esophageal Adenocarcinoma
The 17p (p53) LOH was a strong and significant predictor of progression to esophageal adenocarcinoma (Fig. 2A). Of the 269 patients followed prospectively, 256 had baseline 17p (p53) LOH data; 20 of 54 patients (37%) with 17p (p53) LOH progressed to cancer, compared to only six of 202 patients (3%) with two 17p alleles (RR = 16; 95% CI = 6.2, 39; p < 0.001) (Table 1). The 3-yr cumulative incidence of cancer in those with 17p (p53) LOH was 38% (95% CI = 26, 54) compared to 3.3% (95% CI = 1.4, 8.0) for those with two 17p alleles. Only six of 202 patients (3%) progressed to esophageal adenocarcinoma without a clear 17p (p53) LOH at the baseline endoscopy. Three of the six, including two with “partial” 17p (p53) LOH at baseline, developed clear 17p (p53) LOH at the follow-up endoscopy in which cancer was diagnosed. Two other patients had aneuploid cell populations without 17p (p53) LOH, suggesting that an alternative pathway exists that can also cause the genetic instability that leads to cancer in a minority of patients with Barrett’s esophagus. The final patient had neither 17p (p53) LOH nor aneuploidy at baseline but developed aneuploidy at the follow-up endoscopy in which cancer was diagnosed; 17p (p53) LOH was not evaluated in this endoscopy.
Figure 2.
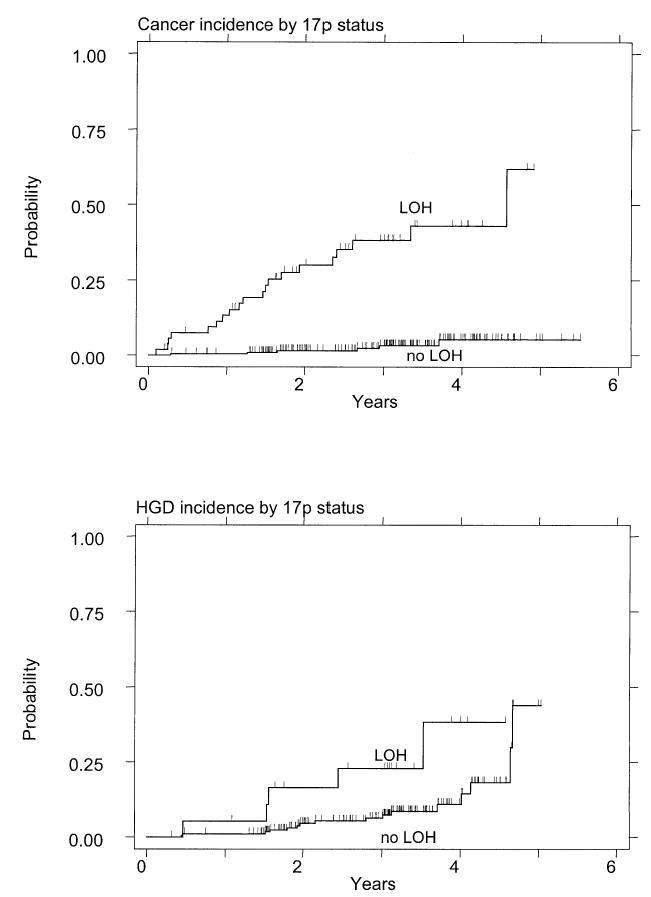
(A) Cumulative incidence of cancer in patients who had Barrett’s esophagus with and without 17p (p53) LOH at the baseline endoscopy. The patients represent all histological grades of dysplasia. Patients with 17p (p53) LOH have a significantly increased rate of progression to esophageal adenocarcinoma. (B) Cumulative incidence of high-grade dysplasia or cancer among patients with and without 17p (p53) LOH at the baseline endoscopy.
Table 1.
17p (p53) LOH and Progression to Neoplastic Endpoints Compared to Patients With Two 17p Alleles
| Patient Subset | Outcome | RR | 95% CI | p |
|---|---|---|---|---|
| All Barrett’s | EA | 16 | 6.2, 39 | <0.001 |
| <HGD | HGD or EA | 3.6 | 1.3, 10 | 0.02 |
| Without 4N | 4N | 6.1 | 3.0, 12 | <0.001 |
| Without aneuploidy | Aneuploidy | 7.5 | 3.5, 16 | <0.001 |
| With HGD | EA | 3 | 1.1, 8.2 | 0.03 |
EA = esophageal adenocarcinoma; HGD = high-grade dysplasia.
17p (p53) LOH and Progression to High-Grade Dysplasia
We then asked whether 17p (p53) LOH was a predictor of progression to high-grade dysplasia or cancer among the 197 informative patients whose biopsy results were negative, indefinite, or low grade at the baseline endoscopy. Again, we found that 17p (p53) LOH was a significant predictor of progression during prospective surveillance (Fig. 2B). Of 19 patients with 17p (p53) LOH, five progressed to high-grade dysplasia or cancer (26%) compared to 16 of 178 patients without 17p LOH (9%) (RR = 3.6; 95% CI = 1.3, 10; p = 0.02) (Table 1).
17p (p53) LOH and Progression From High-Grade Dysplasia to Esophageal Adenocarcinoma
High-grade dysplasia is an advanced stage of neoplastic progression in Barrett’s esophagus with an increased risk for progressing to cancer (18, 51, 52). We therefore asked whether 17p (p53) LOH was associated with an increased risk for progression from high-grade dysplasia to esophageal adenocarcinoma. We found that even at this late stage of neoplasia, patients with 17p LOH had an elevated rate of progression to cancer compared to those with two 17p alleles. In all, 18 of 35 patients with high-grade dysplasia and 17p (p53) LOH progressed to cancer (51%) compared with five of 24 patients who had high-grade dysplasia without 17p LOH (21%) (RR = 3.0; 95% CI = 1.1, 8.2; p = 0.03) (Fig. 3) (Table 1). Among patients with high-grade dysplasia, the 3-yr cumulative incidence of cancer was 55% (95% CI = 37, 54) for patients with 17p (p53) LOH at baseline compared to 33% (95% CI = 14, 64) for patients with two 17p alleles.
Figure 3.
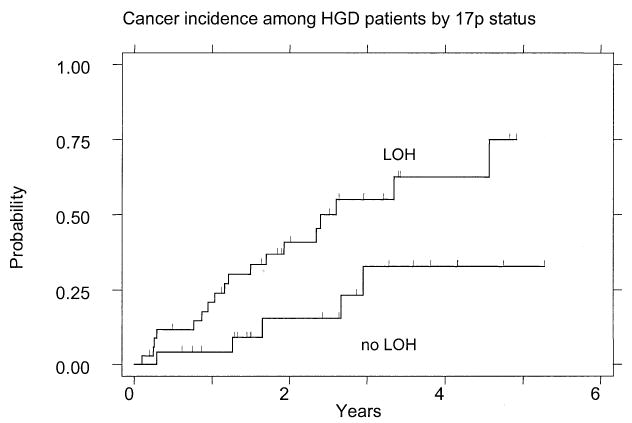
Cumulative incidence of cancer in patients with high-grade dysplasia as a function of 17p (p53) LOH status at the baseline endoscopy.
17p (p53) LOH and Progression to Increased 4N or Aneuploid Cell Populations
We and others have previously reported that patients whose biopsy specimens have increased 4N or aneuploid cell populations are at increased risk for progression to high-grade dysplasia and esophageal adenocarcinoma (19, 21, 47). Therefore, we investigated the relationships between 17p (p53) LOH and progression to these two documented intermediate endpoints of neoplastic progression in Barrett’s esophagus. Again, we found that patients with 17p (p53) LOH were at increased risk for neoplastic progression compared to those with two 17p alleles (Table 1). Thus, among the 211 patients without increased 4N at baseline, 14 of 30 (47%) with 17p (p53) LOH developed elevated 4N fractions at follow-up endoscopies compared to 17 of 181 (9%) with two 17p alleles (RR = 6.1; 95% CI = 3.0, 12; p < 0.001). Similarly, among patients without aneuploidy at the baseline endoscopy, 13 of 27 (48%) with 17p LOH progressed to aneuploidy in follow-up compared to 14 of 181 (8%) with two 17p alleles (RR = 7.5; 95% CI = 3.5,16; p < 0.001).
DISCUSSION
Most patients with Barrett’s esophagus will live out their lifespan without developing esophageal adenocarcinoma (53), and they therefore derive no benefit from expensive and invasive programs for early detection or prevention of malignancy. In contrast, the subset of patients who are at increased risk for progression seem to have markedly improved survival if the cancers are detected at an early, curable stage (5–9). Thus, identification of patient subsets with different risks of progression will likely improve the management of the cancer risk in Barrett’s esophagus by permitting medical resources to be focused on those patients whose increased risk warrants more frequent surveillance or intervention, whereas those at low risk can be monitored less frequently and reassured. Our results demonstrate that 17p (p53) LOH is a strong and significant predictor of progression to esophageal cancer as well as to surrogate endpoints, including increased 4N, aneuploidy, and high-grade dysplasia, in Barrett’s esophagus. Patients whose biopsy specimens had 17p (p53) LOH had a relative risk of 16 (p < 0.001) for progression to cancer compared with those whose biopsy samples had two 17p alleles. The 3-yr cumulative incidence of cancer in those with 17p (p53) LOH was 38% compared to 3.3% for those with two 17p alleles. 17p (p53) LOH seems to arise as an early event in a subset of patients with negative, indefinite or low-grade dysplasia who are at increased risk for progression to high-grade dysplasia or cancer (RR = 3.6). Further, patients with 17p (p53) LOH also had an increased risk for progressing from high-grade dysplasia to esophageal adenocarcinoma (RR = 3.0).
Our results, combined with those of earlier studies and the results in model systems, suggest that 17p (p53) LOH is permissive for the evolution of additional genetic lesions including increased 4N and aneuploid cell populations (20, 25–27, 29, 31). It is likely that lesions in this pathway are the cause of progression to cancer in most patients with Barrett’s esophagus. In our study, 20 of 26 patients (77%) who progressed to cancer had 17p (p53) LOH at the baseline endoscopy. Two others had aneuploidy without 17p (p53) LOH at baseline, suggesting that alternative pathways or lesions in a minority of Barrett’s patients can also lead to the instability that causes esophageal adenocarcinoma. Three patients, including two with “partial” 17p (p53) LOH at baseline, had 17p (p53) LOH detected at the endoscopy in which cancer was diagnosed, and the remaining patient developed an aneuploid cell population at the endoscopy in which cancer was detected. We do not know when these abnormalities developed in patients in whom they were not detected at baseline because intermediate endoscopies were not evaluated. However, it is possible that sampling error may have contributed because we only evaluated one biopsy sample taken at 2-cm intervals in the Barrett’s segment for 17p (p53) LOH. Regardless, 85% of patients (22 of 26) who developed cancer in this study had 17p (p53) LOH or aneuploidy detected at their baseline endoscopy, and 100% had 17p (p53) LOH or aneuploidy detected by the time of the cancer diagnosis.
Neoplastic progression is characterized by evolution of genetically abnormal clones of cells culminating in a “genetic catastrophe” that leads to cancer (31, 32, 54). We have shown previously in retrospective and cross-sectional studies that inactivation of p53 seems to predispose to this “genetic catastrophe,” which is characterized by evolution of increased 4N and aneuploid cell populations, other LOH events, including those affecting chromosomes 5q, 13q, and 18q, and cancer in Barrett’s esophagus (17, 20, 25–27, 29). We have extended these observations in the present study by demonstrating that 17p (p53) LOH is a predictor of progression to increased 4N fractions (RR = 6.1; 95% CI = 3.0, 12.4; p < 0.001), aneuploid cell populations (RR = 7.5; 95% CI = 3.5, 16; p < 0.001) and high-grade dysplasia (RR = 3.6; 95% CI = 1.3, 10. p = 0.02) during prospective endoscopic surveillance. Thus, 17p (p53) LOH, increased 4N and aneuploidy represent manifestations of genomic instability in Barrett’s esophagus that can be used to assess risk for progression to esophageal adenocarcinoma and as surrogate endpoints for prevention trials.
The 17p (p53) LOH, increased 4N, and aneuploidy cannot be considered equivalent to histological grade of dysplasia or to each other. For each histological grade we found subsets of patients with these genetic lesions. For example, 6% of patients whose biopsy specimens were negative or indefinite for dysplasia, 20% of patients with low-grade dysplasia, and 57% of patients with high-grade dysplasia had 17p (p53) LOH at the baseline endoscopy. Similarly, we found increased 4N fractions, aneuploidy or both in 11% of patients whose biopsy results were negative for dysplasia, 8% of indefinite for dysplasia, 13% of low-grade dysplasia, and 54% of high-grade dysplasia. We also found that 17p (p53) LOH was not equivalent to flow cytometric abnormalities because 32% of the patients in our cohort have 17p (p53) LOH without increased 4N or aneuploidy, and two patients in this study developed cancer by an alternative pathway that led to aneuploidy without 17p (p53) LOH. Thus, it is likely that a panel of biomarkers will be required to optimally manage the cancer risk in this condition, and additional follow-up will be required to determine the optimum combination of biomarkers. Nevertheless, clones with 17p (p53) LOH, increased 4N fractions, and aneuploid DNA contents can spread to involve large regions of Barrett’s mucosa (25–28, 30). Thus, it is likely that interventions the only goal of which is to “downstage” grade of dysplasia may leave residual metaplasia with genetically abnormal clones of cells that can progress to cancer (55). Such residual genetically abnormal clones are likely to be responsible for the relatively rapid progression to high-grade dysplasia and cancer that have been reported in some cases after endoscopic ablation (55–58).
In this report, we investigated 17p (p53) LOH, in contrast to many previous studies that have used p53 protein overexpression as a biomarker in Barrett’s esophagus (23, 59–62). Although p53 protein overexpression is frequently considered a surrogate for p53 mutations, it has high false-negative and false-positive (>25%) rates in esophageal cancer compared to the “gold standard” of DNA sequencing (37, 63, 64). We and others have shown previously that 30% of the p53 mutations detected in high-grade dysplasia were nonsense or frameshift mutations that would not be expected to produce a p53 protein (27, 37). However, even given these limitations, three small studies have reported follow-up data suggesting that p53 lesions may predict progression. In one, 5/9 patients with p53 protein overexpression progressed from indefinite/low-grade to HGD or EA compared to 0/16 who were p53 negative (p = 0.01) (65). In the second, four of 11 patients with p53 protein overexpression progressed to EA compared with seven of 41 who were p53 negative (RR = 2.99; 95% CI = 0.57, 15.76; p = 0.197) (16). In the third, three of six patients with low-grade dysplasia and p53 protein overexpression progressed to HGD or EA compared to two of 25 who were p53 negative (p = 0.03) (66). Thus, the weight of evidence indicates that p53 lesions identify risk subsets in Barrett’s esophagus; however, it will be necessary to use diagnostic tests that minimize false positive and negative results to achieve the greatest risk discrimination.
One potential cause of false-negative LOH results is contamination with normal cells that have two 17p alleles (44); for this reason, we have used highly purified flow-sorted neoplastic cells in our studies (25, 26, 35, 46). High levels of genetic instability in advanced esophageal adenocarcinomas can produce a “background” or random frequency of LOH that we have estimated to be 23% (95% CI = 20, 27) at any given chromosome locus (35). For example, one study of chromosome 17 LOH in 12 advanced esophageal adenocarcinomas suggested that there might be five other tumor suppressor genes in addition to p53 on chromosome 17p, but this study was too small to rule out the possibility that the results were simply caused by random instability in advanced cancers (67). Certainly, there is strong evidence that p53 is a target for 17p LOH because up to 92% of esophageal adenocarcinomas have p53 mutations (37, 40–42). We conducted our study at earlier stages of progression in which the background level of LOH is lower and in which the p53 locus is selected for inactivation (25, 27, 35, 68). Further, we have used a platform for LOH analysis that incorporates a panel of robust polymorphic markers, and 17p LOH events that were used in this study included the p53 locus. Finally, we found p53 mutations in exons 5–9 of the remaining allele in 82% of patients (28/34) who had 17p (p53) LOH in high-grade dysplasia, strongly supporting p53 as the target of 17p (p53) LOH in these cases (data not shown). Thus, we believe that under these conditions, 17p (p53) LOH is a specific marker of risk in Barrett’s esophagus.
Most programs of endoscopic biopsy surveillance are based on histological assessment of dysplasia, but this has several shortcomings. The diagnosis of dysplasia is subjective and has well recognized observer variation that has been documented in multiple studies (69–75). Observer variation makes validation of surveillance guidelines difficult because varying histological interpretations may lead to different estimated rates of progression. Even the risk for high-grade dysplasia progressing to cancer varies among centers, and prospective studies have demonstrated that some cases of high-grade dysplasia seem to regress to lesser degrees of biopsy abnormality (47, 51, 52). Most patients with biopsy results that are negative, indefinite, or low-grade dysplasia do not progress to cancer during prolonged follow-up, and we found no statistically significant difference in rates of progression to cancer for these three diagnostic categories in a previous study (47). Further, transient diagnoses of dysplasia seem to increase the cost of endoscopic surveillance in patients with Barrett’s esophagus (76). Thus, there is a need for objective biomarkers that can be used as adjuncts to histological assessment of dysplasia to identify patients at increased risk for progression to cancer so that they can be managed appropriately, whereas patients at low risk can be reassured and undergo surveillance less frequently. Our results indicate that 17p (p53) LOH, increased 4N fractions and aneuploidy are all predictors of progression to esophageal adenocarcinoma in Barrett’s esophagus, providing a panel of objective biomarkers that can be measured in a single endoscopic biopsy and used for risk assessment (20, 25, 26, 46, 47).
Our cohort is high risk, as evidenced by the number of patients who progressed to cancer during the course of this study. Some investigators might suggest that cancers detected during the first year of surveillance should be classified as prevalent cases and excluded from analysis. However, there are no data to support such arbitrary classifications. We have chosen to express our results based on the baseline evaluation and to include all cancers detected subsequently, because exclusion of cancers developing during an arbitrary time interval would convey an inaccurate impression, making it difficult to develop accurate risk stratification based on findings at the baseline endoscopy. Ideally, our results should be confirmed by a prospective study of patients followed in the community, but such a study will require large numbers of patients followed for prolonged periods because of the low risk for progression in most patients with Barrett’s esophagus (10, 11). Until other investigators confirm our results, and certified laboratories are established to measure LOH in clinical samples, we do not recommend any definitive intervention, including surgery or endoscopic ablation, based solely on 17p (p53) LOH status.
Our results illustrate the need for prospective studies to define “early” and “late” events of human neoplastic progression. Cross-sectional results, such as those shown in Figure 1, have been used to conclude that p53 lesions arise as “late” events in neoplastic progression in Barrett’s esophagus and other conditions because their prevalence increases in advanced histological grades. An alternative interpretation is that p53 lesions arise in a small subset of patients at early histological stages, predisposing to progression to more advanced stages of neoplastic progression. Our results, combined with previous studies and those in model systems, are consistent with this latter interpretation and suggest that 17p (p53) LOH arises as an early event that is permissive for the evolution of additional genetic lesions that drive neoplastic progression to more advanced stages (20, 25–27, 29, 31, 32, 77). The 17p (p53) LOH, increased 4N fractions, and aneuploidy can all be assessed on a single endoscopic biopsy, thereby providing a panel of biomarkers that can be used to assess risk for progression and as surrogate endpoints in prevention trials.
Acknowledgments
We thank Christine Karlsen for patient care and David Cowan for database management. This work was supported by National Institutes of Health grant R01 CA61202.
Footnotes
D.S.L. is presently at AstraZeneca Pharmaceuticals (Wayne, PA).
References
- 1.Devesa SS, Blot WJ, Fraumeni JF., Jr Changing patterns in the incidence of esophageal and gastric carcinoma in the United States. Cancer. 1998;83:2049–53. [PubMed] [Google Scholar]
- 2.McKinney A, Sharp L, Macfarlane GJ, Muir CS. Oesophageal and gastric cancer in Scotland 1960–90. Br J Cancer. 1995;71:411–5. doi: 10.1038/bjc.1995.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moller H. Incidence of cancer of oesophagus, cardia and stomach in Denmark. Eur J Cancer Prev. 1992;1:159–64. doi: 10.1097/00008469-199202000-00009. [DOI] [PubMed] [Google Scholar]
- 4.Hansen S, Wiig JN, Giercksky KE, Tretli S. Esophageal and gastric carcinoma in Norway 1958–1992;incidence time trend variability according to morphological subtypes and organ subsites. Int J Cancer. 1997;71:340–4. doi: 10.1002/(sici)1097-0215(19970502)71:3<340::aid-ijc5>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 5.Farrow DC, Vaughan TL. Determinants of survival following the diagnosis of esophageal adenocarcinoma (United States) Cancer Causes Control. 1996;7:322–7. doi: 10.1007/BF00052937. [DOI] [PubMed] [Google Scholar]
- 6.Reid BJ, Blount PL, Feng Z, Levine DS. Optimizing endoscopic biopsy detection of early cancers in Barrett’s high-grade dysplasia. Am J Gastroenterol. 2000;95:3089–96. doi: 10.1111/j.1572-0241.2000.03182.x. [DOI] [PubMed] [Google Scholar]
- 7.Levine DS, Haggitt RC, Blount PL, et al. An endoscopic biopsy protocol can differentiate high-grade dysplasia from early adenocarcinoma in Barrett’s esophagus. Gastroenterology. 1993;105:40–50. doi: 10.1016/0016-5085(93)90008-z. [DOI] [PubMed] [Google Scholar]
- 8.Peters JH, Clark GW, Ireland AP, et al. Outcome of adenocarcinoma arising in Barrett’s esophagus in endoscopically surveyed and nonsurveyed patients. J Thorac Cardiovasc Surg. 1994;108:813–21. [PubMed] [Google Scholar]
- 9.van Sandick JW, van Lanschot JJ, Kuiken BW, et al. Impact of endoscopic biopsy surveillance of Barrett’s oesophagus on pathological stage and clinical outcome of Barrett’s carcinoma. Gut. 1998;43:216–22. doi: 10.1136/gut.43.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drewitz DJ, Sampliner RE, Garewal HS. The incidence of adenocarcinoma in Barrett’s esophagus: A prospective study of 170 patients followed 4.8 years. Am J Gastroenterol. 1997;92:212–5. [PubMed] [Google Scholar]
- 11.O’Connor JB, Falk GW, Richter JE. The incidence of adenocarcinoma and dysplasia in Barrett’s esophagus: Report on the Cleveland Clinic Barrett’s Esophagus Registry. Am J Gastroenterol. 1999;94:2037–42. doi: 10.1111/j.1572-0241.1999.01275.x. [DOI] [PubMed] [Google Scholar]
- 12.Miros M, Kerlin P, Walker N. Only patients with dysplasia progress to adenocarcinoma in Barrett’s oesophagus. Gut. 1991;32:1441–6. doi: 10.1136/gut.32.12.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van der Burgh A, Dees J, Hop WC, van Blankenstein M. Oesophageal cancer is an uncommon cause of death in patients with Barrett’s oesophagus. Gut. 1996;39:5–8. doi: 10.1136/gut.39.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falk GW, Ours TM, Richter JE. Practice patterns for surveillance of Barrett’s esophagus in the United States. Gastrointest Endosc. 2000;52:197–203. doi: 10.1067/mge.2000.107728. [DOI] [PubMed] [Google Scholar]
- 15.Provenzale D, Schmitt C, Wong JB. Barrett’s esophagus: A new look at surveillance based on emerging estimates of cancer risk. Am J Gastroenterol. 1999;94:2043–53. doi: 10.1111/j.1572-0241.1999.01276.x. [DOI] [PubMed] [Google Scholar]
- 16.Bani-Hani K, Martin IG, Hardie LJ, et al. Prospective study of cyclin D1 overexpression in Barrett’s esophagus: Association with increased risk of adenocarcinoma. J Natl Cancer Inst. 2000;92:1316–21. doi: 10.1093/jnci/92.16.1316. [DOI] [PubMed] [Google Scholar]
- 17.Reid BJ, Barrett MT, Galipeau PC, et al. Barrett’s esophagus: Ordering the events that lead to cancer. Eur J Cancer Prev. 1996;5(suppl 2):57–65. doi: 10.1097/00008469-199612002-00009. [DOI] [PubMed] [Google Scholar]
- 18.Reid BJ, Levine DS, Longton G, et al. Predictors of progression to cancer in Barrett’s esophagus: Baseline histology and flow cytometry identify low- and high-risk patient subsets. Am J Gastroenterol. 2000;95:1669–76. doi: 10.1111/j.1572-0241.2000.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid BJ, Blount PL, Rubin CE, et al. Flow-cytometric, and histological progression to malignancy in Barrett’s esophagus: Prospective endoscopic surveillance of a cohort. Gastroenterology. 1992;102:1212–9. [PubMed] [Google Scholar]
- 20.Galipeau PC, Cowan DS, Sanchez CA, et al. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett’s esophagus. Proc Natl Acad Sci USA. 1996;93:7081–4. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teodori L, Gohde W, Persiani M, et al. DNA/protein flow cytometry as a predictive marker of malignancy in dysplasia-free Barrett’s esophagus: Thirteen-year follow-up study on a cohort of patients. Cytometry. 1998;34:257–63. doi: 10.1002/(sici)1097-0320(19981215)34:6<257::aid-cyto3>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 22.Barrett MT, Sanchez CA, Prevo LJ, et al. Early events during neoplastic progression in Barrett’s esophagus. In: Srivastava S, Henson DE, Gazdar G, editors. Molecular pathology of early cancer. Amsterdam: IOS Press; 1999. pp. 179–88. [Google Scholar]
- 23.Kubba AK, Poole NA, Watson A. Role of p53 assessment in management of Barrett’s esophagus. Dig Dis Sci. 1999;44:659–67. doi: 10.1023/a:1026608319881. [DOI] [PubMed] [Google Scholar]
- 24.van Lieshout EM, Jansen JB, Peters WH. Biomarkers in Barrett’s esophagus. Int J Oncol. 1998;13:855–64. doi: 10.3892/ijo.13.4.855. [DOI] [PubMed] [Google Scholar]
- 25.Barrett MT, Sanchez CA, Prevo LJ, et al. Evolution of neoplastic cell lineages in Barrett oesophagus. Nature Genet. 1999;22:106–9. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galipeau PC, Prevo LJ, Sanchez CA, et al. Clonal expansion, and loss of heterozygosity at chromosomes 9p, and 17p in premalignant esophageal (Barrett’s) tissue. J Nat Cancer Inst. 1999;91:2087–95. doi: 10.1093/jnci/91.24.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prevo LJ, Sanchez CA, Galipeau PC, Reid BJ. p53 Mutant clones, and field effects in Barrett’s esophagus. Cancer Res. 1999;59:4784–7. [PubMed] [Google Scholar]
- 28.Reid BJ, Haggitt RC, Rubin CE, Rabinovitch PS. Barrett’s esophagus. Correlation between flow cytometry and histology in detection of patients at risk for adenocarcinoma. Gastroenterology. 1987;93:1–11. [PubMed] [Google Scholar]
- 29.Blount PL, Galipeau PC, Sanchez CA, et al. 17p allelic losses in diploid cells of patients with Barrett’s esophagus who develop aneuploidy. Cancer Res. 1994;54:2292–5. [PubMed] [Google Scholar]
- 30.Rabinovitch PS, Reid BJ, Haggitt RC, et al. Progression to cancer in Barrett’s esophagus is associated with genomic instability. Lab Invest. 1989;60:65–71. [PubMed] [Google Scholar]
- 31.Ko LJ, Prives C. p53: Puzzle and paradigm. Genes Dev. 1996;10:1054–72. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 32.Artandi SE, DePinho RA. Mice without telomerase: What can they teach us about human cancer? Nat Med. 2000;6:852–5. doi: 10.1038/78595. [DOI] [PubMed] [Google Scholar]
- 33.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: Clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–78. [PubMed] [Google Scholar]
- 34.Reid BJ. p53, And neoplastic progression in Barrett’s esophagus. Am J Gastroenterol. 2001;96:1321–23. doi: 10.1111/j.1572-0241.2001.03844.x. [DOI] [PubMed] [Google Scholar]
- 35.Barrett MT, Galipeau PC, Sanchez CA, et al. Determination of the frequency of loss of heterozygosity in esophageal adenocarcinoma by cell sorting, whole genome amplification and microsatellite polymorphisms. Oncogene. 1996;12:1873–8. [PubMed] [Google Scholar]
- 36.Dolan K, Garde J, Gosney J, et al. Allelotype analysis of oesophageal adenocarcinoma: Loss of heterozygosity occurs at multiple sites. Br J Cancer. 1998;78:950–7. doi: 10.1038/bjc.1998.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hamelin R, Flejou JF, Muzeau F, et al. TP53 gene mutations, and p53 protein immunoreactivity in malignant, and premalignant Barrett’s esophagus. Gastroenterology. 1994;107:1012–8. doi: 10.1016/0016-5085(94)90225-9. [DOI] [PubMed] [Google Scholar]
- 38.Hammoud ZT, Kaleem Z, Cooper JD, et al. Allelotype analysis of esophageal adenocarcinomas: Evidence for the involvement of sequences on the long arm of chromosome 4. Cancer Res. 1996;56:4499–4502. [PubMed] [Google Scholar]
- 39.Blount PL, Ramel S, Raskind WH, et al. 17p allelic deletions, and p53 protein overexpression in Barrett’s adenocarcinoma. Cancer Res. 1991;51:5482–6. [PubMed] [Google Scholar]
- 40.Gleeson CM, Sloan JM, McGuigan JA, et al. Base transitions at CpG dinucleotides in the p53 gene are common in esophageal adenocarcinoma. Cancer Res. 1995;55:3406–11. [PubMed] [Google Scholar]
- 41.Muzeau F, Flejou JF, Potet F, et al. Profile of p53 mutations, and abnormal expression of P53 protein in 2 forms of esophageal cancer. Gastroenterol Clin Biol. 1996;20:430–7. (in French) [PubMed] [Google Scholar]
- 42.Neshat K, Sanchez CA, Galipeau PC, et al. Barrett’s esophagus. A model of human neoplastic progression. Cold Spring Harb Symp Quant Biol. 1994;59:577–83. doi: 10.1101/sqb.1994.059.01.065. [DOI] [PubMed] [Google Scholar]
- 43.Blount PL, Meltzer SJ, Yin J, et al. Clonal ordering of 17p and 5q allelic losses in Barrett dysplasia and adenocarcinoma. Proc Natl Acad Sci USA. 1993;90:3221–5. doi: 10.1073/pnas.90.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 45.Barrett MT, Reid BJ, Joslyn G. Genotypic analysis of multiple loci in somatic cells by whole genome amplification. Nucleic Acids Res. 1995;23:3488–92. doi: 10.1093/nar/23.17.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paulson TG, Galipeau PC, Reid BJ. Loss of heterozygosity analysis using whole genome amplification, cell sorting, and fluorescence-based PCR. Genome Res. 1999;9:482–91. [PMC free article] [PubMed] [Google Scholar]
- 47.Reid BJ, Levine DS, Longton G, et al. Predictors of progression to cancer in Barrett’s esophagus: Baseline histology and flow cytometry identify low- and high-risk patient subsets. Am J Gastroenterol. 2000;95:1669–76. doi: 10.1111/j.1572-0241.2000.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haggitt RC. Barrett’s esophagus, dysplasia, and adenocarcinoma. Hum Pathol. 1994;25:982–93. doi: 10.1016/0046-8177(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 49.Gerdes J, Schwab U, Lemke H, Stein H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31:13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- 50.Gerdes J, Lemke H, Baisch H, et al. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–5. [PubMed] [Google Scholar]
- 51.Sontag SJ, Schnell TG, Chejfec G, et al. High grade dysplasia: Surveillance endoscopy once a year is sufficient in most patients. Gastroenterology. 1999;116:G1334. [Google Scholar]
- 52.Weston AP, Sharma P, Topalovski M, et al. Prospective long-term follow-up of Barrett’s high-grade dysplasia: Risky business. Gastroenterology. 1999;116:A352–3. doi: 10.1111/j.1572-0241.2000.02234.x. [DOI] [PubMed] [Google Scholar]
- 53.Cameron AJ, Zinsmeister AR, Ballard DJ, Carney JA. Prevalence of columnar-lined (Barrett’s) esophagus. Comparison of population-based clinical and autopsy findings. Gastroenterology. 1990;99:918–22. doi: 10.1016/0016-5085(90)90607-3. [DOI] [PubMed] [Google Scholar]
- 54.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 55.Krishnadath KK, Wang KK, Taniguchi K, et al. Persistent genetic abnormalities in Barrett’s esophagus after photodynamic therapy. Gastroenterology. 2000;119:624–30. doi: 10.1053/gast.2000.18012. [DOI] [PubMed] [Google Scholar]
- 56.Overholt BF, Panjehpour M, Haydek JM. Photodynamic therapy for Barrett’s esophagus: Follow-up in 100 patients. Gastrointest Endosc. 1999;49:1–7. doi: 10.1016/s0016-5107(99)70437-2. [DOI] [PubMed] [Google Scholar]
- 57.Van Laethem JL, Peny MO, Salmon I, et al. Intramucosal adenocarcinoma arising under squamous re-epithelialisation of Barrett’s oesophagus. Gut. 2000;46:574–7. doi: 10.1136/gut.46.4.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ertan A, Zimmerman M, Younes M. Esophageal adenocarcinoma associated with Barrett’s esophagus: Long-term management with laser ablation. Am J Gastroenterol. 1995;90:2201–3. [PubMed] [Google Scholar]
- 59.Krishnadath KK, Tilanus HW, van Blankenstein M, et al. Accumulation of p53 protein in normal, dysplastic, and neoplastic Barrett’s oesophagus. J Pathol. 1995;175:175–80. doi: 10.1002/path.1711750204. [DOI] [PubMed] [Google Scholar]
- 60.Jones DR, Davidson AG, Summers CL, et al. Potential application of p53 as an intermediate biomarker in Barrett’s esophagus. Ann Thorac Surg. 1994;57:598–603. doi: 10.1016/0003-4975(94)90551-7. [DOI] [PubMed] [Google Scholar]
- 61.Gimenez A, Minguela A, Parrilla P, et al. Flow cytometric DNA analysis, and p53 protein expression show a good correlation with histologic findings in patients with Barrett’s esophagus. Cancer. 1998;83:641–51. doi: 10.1002/(sici)1097-0142(19980815)83:4<641::aid-cncr3>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 62.Younes M, Lebovitz RM, Lechago LV, Lechago J. p53 Protein accumulation in Barrett’s metaplasia, dysplasia, and carcinoma: A follow-up study. Gastroenterology. 1993;105:1637–42. doi: 10.1016/0016-5085(93)91058-p. [DOI] [PubMed] [Google Scholar]
- 63.Coggi G, Bosari S, Roncalli M, et al. A p53 protein accumulation and p53 gene mutation in esophageal carcinoma. A molecular and immunohistochemical study with clinicopathologic correlations. Cancer. 1997;79:425–32. doi: 10.1002/(sici)1097-0142(19970201)79:3<425::aid-cncr1>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 64.Moore JH, Lesser EJ, Erdody DH, et al. Intestinal differentiation and p53 gene alterations in Barrett’s esophagus and esophageal adenocarcinoma. Int J Cancer. 1994;56:487–93. doi: 10.1002/ijc.2910560406. [DOI] [PubMed] [Google Scholar]
- 65.Younes M, Ertan A, Lechago LV, et al. p53: Protein accumulation is a specific marker of malignant potential in Barrett’s metaplasia. Dig Dis Sci. 1997;42:697–701. doi: 10.1023/a:1018828207371. [DOI] [PubMed] [Google Scholar]
- 66.Weston AP, Banerjee SK, Persons DL, et al. p53 Positivity in low-grade dysplasia (LGD) in Barrett’s esophagus. Marker predictive of progression to cancer or multifocal high-grade dysplasia (mHGD) Am J Gastroenterol. 1999;94:2603. doi: 10.1111/j.1572-0241.2001.03851.x. [DOI] [PubMed] [Google Scholar]
- 67.Dunn J, Garde J, Dolan K, et al. Multiple target sites of allelic imbalance on chromosome 17 in Barrett’s oesophageal cancer. Oncogene. 1999;18:987–93. doi: 10.1038/sj.onc.1202371. [DOI] [PubMed] [Google Scholar]
- 68.Mei R, Galipeau PC, Prass C, et al. Genome-wide detection of loss of heterozygosity with single nucleotide polymorphisms, and high density DNA arrays. Genome Res. 2000;10:1126 –37. doi: 10.1101/gr.10.8.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reid BJ, Haggitt RC, Rubin CE, et al. Observer variation in the diagnosis of dysplasia in Barrett’s esophagus. Hum Pathol. 1988;19:166–78. doi: 10.1016/s0046-8177(88)80344-7. [DOI] [PubMed] [Google Scholar]
- 70.Sagan C, Flejou JF, Diebold MD, et al. Observer variation in the diagnosis of dysplasia in Barrett’s mucosa. Gastroenterol Clin Biol. 1994;18:D31–4. [PubMed] [Google Scholar]
- 71.van Sandick JW, Baak JP, van Lanschot JJ, et al. Computerized quantitative pathology for the grading of dysplasia in surveillance biopsies of Barrett’s oesophagus. J Pathol. 2000;190:177–83. doi: 10.1002/(SICI)1096-9896(200002)190:2<177::AID-PATH508>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 72.Polkowski W, Baak JP, van Lanschot JJ, et al. Clinical decision making in Barrett’s oesophagus can be supported by computerized immunoquantitation and morphometry of features associated with proliferation and differentiation. J Pathol. 1998;184:161–8. doi: 10.1002/(SICI)1096-9896(199802)184:2<161::AID-PATH971>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 73.Ormsby AH, Perras RE, Henricks WH, et al. Interobserver variation in the diagnosis of superficial adenocarcinoma in Barrett’s esophagus (BE) Mod Pathol. 1998;11:A386. [Google Scholar]
- 74.Ormsby AH, Perras RE, Henricks WH, et al. Interobserver variation in Barrett’s (BE) related high-grade dysplasia (HGD) and superficial adenocarcinoma: Can it be improved using uniform pathologic criteria? Gastroenterology. 2000;118:A3764. [Google Scholar]
- 75.Alikhan M, Rex D, Khan A, et al. Variable pathologic interpretation of columnar lined esophagus by general pathologists in community practice. Gastrointest Endosc. 1999;50:23–6. doi: 10.1016/s0016-5107(99)70339-1. [DOI] [PubMed] [Google Scholar]
- 76.Ofman JJ, Lewin K, Ramers C, et al. The economic impact of the diagnosis of dysplasia in Barrett’s esophagus. Am J Gastroenterol. 2000;95:2946–52. doi: 10.1111/j.1572-0241.2000.03209.x. [DOI] [PubMed] [Google Scholar]
- 77.Artandi SE, Chang S, Lee SL, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–5. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]


