Abstract
The important identity elements in tRNAGln and tRNAAsn for bacterial GatCAB and in tRNAGln for archaeal GatDE are the D-loop and the first base pair of the acceptor stem. Here we show that Methanothermobacter thermautotrophicus GatCAB, the archaeal enzyme, is different as it discriminates Asp-tRNAAsp and Asp-tRNAAsn by use of U49, the D-loop and to a lesser extent the variable loop. Since archaea possess the tRNAGln-specific amidotransferase GatDE, the archaeal GatCAB enzyme evolved to recognize different elements in tRNAAsn than those recognized by GatDE or by the bacterial GatCAB enzyme in their tRNA substrates.
Keywords: tRNA, transamidation, Asn-tRNA, Gln-tRNA, evolution
1. Introduction
Attaching the correct amino acid to its cognate tRNA species is an essential step for the fidelity of protein synthesis, catalyzed usually by the aminoacyl-tRNA synthetases (aaRS) [1]. However, many bacterial and archaeal genomes do not encode an asparaginyl-tRNA synthetase (AsnRS) and instead use an indirect pathway for Asn-tRNA synthesis. These organisms take advantage of a non-discriminating aspartyl-tRNA synthetase (ND-AspRS) to form Asp-tRNAAsn and an Asp-tRNAAsn-dependent amidotransferase (Asp-AdT) that converts the mischarged tRNA by amidation to Asn-tRNAAsn [2-8]. Prokaryotes lacking a glutaminyl-tRNA synthetase use a similar two-step tRNA-dependent transamidation pathway to form Gln-tRNA with the aid of a ND-glutamyl-tRNA synthetase and a Glu-AdT [3,9,10].
Two AdTs exist in nature, the heterotrimeric GatCAB [10] and the heterodimeric GatDE [3]. The former is found in both archaea and bacteria [3]. In vitro GatCAB is capable of acting as both a Glu-AdT and an Asp-AdT [3,5,7]. GatDE occurs only in archaea and functions solely as a Glu-AdT [3]. Given the presence of GatDE in every archaeon the archaeal GatCAB enzyme may function in vivo as an Asp-AdT; this is a plausible but still unproven assumption. The GatC protein is small, about 100 aa long, and is likely a chaperone for GatA [10], helping it bind to GatB [11]. GatA turned out to be an amidase [12] whereas GatD was shown to be an asparaginase [3,13]. GatE and GatB belong to an isolated protein family and have been implicated as tRNA binding domains catalyzing the ATP-dependent activation of the mischarged tRNA and subsequent amidation into the correct aa-tRNA [11,13-15].
AdTs must recognize specific elements of a tRNA to discriminate the mischarged species (Asp-tRNAAsn or Glu-tRNAGln) from the correctly charged ones (Asp-tRNAAsp or Glu-tRNAGlu). Studies with Staphylococcus aureus GatCAB and Methanothermobacter thermautotrophicus GatDE showed that the D-loop and the first base-pair of the acceptor stem are important recognition elements for tRNAGln identity [10,15]. Recent work revealed similar recognition elements in use by bacterial GatCAB to discriminate tRNAAsn from tRNAAsp [16]. The same work suggested the variable loop would be the major identity element recognized by the archaeal GatCAB to differentiate tRNAAsn from tRNAAsp [16]. In M. thermautotrophicus, the first base pair of the acceptor stem is invariant between tRNAAsp and tRNAAsn. Given that the AdT tRNA recognition work to date focused on the bacterial GatCAB enzyme and on the archaeal specific Glu-AdT, GatDE, we decided to study the M. thermautotrophicus GatCAB as an Asp-AdT. We demonstrate a new assay for measuring aa-tRNA recognition by AdTs and report on the identity elements used by GatCAB to discriminate tRNAAsn from tRNAAsp.
2. Materials and Methods
2.1 General
Oligonucleotide synthesis and DNA sequencing were performed by the Keck Foundation Biotechnology Resource Laboratory at Yale University. [14C]Asp (213 mCi/mmol) and [3H]Asp (26 Ci/mmol) were purchased from Amersham Biosciences (Piscataway, NJ) and [14C]Asn (228.4 mCi/mmol) was purchased from Perkin-Elmer Life Sciences (Boston, NE).
2.2 Preparation of enzymes and tRNAs
The heterotrimeric M. thermautotrophicus GatCAB enzyme was overexpressed as a C-terminal His6-tag fusion at the GatB subunit, purified by nickel affinity chromatography as previously described [17] föllowed by size-exclusion chromatography on a Superdex HR200 column (Amersham Biosciences) in buffer A (50 mM HEPES-KOH (pH 7.4), 10 mM MgCl2, 1 mM DTT, 100 mM NaCl) with 10% glycerol, then dialyzed in buffer A containing 50 % glycerol and stored at −20 °C before use.
For preparation of in vitro tRNA transcripts, chemically synthesized oligonucleotides corresponding to wild-type and mutant tRNAAsn and tRNAAsp sequences were cloned into pUC19. The transcription templates were digested with BstNI (tRNAAsn) or NsiI (tRNAAsp) at 55 °C for 16 hr. In vitro T7 RNA polymerase run-off transcriptions were conducted as described [18] and the RNA purified using a Qiagen (Valencia, CA) RNA-DNA kit according to manufacturer's instructions.
2.3 tRNA aminoacylation
For amidotransferase assays, tRNAAsp and tRNAAsn generated by in vitro transcription was charged with [14C]Asp (50 μM) or [3H]Asp (50 μM) by 4 μg purified M. thermautotrophicus AspRS as described [17]. After aminoacylation, phenol:chloroform (1:1) extraction, and precipitation with two volumes of ethanol the Asp-tRNA preparations were stored at −20 °C.
2.4 Amidotransferase assay
GatCAB activity was measured as described earlier [5] with slight modifications. [14C] or [3H] labeled Asp-tRNAs were suspended in 100 μl amidation buffer (50 mM HEPES-KOH, pH 7.4, 25 mM KCl, 10 mM MgCl2, 2 mM dithiothreitol) and 10 nM of M. thermautotrophicus GatCAB enzyme was added and incubated at 37 °C for 5 min. Aliquots of 20 μl were removed every 60 seconds and quenched by the addition of 20 μl of 50 mM KOH. The tRNAs were deacylated by a 15 min incubation at 25 °C, dried in a Speedvac, and then resuspended in 3 μl of 50 nM of unlabeled Asp and Asn. An aliquot (1.5 μl) was spotted onto a cellulose TLC plate (Sigma-Aldrich, St. Louis, MO) and developed in ammonia: water: chloroform: methanol (2:1:6:6). The [14C]-radioactive amino acids were detected in a Storm 860 Bioimager and analyzed with ImageQuaNT V4.0 program (Amersham Biosciences). When [3H]-aminoacyl-tRNAs were used, the [3H] amino acids were detected after spraying with 0.02 % ninhydrin and excising the portions of the TLC plate corresponding to the Asp and Asn spots, and finally liquid scintillation counting.
2.5 RNase Protection Assay
To investigate binding between Asp-tRNAs and GatCAB, an RNase protection assay [19] was used. 40 nM of [3H] Asp-tRNA was preincubated for 30 min at 4 °C in the presence of 40-2000 nM GatCAB, 50 mM HEPES-KOH (pH 7.4), 10 mM MgCl2, 25 mM KCl and 20 % glycerol to form a ternary complex of Asp-tRNA and GatCAB protein. RNase digestion was started by the addition of 1 μg pancreatic RNase mix (Roche Biochemical, Indianapolis, IN) and was stopped by withdrawing 20 μl and spotting onto a 3mm paper filter (Whatman, NJ) immersed with 10 % of trichloroacetic acid (TCA) solution. The paper filters were washed twice with 10 % and 5 % of TCA solution respectively and residual radioactivity was counted by liquid scintillation counting.
3. Results
3.1 Modified bases in tRNAAsn and tRNA Asp do not play significant roles in recognition by GatCAB
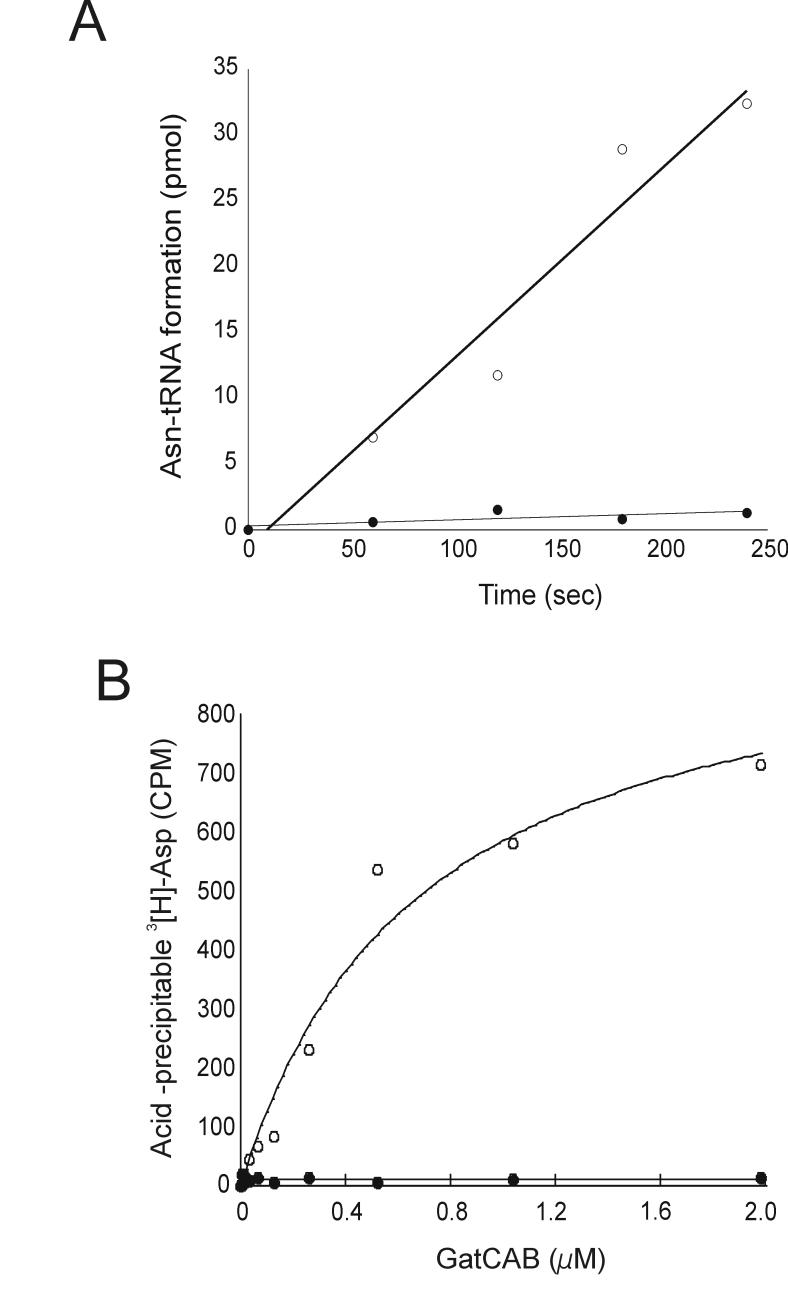
Previously, it was shown that Asp-tRNAAsn derived from in vitro transcription could be transamidated by recombinant M. thermautotrophicus GatCAB enzyme [3], suggesting that modified bases in tRNAAsn transcripts do not play significant roles in the recognition of Asp-tRNAAsn by GatCAB. In order to determine whether the same enzyme still discriminates against the unmodified tRNAAsp transcript we employed two methods. The first, a standard in vitro transamidation assay [10] based on the separation of amino acids by TLC was carried out with in vitro transcribed Asp-tRNAAsn and Asp-tRNAAsp (Fig. 1A). The second technique was an RNase protection assay [18], which has been used for investigation of binding properties of aa-tRNAs with EF-Tu, adapted for the investigation of interactions between GatCAB and Asp-tRNA. EF-Tu protects bound aa-tRNAs from RNase digestion, and the amount of TCA-insoluble radioactivity correlates with the level of ternary complex between EF-Tu and radiolabed aa-tRNAs [18]. This was used to measure Asp-tRNAAsn:GatCAB complex formation. As expected, Asp-tRNAAsn (as transcript) was converted to Asn-tRNAAsn by the addition of M. thermautotrophicus GatCAB [3], while the Asp-tRNAAsp transcript was not transamidated (Fig. 1A). The amount of [3H]Asp-tRNAAsn protected in this assay increased with increasing GatCAB concentration (Fig. 1B). Using linear regression analysis, we estimated the KD of M. thermautotrophicus GatCAB for Asp-tRNAAsn to be approximately 0.6 μM. Asp-tRNAAsp was not protected from RNase digestion by any amount of GatCAB tested. These results suggest that GatCAB specifically binds Asp-tRNAAsn. Therefore, modified nucleotides do not play an essential role in GatCAB discrimination of Asp-tRNAAsn from Asp-tRNAAsp.
Figure 1.
Recognition of Asp-tRNAAsn and Asp-tRNAAsp by GatCAB. (A) Time course of transamidation of Asp-tRNAAsn (o) and Asp-tRNAAsp (●). [14C]Asp-tRNAAsn or [14C]Asp-tRNAAsp (0.5 μM) were incubated with M. thermautotrophicus GatCAB (10 nM) at 37°C. (B) RNase A protection analysis of GatCAB:Asp-tRNA complexes. [3H]Asp-tRNAAsn (o) (10 nM) and [3H]Asp-tRNAAsp (●) (10 nM) were mixed with 0-2000 nM of GatCAB (A) and treated with RNase mix (10μg) for 30 seconds at 4°C. The residual radioactivity on the filter after TCA washing was determined as described in Materials and Methods.
D-loop and T-arm are important for GatCAB recognition of Asp- tRNAAsn
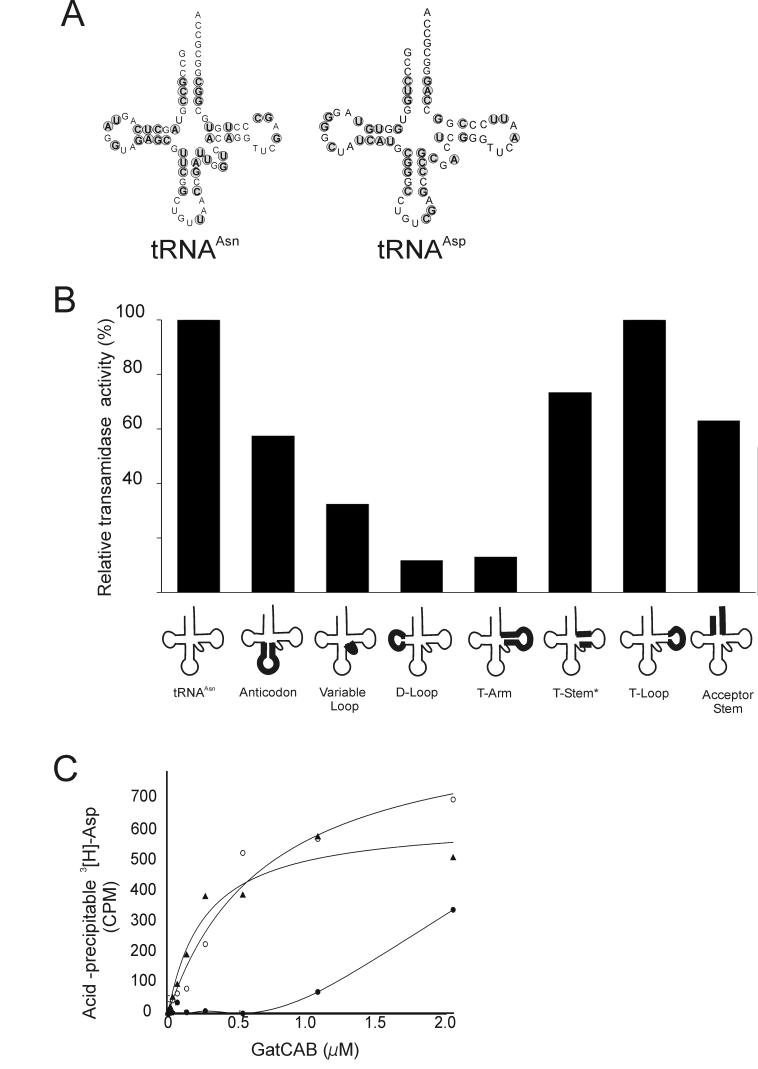
We constructed eight chimeric tRNAAsn variants (tRNAAsx) in which different structural portions of tRNAAsp replaced the corresponding ones in tRNAAsn (sequences in Fig. 2A). These tRNAAsx variants (depicted schematically in Fig. 2B) were then aspartylated with M. thermautotrophicus AspRS. Seven out of eight of these tRNAAsx mutants could be aspartylated; exchange of the tRNAAsn D-arm for that of tRNAAsp gave rise to an unchargeable tRNA. The remaining Asp-tRNAAsx variants were tested for GatCAB-catalyzed amidation.
Figure 2.
Effect of domain swap tRNAAsn mutants on transamidation activity and binding to GatCAB. (A) Clover-leaf representation of M. thermautotrophicus tRNAAsn and tRNAAsp. Sequence differences between the two tRNAs are circled. (B) Relative transamidation activities of various Asp-tRNAAsx mutants and wild-type Asp-tRNAAsn. Transamidation activity was measured as described in Materials and Methods with 1 μM wild-type Asp-tRNAAsn or mutant Asp-tRNAAsx and relative activity was expressed as ratio of conversion rate of wild-type Asp-tRNAAsn to Asn-tRNAAsn. The T-stem* swap did not include the first base pair of the stem. (C) RNase A protection analysis of GatCAB:Asp-tRNA complexes. Wild type Asp-tRNAAsn (o) and swap tRNAAsn mutant with T-arm (●) or anticodon arm (▲) portion of tRNAAsp. The curves were fitted from the experimental points.
For five of the other Asp-tRNAAsx swap mutants, a 0-4 fold decrease of conversion efficiency was observed. The T-stem swap mutant retained the A49-U65 base pair found in tRNAAsn (see below). The largest effects were seen with the T-arm and D-loop Asp-tRNAAsx mutants; they supported the amidotransferase 10-fold less than did wild-type Asn-tRNAAsn.
A similar effect was seen when the interaction of selected Asp-tRNAs was investigated with the RNase protection assay. While wild-type Asp-tRNAAsn and anticodon swapping mutants had nearly similar binding affinity to GatCAB, the protection level of the T stem-loop mutant was significantly reduced (Fig. 2C), indicating the tRNAAsn T-arm is critical for specific recognition by GatCAB.
3.2 U49 in tRNAAsp is a major antideterminant for GatCAB recognition
To reveal a more detailed picture of Asp-tRNAAsn recognition by GatCAB, fifteen tRNAAsn mutants were generated based on the sequence differences between M. thermautotrophicus tRNAAsp and tRNAAsn (Fig. 2A). After aspartylation their substrate qualities were determined in initial velocity experiments with GatCAB (Table 1). Most mutations did not significantly impact the ability of GatCAB to use the tRNA variants as substrates. Confirming our anticodon swap experiment (Fig. 2B), point mutations in the anticodon loop did not appreciably alter the ability of GatCAB to productively bind to the tRNA. Two tRNA mutants could not be transamidated; they had replacements of the A49-U65 base pair with a U49-G65 wobble pair (present in M. thermautotrophicus tRNAAsp) or with a U49-A65 pair. Base pair changes in this position (to C49-G65 or G49-U65) led to tRNAs that were much less affected in their ability to be substrates for GatCAB. Therefore we conclude that U49 may act as an antideterminant for GatCAB recognition of tRNAAsp. Three other tRNAAsn mutants in our collection showed marked intermediate effects on transamidation; position 9 located between the acceptor and D-stems, position 47 in the variable loop and position 56 in the T-loop. Nucleotides in these positions should contribute to the overall tertiary structure of the tRNA.
Table 1.
GatCAB activity with Asp-tRNAAsn mutantsa
| Base change | Region in tRNA | Relative Transamidase Activity (%) |
|---|---|---|
| Wild-type tRNAAsn | 100.0 | |
| A9 → G9 | D-stem | 29.8 |
| G10·C25 →A10·U25 | 98.7 | |
| C11·G24 →U11·A24 | 91.3 | |
| G13·C22 →U13·U22 | 70.9 | |
| U27·U43 →C27·G43 | Anticodon stem | 55.4 |
| U36→A36 | Anticodon loop | 68.9 |
| U36→C36 | 98.7 | |
| A37→G37 | 66.5 | |
| U47→Δ47 | Variable loop | 40.1 |
| A49·U65→U49·G65 | T-stem | < 0.5 |
| A49·U65→C49·G65 | 62.8 | |
| A49·U65→G49·U65 | 49.9 | |
| A49·U65→U49·A65 | < 0.5 | |
| G56→A56 | T-loop | 38.1 |
| C5·G68→U5·A68 | Acceptor stem | 51.1 |
| Wild-type tRNAAsp | < 0.5 |
[14C]Asp-tRNA (0.5 μM) was incubated with M. thermautotrophicus GatCAB (10 nM) at 37°C. The enzyme activity with each variant was expressed as relative percentage of conversion rate for wild type Asp-tRNAAsn to Asn-tRNAAsn.
A comparison of position 49 in archaeal tRNAAsn and tRNAAsp species makes it apparent that U49 is not conserved as an anti-determinant (Table 2). In most archaea encoding a GatCAB position 49 differs in tRNAAsp and tRNAAsn. When purine at position 49 occurs in tRNAAsn, tRNAAsp has a pyrmidine or vice versa; this suggests that position 49 may still serve as anti-determinant for GatCAB recognition in many organisms (Table 2). Nanoarchaeum equitans and Methanopyrus kandleri are notable exceptions. Position 49 is invariant between tRNAAsp and tRNAAsn in archaea with an AsnRS (Table 2).
Table 2.
Alignment of the archaeal tRNAAsn and tRNAAsp isoacceptors at positions 49 and 65 in the T-stem.a
3.3 Asp-AdT active across Domains of Life
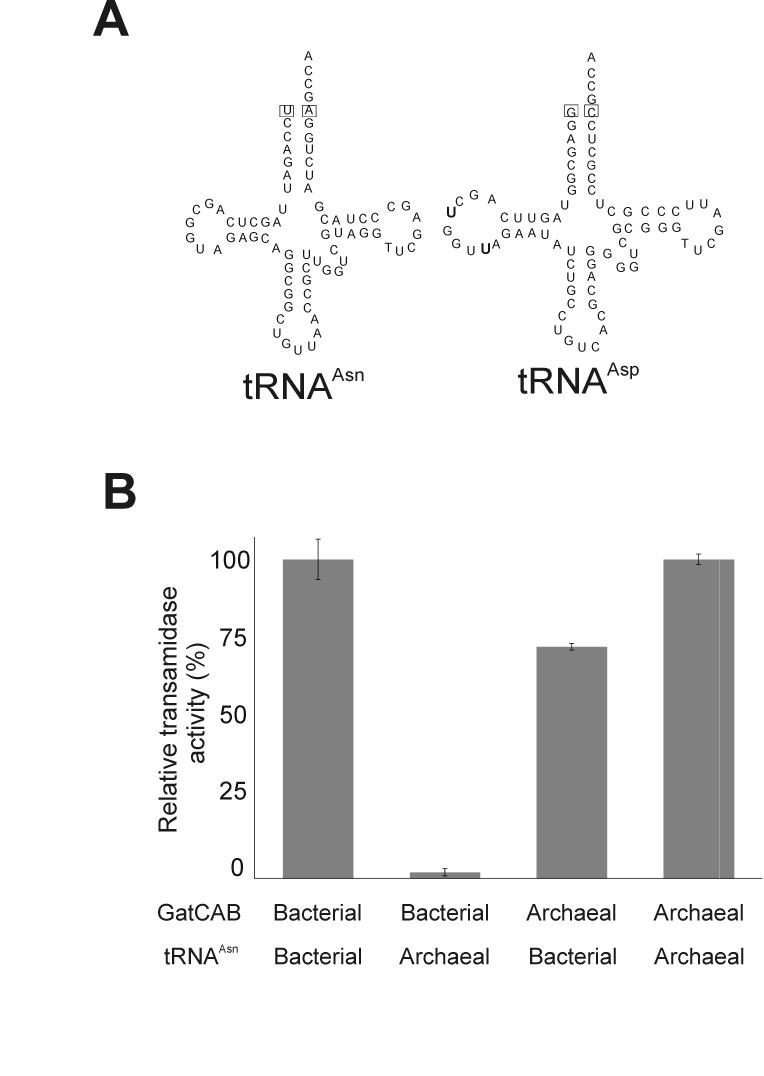
In bacteria that use the two-step transamidation pathway tRNAAsn and tRNAGln have a U1-A72 base pair, while a G1-C72 pair is common in tRNAAsp and tRNAGlu; these are important identity elements for the bacterial GatCAB (Fig. 3A) [16]. Mutating the U1-A72 pair to G1-C72 in S. aureus tRNAGln significantly decreased the ability of GatCAB to recognize the tRNA [11]. Similar results were obtained for the N. meningitidis GatCAB and tRNAAsn [16]. In the case of GatDE, the archaeal Glu-AdT, the first base pair of the acceptor stem is also important to achieve productive tRNA binding [15]. The archaeal tRNAAsn and tRNAAsp species contain a G1-C72 base pair (Fig. 2A); therefore this position in the tRNA is not used for discriminating tRNAAsn from tRNAAsp by the archaeal GatCAB. Since we are also working with the purified bacterial GatCAB enzyme from Helicobacter pylori, we compared the ability of this bacterial AdT with that of the archaeal M. thermautotrophicus enzyme to use either archaeal Asp-tRNAAsn or H. pylori Asp-tRNAAsn as a substrate. The results show (Fig. 3B) that the archaeal enzyme could transamidate the heterologous bacterial Asp-tRNAAsn whereas the bacterial enzyme could only transamidate its own Asp-tRNAAsn. A similar finding with the N. meningitidis and Methanoscarcina bakeri enzymes was very recently reported [16].
Figure 3.
Transamidation by H. pylori or M. thermautotrophicus GatCAB with homologous and heterologous Asp-tRNAAsn. (A) Clover-leaf presentation of H. pylori tRNAAsn and tRNAAsp species. The important identity elements [16] are boxed. The two additional U residues in the D-loop are shown in bold. (B) Transamidation of M. thermautotrophicus and H. pylori Asp-tRNAAsn by a bacterial (H. pylori) or archaeal (M. thermautotrophicus) GatCAB. Reactions were carried out over 5 minutes with 10 nM enzyme. Activities are plotted relative to the homologous reaction; e.g., the activity of the H. pylori GatCAB with the M. thermautotrophicus Asp-tRNAAsn was relative to the activity of the H. pylori with its own Asp-tRNAAsn.
4. Discussion
Studies of specific tRNA recognition by aminoacyl-tRNA synthetases by genetic, biochemical and structural techniques [20] have led to a detailed understanding of the set of discrete tRNA identity elements that distinguish an isoacceptor group from all the other tRNA species [21]. Such investigations are now underway to characterize the enzymes that form aa-tRNA by tRNA-dependent transamidation (e.g., [11,15,16]). Therefore it is appropriate to ask whether the tRNA recognition process of tRNA-dependent amidotransferases is the same as that for aaRSs. In theory, these processes could operate differently. While the aaRS selects its cognate tRNA from amongst a large number of cellular tRNAs, M. thermautotrophicus GatCAB, for example, may first recognize the Asp moiety on the 3′ end of the aa-tRNA, narrowing the pool to Asp-tRNAAsn and Asp-tRNAAsp. In a second step only the rejection of the perfect cognate aa-tRNA (Asp-tRNAAsp) is required. Following this reasoning we synthesized ‘transplantation’ tRNA mutants (Fig. 2).
It is more difficult to establish a complete list of tRNA identity elements for such enzymes, as their substrates need to be synthesized by another enzyme, an aaRS. The generation of substrates for M. thermautotrophicus GatCAB, a collection of mischarged Asp-tRNAAsn species, therefore relies on at least partial preservation of the tRNAAsn identity elements used by AspRS. Indeed, many tRNAAsn mutants that were constructed in this study could not be aspartylated satisfactorily; however a sufficient number was obtained for our studies (Table 1). Although some of the tRNAAsx mutants used in this study (including the D-loop and T-arm swap mutants) could be aspartylated only to half the level obtained with the wild-type tRNAAsn, we do not believe that inhibition by uncharged tRNA played a significant effect in our transamidation results. When we added a 64-fold excess of uncharged tRNAAsn, we were unable to compete GatCAB away from Asp-tRNAAsn. We were able to show that the D-loop and the T-stem/loop of tRNAAsn are important for GatCAB recognition. Since no tRNAAsn mutants with single base changes in the D-loop could satisfactorily be aspartylated, it is currently unclear, if size of the D-loop or specific bases are important for GatCAB recognition. However, the data clearly show that a U49 in the T-arm (as found in M. thermautotrophicus tRNAAsp) is sufficient for GatCAB rejection (Table 2). Given the tRNA sequences of archaeal tRNAAsp species (Table 2), position 49 is probably used as an anti-determinant by many archaeal GatCAB enzymes to discriminate against Asp-tRNAAsp. The importance of the variable loop for GatCAB recognition is also evident from the significant activity loss observed in the domain-swap mutant (Fig. 2B) and in the U47 → Δ47 tRNAAsn (Table 1). This region was also predicted to be important based on detailed tRNA sequence comparisons [16]. In N. equitans and M. kandleri, whose tRNAAsp and tRNAAsn species are invariant at position 49, discrimination against Asp-tRNAAsp is probably accomplished either by use of the D and variable loops alone or by recognition of additional elements in the tRNAs.
In bacteria, a single AdT, GatCAB, exists to transamidate Asp-tRNAAsn and Glu-tRNAGln [7]. It is not too surprising then that the major recognition elements, the U1-A72 pair and the smaller D-loop, are conserved between bacterial tRNAGln and tRNAAsn species [11,16]. In archaea, two AdTs exist, GatDE and GatCAB [3]. The former is solely a Glu-AdT discriminating Glu-tRNAGln from Glu-tRNAGlu, Asp-tRNAAsp, and Asp-tRNAAsn [3]. One of the major elements GatDE recognizes in tRNAGln is the A1-U72 pair in the acceptor stem [15]. The other three tRNA species mentioned have a G-C pair at that position. It appears, one of the ways archaea may have evolved an AdT specific for Glu-tRNAGln, was to encode tRNAAsn with the G1-C72 pair found in tRNAGlu and tRNAAsp making Asp-tRNAAsn a poorer substrate for GatDE. Evolution of the specific Glu-AdT, GatDE, in archaea, therefore, enacted a selective pressure on the archaeal GatCAB, forcing its coevolution with tRNAAsn in order to maintain its Asp-AdT function. This same pressure may have been responsible for the evolution of new antideterminants, such as position 49, in tRNAAsp.
Acknowledgments
We would like to thank Patrick, O'Donoghue, Liang Feng and Debra Tumbula-Hansen for critical discussions and Joanne Ho for experimental assistance. This work was supported by grants from the National Institute of General Medical Sciences (GM22854) and the Department of Energy (DE-FG02-98ER20311).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–50. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Curnow AW, Ibba M, Söll D. tRNA-dependent asparagine formation. Nature. 1996;382:589–590. doi: 10.1038/382589b0. [DOI] [PubMed] [Google Scholar]
- 3.Tumbula DL, Becker HD, Chang WZ, Söll D. Domain-specific recruitment of amide amino acids for protein synthesis. Nature. 2000;407:106–110. doi: 10.1038/35024120. [DOI] [PubMed] [Google Scholar]
- 4.Curnow AW, Tumbula DL, Pelaschier JT, Min B, Söll D. Glutamyl-tRNAGln amidotransferase in Deinococcus radiodurans may be confined to asparagine biosynthesis. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12838–12843. doi: 10.1073/pnas.95.22.12838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker HD, et al. The heterotrimeric Thermus thermophilus Asp-tRNAAsn amidotransferase can also generate Gln-tRNAGln. FEBS Lett. 2000;476:140–144. doi: 10.1016/s0014-5793(00)01697-5. [DOI] [PubMed] [Google Scholar]
- 6.Becker HD, Roy H, Moulinier L, Mazauric MH, Keith G, Kern D. Thermus thermophilus contains an eubacterial and an archaebacterial aspartyl-tRNA synthetase. Biochemistry. 2000;39:3216–3230. doi: 10.1021/bi992573y. [DOI] [PubMed] [Google Scholar]
- 7.Raczniak G, Becker H, Min B, Söll D. A single amidotransferase forms asparaginyl-tRNA and glutaminyl-tRNA in Chlamydia trachomatis. J. Biol. Chem. 2001;276:45862–45867. doi: 10.1074/jbc.M109494200. [DOI] [PubMed] [Google Scholar]
- 8.Akochy PM, Bernard D, Roy PH, Lapointe J. Direct glutaminyl-tRNA biosynthesis and indirect asparaginyl-tRNA biosynthesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 2004;186:767–76. doi: 10.1128/JB.186.3.767-776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilcox M, Nirenberg M. Transfer RNA as a cofactor coupling amino acid synthesis with that of protein. Proc. Natl. Acad. Sci. U. S. A. 1968;61:229–236. doi: 10.1073/pnas.61.1.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curnow AW, Hong K, Yuan R, Kim S, Martins O, Winkler W, Henkin TM, Söll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. U. S. A. 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura A, Yao M, Chimnaronk S, Sakai N, Tanaka I. Ammonia channel couples glutaminase with transamidase reactions in GatCAB. Science. 2006;312:1954–8. doi: 10.1126/science.1127156. [DOI] [PubMed] [Google Scholar]
- 12.Harpel MR, et al. Mutagenesis and mechanism-based inhibition of Streptococcus pyogenes Glu-tRNAGln amidotransferase implicate a serine-based glutaminase site. Biochemistry. 2002;41:6398–6407. doi: 10.1021/bi012126u. [DOI] [PubMed] [Google Scholar]
- 13.Feng L, Sheppard K, Tumbula-Hansen D, Söll D. Gln-tRNAGln formation from Glu-tRNAGln requires cooperation of an asparaginase and a Glu-tRNAGln kinase. J. Biol. Chem. 2005;280:8150–5. doi: 10.1074/jbc.M411098200. [DOI] [PubMed] [Google Scholar]
- 14.Wilcox M. Gamma-glutamyl phosphate attached to glutamine-specific tRNA. A precursor of glutaminyl-tRNA in Bacillus subtilis. Eur. J. Biochem. 1969;11:405–412. doi: 10.1111/j.1432-1033.1969.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 15.Oshikane H, et al. Structural basis of RNA-dependent recruitment of glutamine to the genetic code. Science. 2006;312:1950–4. doi: 10.1126/science.1128470. [DOI] [PubMed] [Google Scholar]
- 16.Bailly M, Giannouli S, Blaise M, Stathopoulos C, Kern D, Becker HD. A single tRNA base pair mediates bacterial tRNA-dependent biosynthesis of asparagine. Nucleic Acids Res. 2006;34:6083–6094. doi: 10.1093/nar/gkl622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tumbula-Hansen D, Feng L, Toogood H, Stetter KO, Söll D. Evolutionary divergence of the archaeal aspartyl-tRNA synthetases into discriminating and nondiscriminating forms. J. Biol. Chem. 2002;277:37184–90. doi: 10.1074/jbc.M204767200. [DOI] [PubMed] [Google Scholar]
- 18.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–98. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knowlton RG, Yarus M. Discrimination between aminoacyl groups on su+ 7 tRNA by elongation factor Tu. J. Mol. Biol. 1980;139:721–32. doi: 10.1016/0022-2836(80)90057-1. [DOI] [PubMed] [Google Scholar]
- 20.Ibba M, Francklyn CS, Cusack S. The Aminoacyl-tRNA Synthetases, Landes Bioscience. Georgetown, Texas: 2005. [Google Scholar]
- 21.Giege R, Sissler M, Florentz C. Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res. 1998;26:5017–35. doi: 10.1093/nar/26.22.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]