Abstract
Background
Because of its recent identification, few multi-year epidemiologic studies of hMPV infection have been reported.
Objective
We sought to retrospectively describe hMPV infections among patients evaluated by a large US Midwestern referral laboratory.
Study design
Clinical specimens were submitted to a large US Midwest referral hospital from 1 October 2001 to 18 May 2004. RT-PCR was used to retrospectively screen the clinical specimens for human metapneumovirus. Demographic and clinical data were retrieved.
Results
34 (2.6%) of 1294 specimens were hMPV positive. Among these, 21 (62%) were culture positive and available for genetic typing. A previously considered rare genotype of hMPV, B1, was the most common single genotype identified, comprising 9 (43%) of the 21 isolates. Multivariate logistic regression modeling identified patients aged 0.4–9 years (OR = 8.9; 95% CI = 2.0–38.5) and those under intensive care (OR = 3.2; 95% CI = 1.1–8.7) as more likely to have hMPV infection than their peers.
Conclusion
In this large referral hospital viral assays more often had evidence of hMPV when they were collected from children receiving intensive care.
Keywords: Human metapneumovirus, Respiratory viruses, Epidemiology, Genotyping
1. Introduction
Since hMPV was first reported (van den Hoogen et al., 2001) in 2001, much progress has been made in its detection (Cote et al., 2003; Ebihara et al., 2003, 2004; Hamelin and Boivin, 2005; Leung et al., 2005; Mackay et al., 2003; Maertzdorf et al., 2004), in understanding its genetic diversity (Boivin et al., 2002; Falsey et al., 2003; Ludewick et al., 2005; Mackay et al., 2003) and in documenting its varied clinical presentation (Falsey et al., 2003; Principi et al., 2006). By linking demographic and clinical data to a large, 3-year, viral culture specimen repository, we sought to describe hMPV infections among patients evaluated by a large US Midwestern referral laboratory.
2. Methods
2.1. Specimen collection and clinical data
We retrospectively studied a convenient sample of 1500 available, archived respiratory specimens, collected from 1 October 2001 to 18 May 2004, at the at the University of Iowa Hospitals and Clinics' Clinical Microbiology Laboratory. Respiratory specimen data (75% collected by nasal wash) were linked to patient demographic and clinical data: gender, age, race/ethnicity, clinic/ward source, date and site of culture, clinical diagnoses, patient disposition, admission/discharge date, and co-infections with other respiratory pathogens such as adenovirus, influenza A, influenza B, parainfluenza (1, 2, 3) and respiratory syncytial virus (RSV).
2.2. Reverse-transcription polymerase chain reaction (RT-PCR)
Viral specimens were evaluated for molecular evidence of hMPV as per our previous report (Gray et al., 2006). Briefly, specimens were screened with a one-step RT-PCR procedure using primer set F2 (Table 1). Screened specimens that yielded bands within 50 bp of the expected 347 bp product, were further tested with a two-step RT-PCR, using the F1, F2 and N-gene primer sets (Table 1). Specimens were designated as “RT-PCR+” if the N-gene primer set and at least one of the confirmatory F-gene primer sets yielded appropriately sized bands.
Table 1.
Oligonucleotide primers used for amplification and sequencing of hMPV
| Primer set name | Target | Amplicon size (bp) | Sequence |
|---|---|---|---|
| F1 (Peret et al., 2002) | F gene | 450 | Forward: 5′-CTTTGGACTTAATGACAGATG-3′; |
| Reverse: 5′-GTCTTCCTGTGCTAACTTTG-3′ | |||
| F2 (Falsey et al., 2003) | F gene | 347 | Forward: 5′-GAGCAAATTGAAAATCCCAGACA-3′; |
| Reverse: 5′-GAAAACTGCCGCACAACATTTAG-3′ | |||
| N1 (Mackay et al., 2003) | N gene | 210 | Forward: 5′-AACCGTGTACTAAGTGATGCACTC-3′; |
| Reverse: 5′-CATTGTTTGACCGGCCCCATAA-3′ | |||
| Guniv (Ludewick et al., 2005) | G gene | 800–1000 | Forward: 5′-GAGAACATTCGRRCRATAGAYATG-3′; |
| Reverse: 5′-AGATAGACATTRACAGTGGATTCA-3′ |
2.3. hMPV isolation
RT-PCR+ specimens were further studied with shell-vial LLC-MK2 cell culture for the presence of viable hMPV. Shell vials were incubated for 3–4 weeks at or until cell disruption occurred. Infected cell supernatant media was harvested each week upon cell media replacement. From an aliquot of the infected media, RNA was extracted and a subsequent RT-PCR was performed via the hMPV F2 gene one-step protocol.
2.4. Sequencing
Amplification products obtained using Guniv primers (Table 1) adapted for amplification of a 800–1000 bp region of the hMPV G protein gene were electrophoresed on 1.0% agarose gels and purified. The purified DNA was then sequenced in both directions.
2.5. Phylogenetic analysis
Alignments of the partial nucleotide sequences of the hMPV G protein gene were generated with the program ClustalW (National Center for Biotechnology Information, Bethesda, MD).
2.6. Statistical analysis
The Cochan–Armitage test was used to verify prevalence trends among infants. Binomial confidence intervals were computed about crude prevalence statistics. Unadjusted and adjusted odds ratios were computed for each risk factor using logistic regression. The final multivariable model was identified by using a saturated model and manual backwards elimination.
3. Results
3.1. Diagnostics and genetic characterization
Considering only the first specimen per medical event and good linkage data, 1294 specimens were available for study. Thirty-four (2.6%) of these specimens were positive by the RT-PCR screening assay and all were confirmed by the additional RT-PCR assays. Of the RT-PCR positive specimens, 21 (62%) grew hMPV virus in LLC-MK2 cell cultures. CPE was detectable at 14 days post-inoculation.
3.2. Clinical features
Among the 34 hMPV+ patients, seven presented with respiratory failure (Table 2). One of these, a 3-year-old Caucasian boy was respirator-dependent. Twenty-seven of the 34 hMPV+ patients were hospitalized at the time of culture (median stay 8 days). Frequent diagnoses among the hMPV+ patients included pneumonia, bronchiolitis, and nonspecific acute upper respiratory infection. Among the patients from whom hMPV genotype B was detected, 42% (5 isolates) had leukemia, in contrast to 11% (1 isolate) of patients from whom hMPV genotype A was grown. Among these five hMPV genotype B isolates, four (80%) were of subtype B1 (Table 2).
Table 2.
Demographics and clinical features of patients RT-PCR positive for human metapneumovirus (hMPV)
| Case | Date (month/year) | hMPV isolate, genotype (Peret et al., 2004), GenBank accession number | Race | Gender | Age (year) | Days in hospital | Co-infectiona | Reason for assessment (diagnosis) | Other diagnoses of interest |
|---|---|---|---|---|---|---|---|---|---|
| 1 | March 2002 | – | Caucasian | F | 54.7 | 14 | – | Obstructive bronchitis without exacerbation | Heart replaced by transplant |
| 2 | March 2002 | IA3-2002, A1, DQ312444 | Caucasian | M | 0.6 | 8 | – | Congestive heart failure, unspecified | Congenital mitral insufficiency |
| 3 | March 2002 | – | Asian | F | 4.6 | 3 | – | Bronchitis, not specified as acute or chronic | |
| 4 | March 2002 | – | Caucasian | F | 8.9 | 2 | – | Pneumonia, organism unspecified | Congenital cytomegalovirus infection |
| 5 | December 2002 | IA4-2002, B1, DQ312445 | Caucasian | F | 6.3 | 3 | – | Fever | Acute lymphoid leukemia in remission; anemia, unspecified |
| 6 | December 2002 | IA5-2002, B1, DQ312446 | Caucasian | F | 5.8 | – | – | Osteopetrosis | Bone marrow transplant |
| 7 | January 2003 | – | Caucasian | F | 78.5 | 10 | – | Obstructive bronchitis with (acute) exacerbation | Respiratory failure |
| 8 | January 2003 | IA6-2003, B1, DQ312447 | Caucasian | F | 9.5 | – | – | Chronic myeloid leukemia without mention of remission | |
| 9 | January 2003 | – | Caucasian | M | 2.7 | 2 | – | Upper respiratory infection (acute), unspecified site | Chronic respiratory disease arising in the perinatal period |
| 10 | January 2003 | IA7-2003, B1, DQ312448 | Caucasian | M | 17.8 | 9 | – | Respiratory failure | Congenital quadriplegic |
| 11 | January 2003 | – | Caucasian | M | 3.5 | 6 | – | Respiratory failure | Chronic respiratory disease arising in the perinatal period; dependence on respirator; pulmonary collapse, other chronic pulmonary heart disease |
| 12 | January 2003 | – | African American | M | 0.8 | 1 | – | Acute bronchiolitis due to other infectious organisms | Pulmonary collapse, congenital anomaly of lung unspecified |
| 13 | January 2003 | IA8-2003, A2, DQ312449 | African American | F | 2.7 | 3 | – | Pneumonia, organism unspecified | Prader–Willi syndrome |
| 14 | January 2003 | IA9-2003, B1, DQ312450 | Caucasian | F | 5.4 | 5 | – | Bacteremia (micrococcus species) | Acute lymphoid leukemia without remission, agranulocytosis |
| 15 | January 2003 | IA10-2003, A1, DQ312451 | Caucasian | F | 22.8 | 8 | – | Respiratory failure | Acute renal failure; systemic lupus erythematosus; nephritis and nephropathy, not specified as acute or chronic, in diseases classified elsewhere |
| 16 | January 2003 | IA11-2003, B1, DQ312452 | Caucasian | F | 18.6 | – | – | Influenza with other respiratory manifestations | |
| 17 | January 2003 | – | Caucasian | F | 0.8 | 11 | – | Respiratory failure | Pneumonia, organism unspecified; agenesis of lung |
| 18 | February 2003 | IA13-2003, B1, DQ312454 | Unknown | F | 0.9 | – | – | Cough | |
| 19 | February 2003 | IA14-2003, A2, DQ312455 | Caucasian | M | 57.7 | 18 | – | Pneumonia due to streptococcus goup A | |
| 20b | February 2003 | – | Caucasian | M | 61.4 | – | – | Myeloid leukemia | Pneumonia; aspergillosis |
| 21 | February 2003 | IA15-2003, A1, DQ312456 | Caucasian | F | 0.3 | 20 | – | Acute bronchiolitis due to other infectious organisms | Tetralogy of fallot; infundibular pulmonic stenosis; ostium secundum type atrial septal defect |
| 22 | February 2003 | IA16-2003, B2, DQ312457 | Caucasian | F | 6.3 | 4 | Influenza B | Other specified aplastic anemias | Acute lymphoid leukemia in remission |
| 23 | February 2003 | IA17-2003, A2, DQ312458 | Unknown | F | 45.0 | – | – | Screening for unspecified condition | |
| 24 | February 2003 | IA-18-2003, B1, DQ312459 | Caucasian | M | 3.9 | 8 | – | Unspecified infectious and parasitic diseases | Postinfectious encephalitis; coma |
| 25b | February 2003 | – | Caucasian | M | 68.7 | 14 | – | Malignant neoplasm lower lobe, bronchus or lung | Pneumococcal pneumonia; abscess of lung; respiratory failure |
| 26 | February 2003 | – | Caucasian | M | 0.1 | 6 | – | Acute bronchiolitis due to respiratory syncytial virusc | Pulmonary collapse; respiratory failure |
| 27 | March 2003 | – | Unknown | M | 1.4 | – | RSV | Acute bronchiolitis due to respiratory syncytial virus | Chronic maxillary sinusitis |
| 28 | March 2003 | IA19-2003, B2, DQ312460 | Caucasian | M | 3.9 | 13 | – | Respiratory failure | Hypertrophic obstructive cardiomyopathy; complications of transplanted liver |
| 29 | March 2003 | – | Caucasian | F | 30.6 | 17 | – | Other pulmonary insufficiency, not elsewhere classified | Other shock without mention of trauma; pneumonia, organism unspecified; anoxic brain damage |
| 30 | April 2003 | IA20-2003, B1, DQ312461 | Caucasian | M | 4.3 | 45 | – | Acute lymphoid leukemia without remission | Other specified aplastic anemias; bacteremia; pseudomonas; hyperpotassemia; other pulmonary insufficiency, not elsewhere classified; hypopotassemia; abnormal liver scan |
| 31 | January 2004 | IA21-2003, A2, DQ312462 | Unknown | F | 4.4 | 3 | – | Fever | Acute lymphoid leukemia in remission; other diseases of nasal cavity and sinuses; agranulocytosis; Other nonspecific abnormal serum enzyme levels |
| 32 | February 2004 | IA22-2003, A2, DQ312463 | Unknown | M | 10.2 | 6 | – | Other specified aplastic anemias | Hypopotassemia; Hodgkin's disease, unspecified |
| 33 | March 2004 | IA23-2004, A2, DQ312464 | Hispanic | M | 1.2 | 1 | – | Foreign body in esophagus | Food/vomit pneumonitis; pulmonary insufficiency following trauma and surgery |
| 34 | March 2004 | IA24-2004, B2, DQ312465 | Unknown | F | 2.1 | 8 | – | Epilepsy, grand mal status | Pneumonitis due to inhalation of food or vomitus; periventricular leukomalacia; unspecified otitis media; esophageal reflux |
Diagnoses were recorded using the International Classification of Diseases, Ninth Revision.
Another virus detected in the same culture specimen where hMPV was detected.
Patient died.
Not confirmed by laboratory microbiological tests.
3.3. Risk factor modeling
Data from the 1294 patients were analyzed. The median patient age was 5.3 years: 52% were male and 66.8% were Caucasian (Table 3). Most specimens, 83.5%, were collected during the months of December–April.
Table 3.
Characteristics of clinical specimens tested for human metapneumovirus in comparison to other respiratory viruses, without duplicatesa
| Risk factor | Number of specimens (%) |
|||||
|---|---|---|---|---|---|---|
| Total sample (n = 1294) | Human metapneumovirus-positive (n = 34) | Respiratory syncytial virus-positive (n = 174) | Parainfluenza virus-positive (n = 22) | Influenza virusb-positive (n = 120) | Adenovirus-positive (n = 11) | |
| Age group (year) | ||||||
| <0.4 | 288 (22.3) | 2 (5.9) | 59 (33.9) | 3 (13.6) | 5 (4.2) | 0 (0.0) |
| 0.4–9 | 415 (32.1) | 21 (61.8) | 97 (55.8) | 17 (77.3) | 50 (41.7) | 6 (54.6) |
| 10–40 | 261 (20.2) | 5 (14.7) | 7 (4) | 0 (0.0) | 41 (34.2) | 2 (18.2) |
| >40 | 329 (25.4) | 6 (17.7) | 10 (5.8) | 2 (9.1) | 24 (20) | 3 (27.3) |
| Unkown | 1 (0.1) | 0 (0.0) | 1 (0.6) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Gender | ||||||
| Female | 621 (48) | 19 (55.9) | 78 (44.8) | 11 (50) | 61 (50.8) | 5 (45.5) |
| Male | 673 (52) | 15 (44.1) | 96 (55.2) | 11 (50) | 59 (49.2) | 6 (54.6) |
| Race/ethnicity | ||||||
| Caucasian | 864 (66.8) | 24 (70.6) | 108 (62.1) | 13 (59.1) | 69 (57.5) | 7 (63.6) |
| African American | 68 (5.3) | 2 (5.9) | 9 (5.2) | 3 (2.5) | 1 (9.1) | |
| Hispanic | 45 (3.5) | 1 (2.9) | 5 (2.9) | 1 (4.6) | 8 (6.7) | 0 (0.0) |
| Asian | 27 (2.1) | 1 (2.9) | 3 (1.7) | 1 (4.6) | 4 (3.3) | 0 (0.0) |
| Othersc | 43 (3.4) | 0 (0.0) | 6 (3.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Unkown | 247 (19.1) | 6 (17.7) | 43 (24.7) | 7 (31.8) | 36 (30) | 3 (27.3) |
| Patient group | ||||||
| Intensive care unit | 223 (17.2) | 9 (26.5) | 16 (9.2) | 5 (22.7) | 4 (3.3) | 0 (0.0) |
| Observationd | 70 (5.4) | 2 (5.9) | 8 (4.6) | 1 (4.6) | 2 (1.7) | 0 (0.0) |
| Hospital ward | 607 (46.9) | 16 (47.1) | 73 (42) | 12 (54.6) | 32 (26.7) | 5 (45.5) |
| Outpatient | 394 (30.5) | 7 (20.6) | 77 (44.3) | 4 (18.2) | 82 (68.3) | 6 (54.6) |
| Month | ||||||
| January–March | 759 (58.7) | 31 (91.2) | 144 (82.8) | 5 (22.7) | 69 (57.5) | 9 (81.8) |
| April–June | 116 (9.0) | 1 (2.9) | 13 (7.5) | 4 (18.2) | 0 (0.0) | 0 (0.0) |
| July–September | 54 (4.2) | 0 (0.0) | 0 (0.0) | 4 (18.2) | 0 (0.0) | 0 (0.0) |
| October–December | 365 (28.2) | 2 (5.9) | 17 (9.8) | 9 (40.9) | 51 (42.5) | 2 (18.2) |
| Culture sites | ||||||
| Nasopharyngeal wash | 11 (0.9) | 1 (2.9) | 1 (0.6) | 0 (0.0) | 1 (0.8) | 0 (0.0) |
| Nasal wash | 972 (75.1) | 24 (70.6) | 164 (94.3) | 16 (72.7) | 111 (92.5) | 10 (90.9) |
| Bronch wash | 30 (2.3) | 1 (2.9) | 0 (0.0) | 1 (4.6) | 0 (0.0) | 0 (0.0) |
| Bronchoalveolar lavage | 135 (10.4) | 2 (5.9) | 2 (1.2) | 0 (0.0) | 1 (0.8) | 1 (9.1) |
| Tracheal aspirate | 101 (7.8) | 6 (17.7) | 7 (4) | 5 (22.7) | 2 (1.7) | 0 (0.0) |
| Others | 45 (3.5) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 5 (4.2) | 0 (0.0) |
Duplicates were defined as multiple specimens with the same laboratory results taken from the same patient during a 2-week period.
Influenza A, influenza B or both.
The other race/ethnicity category includes Native Americans.
Patients held for observation but not hospitalized.
Most (61.8%) of the hMPV+ specimens were from patients with ages of 0.4–9 years (Table 3). In fact, a significant increasing trend (p < .001) of hMPV+ prevalence by age was noticed in infants up to 6 years old (Fig. 1). Specimens that were positive for parainfluenza viruses were similarly more likely to be statistically associated with this age group, in contrast to the age distributions and influenza A virus, which were not statistically associated (Table 3).
Fig. 1.
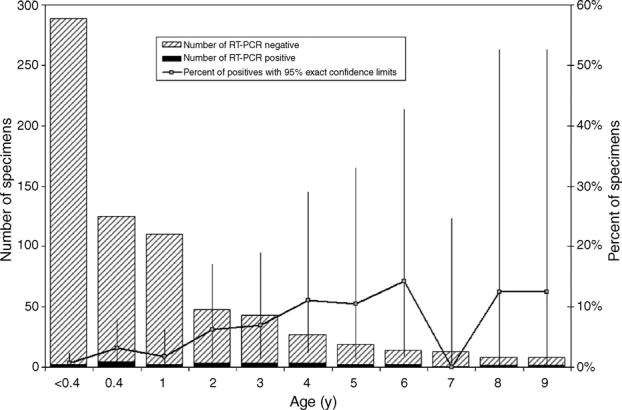
Distribution of human metapneumovirus RT-PCR+ specimens from patients with ages 0–9 years old.
All 34 hMPV+ specimens were also screened for RSV, and one was positive. Similarly, 33 of the hMPV+ specimens were screened for influenza A and B infection, and only one yielded evidence of co-infection with influenza B (Table 2). Ninety-three percent of the RSV+ specimens were collected during the same “high-risk” period (December–April) as were many of the hMPV+ specimens (Table 3).
3.4. Multivariable modeling
Logistic regression modeling revealed that only age group and patient group were associated with hMPV+ specimens (Table 4). Gender was not an important predictor of hMPV infection. The final multivariate logistic model indicated that children between 0.4 and 9 years of age had 8.9 times the odds of having hMPV infection compared with infants 0.4 years old or younger (Table 4). In addition, when adjusting for age group, intensive care unit patients had a higher odds (OR = 3.2) of being hMPV+ as comparing to outpatients.
Table 4.
Prevalence and odds ratio of RT-PCR positivity for human metapneumovirus by risk factor, without duplicatesa
| Risk factor | Crude prevalence %hMPV PCR+ (95% CI) | Bivariate analyses OR (95% CI) | Multivariable logistic regression OR (95% CI) |
|---|---|---|---|
| Age group (year) | |||
| <0.4 | 0.7 (0.1–2.5) | Reference | Reference |
| 0.4–9 | 5.1 (3.2–7.6) | 7.6 (1.8–32.8) | 8.9 (2.0–38.5) |
| 10–40 | 1.9 (0.6–4.4) | 2.8 (0.5–14.5) | 3.2 (0.6–16.9) |
| >40 | 1.8 (0.7–3.9) | 2.7 (0.5–13.3) | 2.7 (0.5–13.4) |
| Gender | |||
| Female | 3.1 (1.9–4.7) | 1.4 (2.7–0) | – |
| Male | 2.2 (1.3–3.7) | Reference | – |
| Patient group | |||
| Intensive care unit | 96 (1.9–7.5) | 2.3 (0.9–6.3) | 3.2 (1.1–8.7) |
| Observationb | 2.9 (0–6.8) | 1.6 (0.3–8) | 1.9 (0.4–9.3) |
| Hospital ward | 2.6 (1.5–4.3) | 1.5 (0.6–3.7) | 1.9 (0.8–4.6) |
| Outpatient | 1.8 (0.7–3.6) | Reference | Reference |
Due to missing data the total counts per risk factor are not identical. OR: odds ratio; CI: confidence interval.
Duplicates were defined as multiple specimens with the same laboratory results taken from the same patient during a 2-week period.
Patients held for observation but not hospitalized.
3.5. Phylogenetic analyses of hMPV strains
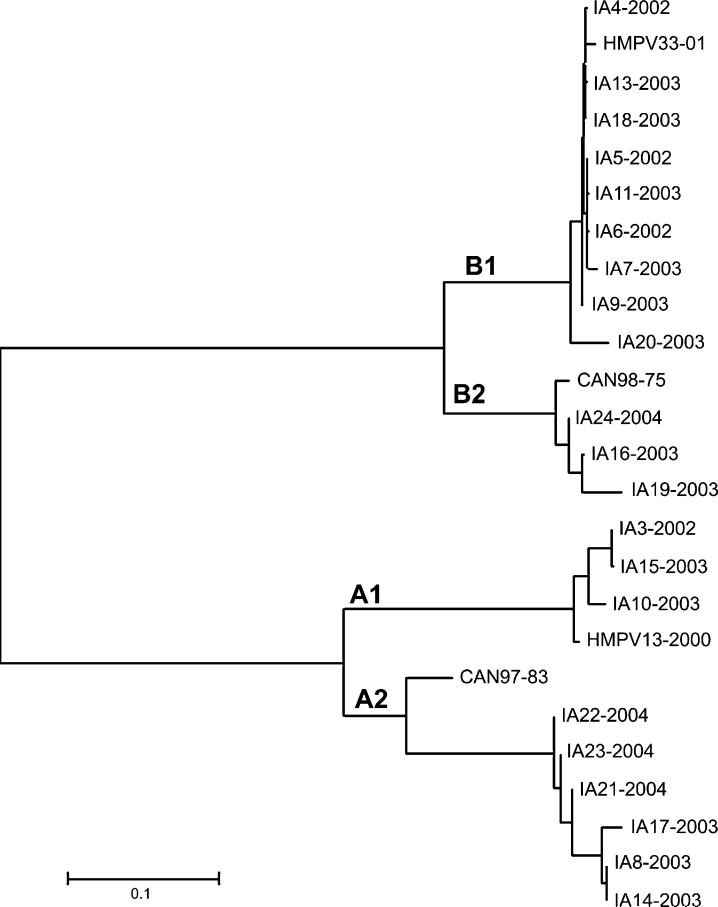
The 21 specimens that were culture positive for hMPV were successfully sequenced. Phylogenetic studies (Fig. 2) revealed a high prevalence of subtypes B1 (nine isolates) and A2 (six isolates).
Fig. 2.
Neighbor joining tree computed from alignment of partial nucleotide sequences of the hMPV G protein gene using the MEGA 2 program (University of Pittsburgh, Pittsburgh, PA). Sequences obtained from hMPV isolates in this study (GenBank accession numbers are listed in Table 2) are compared to prototypic Canadian viruses: HMPV-13-00 (genotype A1), Can97-83 (genotype A2), HMPV-33-01 (genotype B1), and CAN98-75 (genotype B2), GenBank accession numbers AY485232, AY485253, AY485242, and AY485245, respectively.
4. Discussion
Previous studies have associated hMPV infection with rhinorrhea, pharyngitis, otitis media, bronchiolitis, bronchitis, pneumonia, respiratory failure, and death (Pelletier et al., 2002; Williams et al., 2004). hMPV infections may exacerbate reactive airway disease among both the young and the old (Boivin et al., 2002; Greensill et al., 2003). There is evidence that some patients suffer repeat hMPV infections (Boivin et al., 2003), that mixed hMPV infections may be common (Schildgen et al., 2005), that many hMPV infections occur in winter months, and that the immunocompromised may be at greater risk of hMPV disease (van den Hoogen et al., 2001). Our data are consistent with many of these observations but offer some new possible risk factor associations. Our 34 hMPV+ specimens were obtained from patients with a wide array of clinical conditions, many of whom were immunocompromised. Among the 34 hMPV+ specimens, two (Table 2, patients 20 and 25) were taken from patients >60 years of age who died; however, their deaths occurred 2 months after the specimens were collected and had underlying cancers that could explain their deaths. A third severely ill patient who survived, a 3-year-old boy (Table 2, patient 24) diagnosed with acute encephalomyelitis and coma, also had hMPV detected (genotype B1). Again while his respiratory culture yielded hMPV, it is difficult to clearly implicate hMPV as the etiology for his encephalitis. However, encephalitis has been previously found to be associated with hMPV (Boivin et al., 2002).
Although it is difficult to determine the primary etiology of many of the patients' diagnoses through this retrospective review of medical records, it seems safe to assume that clinicians were at least concerned about a viral cofactor, as they took the initiative to order viral cultures. Thus far asymptomatic carriage of hMPV has been only rarely reported (Boivin et al., 2002; Williams et al., 2004). Among the 34 hPMV+ patients, it is interesting to note that four (Table 2, patients 14, 19, 25, 30) had bacterial infections, four had other viral infections (Table 2, patients 4, 16, 26, 27), and one had a fungal infection (Table 2, patient 20).
Consistent with previous reports, our hMPV+ patients ranged in age from less than a year to 79 years (Boivin et al., 2003) and most infections occurred during the winter months and early spring (van den Hoogen et al., 2004). Also consistent with a previous report, we found that patients from the 0.4 to 9 years had a markedly increased risk (OR = 8.9, Table 4) of infection compared to those less than 0.4 years of age (Ludewick et al., 2005). This observation is likely due to waning maternal immunity. The increased risk and the increasing trend in hMPV prevalence (Fig. 1) up to age six suggests hMPV surveillance and preventive measures may be most efficiently focused upon patients less than 6 years of age.
Recent studies suggest that hMPV infection is not without clinical morbidity. Many of our hMPV+ patients and those previously reported (Boivin et al., 2004; Ludewick et al., 2005; Peret et al., 2004) have been hospitalized with severe disease. While severe disease association observations in clinical case series studies may represent a bias in sample selection (e.g. only the most severely ill are cultured), in our study we were able to compare the odds of a specimen being hMPV+ through comparisons of patient groups' clinical status (e.g. outpatient, observation, hospitalization ward, intensive care unit). This comparison revealed that after adjusting for age differences, intensive care unit patients had increased odds of being hMPV+ compared to similarly tested outpatients, suggesting that hMPV infections may be more common among intensive care unit patients. This is likely due to the patients' underlying conditions, but it might also be explained by better clinical diagnosis or nosocomial transmission.
Our data were also unique in that we had a relative high proportion of isolates (43% of 21 sequenced) that were classified in the B1 genome subgroup (Fig. 2) (Ludewick et al., 2005; Mackay et al., 2004). This subgroup has previously been found to be rather uncommon in Europe, Canada, and South Africa (Fox et al., 1977, 1982; Hall et al., 1971). Some have speculated that the prevalence of circulating genome groups may change over time reflecting community immunity rather than viral emergence (Peret et al., 2002).
Like most previously published studies of hMPV, our analysis was limited to archived specimens collected for clinical purposes. This of course results in potential sampling biases and does not provide accurate community acquired incidence or risk factor data. However, because hMPV infections are rather rare, collecting such data would be difficult and very expensive.
Acknowledgements
Approved by the University of Iowa's institutional review board, this work was funded by the National Institute of Allergy and Infectious Diseases (R03 AI054570-01A1). We thank Mark LeBeck and Troy McCarthy of the Center for Emerging Infectious Diseases for their help with hMPV sequencing and the phylogenetic analysis.
Abbreviations
- hMPV
human metapneumovirus
- RT-PCR
reverse-transcription polymerase chain reaction
- E-MEM
Eagle's Minimum Essential Media
- CPE
cytopathic effect
References
- Boivin G, Abed Y, Pelletier G, Ruel L, Moisan D, Cote S, et al. Virological features and clinical manifestations associated with human metapneumovirus: a new paramyxovirus responsible for acute respiratory-tract infections in all age groups. J Infect Dis. 2002;186:1330–4. doi: 10.1086/344319. [DOI] [PubMed] [Google Scholar]
- Boivin G, De Serres G, Cote S, Gilca R, Abed Y, Rochette L, et al. Human metapneumovirus infections in hospitalized children. Emerg Infect Dis. 2003;9:634–40. doi: 10.3201/eid0906.030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boivin G, Mackay I, Sloots TP, Madhi S, Freymuth F, Wolf D, et al. Global genetic diversity of human metapneumovirus fusion gene. Emerg Infect Dis. 2004;10:1154–7. doi: 10.3201/eid1006.031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote S, Abed Y, Boivin G. Comparative evaluation of real-time PCR assays for detection of the human metapneumovirus. J Clin Microbiol. 2003;41:3631–5. doi: 10.1128/JCM.41.8.3631-3635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara T, Endo R, Kikuta H, Ishiguro N, Ishiko H, Kobayashi K. Comparison of the seroprevalence of human metapneumovirus and human respiratory syncytial virus. J Med Virol. 2004;72:304–6. doi: 10.1002/jmv.10572. [DOI] [PubMed] [Google Scholar]
- Ebihara T, Endo R, Kikuta H, Ishiguro N, Yoshioka M, Ma X, et al. Seroprevalence of human metapneumovirus in Japan. J Med Virol. 2003;70:281–3. doi: 10.1002/jmv.10391. [DOI] [PubMed] [Google Scholar]
- Falsey AR, Erdman D, Anderson LJ, Walsh EE. Human metapneumovirus infections in young and elderly adults. J Infect Dis. 2003;187:785–90. doi: 10.1086/367901. [DOI] [PubMed] [Google Scholar]
- Fox JP, Cooney MK, Hall CE, Foy HM. Influenza virus infections in Seattle families, 1975–1979. II. Pattern of infection in invaded households and relation of age and prior antibody to occurrence of infection and related illness. Am J Epidemiol. 1982;116:228–42. doi: 10.1093/oxfordjournals.aje.a113408. [DOI] [PubMed] [Google Scholar]
- Fox JP, Hall CE, Cooney MK. The Seattle Virus Watch. VII. Observations of adenovirus infections. Am J Epidemiol. 1977;105:362–86. doi: 10.1093/oxfordjournals.aje.a112394. [DOI] [PubMed] [Google Scholar]
- Gray G, Capuano A, Setterquist S, Sanchez J, Neville J, Olson J, et al. Human metapneumovirus. Peru Emerg Infect Dis. 2006;12:347–50. doi: 10.3201/eid1202.051133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greensill J, McNamara PS, Dove W, Flanagan B, Smyth RL, Hart CA. Human metapneumovirus in severe respiratory syncytial virus bronchiolitis. Emerg Infect Dis. 2003;9:372–5. doi: 10.3201/eid0903.020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall CE, Brandt CD, Frothingham TE, Spigland I, Cooney MK, Fox JP. The virus watch program: a continuing surveillance of viral infections in metropolitan New York families. IX. A comparison of infections with several respiratory pathogens in New York and New Orleans families. Am J Epidemiol. 1971;94:367–85. doi: 10.1093/oxfordjournals.aje.a121332. [DOI] [PubMed] [Google Scholar]
- Hamelin ME, Boivin G. Development and validation of an enzyme-linked immunosorbent assay for human metapneumovirus serology based on a recombinant viral protein. Clin Diagn Lab Immunol. 2005;12:249–53. doi: 10.1128/CDLI.12.2.249-253.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung J, Esper F, Weibel C, Kahn JS. Seroepidemiology of human metapneumovirus (hMPV) on the basis of a novel enzyme-linked immunosorbent assay utilizing hMPV fusion protein expressed in recombinant vesicular stomatitis virus. J Clin Microbiol. 2005;43:1213–9. doi: 10.1128/JCM.43.3.1213-1219.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewick HP, Abed Y, van Niekerk N, Boivin G, Klugman KP, Madhi SA. Human metapneumovirus genetic variability, South Africa. Emerg Infect Dis. 2005;11:1074–8. doi: 10.3201/eid1107.050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IM, Bialasiewicz S, Waliuzzaman Z, Chidlow GR, Fegredo DC, Laingam S, et al. Use of the P gene to genotype human metapneumovirus identifies 4 viral subtypes. J Infect Dis. 2004;190:1913–8. doi: 10.1086/425013. [DOI] [PubMed] [Google Scholar]
- Mackay IM, Jacob KC, Woolhouse D, Waller K, Syrmis MW, Whiley DM, et al. Molecular assays for detection of human metapneumovirus. J Clin Microbiol. 2003;41:100–5. doi: 10.1128/JCM.41.1.100-105.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertzdorf J, Wang CK, Brown JB, Quinto JD, Chu M, de Graaf M, et al. Real-time reverse transcriptase PCR assay for detection of human metapneumoviruses from all known genetic lineages. J Clin Microbiol. 2004;42:981–6. doi: 10.1128/JCM.42.3.981-986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier G, Dery P, Abed Y, Boivin G. Respiratory tract reinfections by the new human metapneumovirus in an immunocompromised child. Emerg Infect Dis. 2002;8:976–8. doi: 10.3201/eid0809.020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peret TC, Abed Y, Anderson LJ, Erdman DD, Boivin G. Sequence polymorphism of the predicted human metapneumovirus G glycoprotein. J Gen Virol. 2004;85:679–86. doi: 10.1099/vir.0.19504-0. [DOI] [PubMed] [Google Scholar]
- Peret TC, Boivin G, Li Y, Couillard M, Humphrey C, Osterhaus AD, et al. Characterization of human metapneumoviruses isolated from patients in North America. J Infect Dis. 2002;185:1660–3. doi: 10.1086/340518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Principi N, Bosis S, Esposito S. Human metapneumovirus in paediatric patients. Clin Microbiol Infect. 2006;12:301–8. doi: 10.1111/j.1469-0691.2005.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildgen O, Glatzel T, Geikowski T, Scheibner B, Matz B, Bindl L, et al. Human metapneumovirus RNA in encephalitis patient. Emerg Infect Dis. 2005;11:467–70. doi: 10.3201/eid1103.040676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–24. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hoogen BG, Osterhaus DM, Fouchier RA. Clinical impact and diagnosis of human metapneumovirus infection. Pediatr Infect Dis J. 2004;23:S25–32. doi: 10.1097/01.inf.0000108190.09824.e8. [DOI] [PubMed] [Google Scholar]
- Williams JV, Harris PA, Tollefson SJ, Halburnt-Rush LL, Pingsterhaus JM, Edwards KM, et al. Human metapneumovirus and lower respiratory tract disease in otherwise healthy infants and children. N Engl J Med. 2004;350:443–50. doi: 10.1056/NEJMoa025472. [DOI] [PMC free article] [PubMed] [Google Scholar]