Abstract
BC1 RNA is a dendritic untranslated RNA that has been implicated in local translational control mechanisms in neurons. Prerequisite for a functional role of the RNA in synaptodendritic domains is its targeted delivery along the dendritic extent. We report here that the targeting-competent 5′ BC1 domain carries two dendritic targeting codes. One code, specifying somatic export, is located in the medial-basal region of the 5′ BC1 stem-loop structure. It is defined by an export-determinant stem-bulge motif. The second code, specifying long-range dendritic delivery, is located in the apical part of the 5′ stem-loop domain. This element features a GA kink-turn (KT) motif that is indispensable for distal targeting. It specifically interacts with heterogeneous nuclear ribonucleoprotein A2, a trans-acting targeting factor that has previously been implicated in the transport of MBP mRNA in oligodendrocytes and neurons. Our work suggests that a BC1 KT motif encodes distal targeting via the A2 pathway and that architectural RNA elements, such as KT motifs, may function as spatial codes in neural cells.
Introduction
The targeted delivery of select RNAs to synaptodendritic domains in neurons is an elemental mechanism in the spatiotemporal regulation of postsynaptic gene expression (for reviews see Smith, 2004; Kindler et al., 2005). Activity-dependent on-site control of protein synthesis is now seen as one of the key underpinnings of input-specific long-term synaptic plasticity, with dendritic RNA transport in turn serving as a functional prerequisite for the local translation of mRNAs at the synapse (Wells et al., 2000; Job and Eberwine, 2001; Richter, 2001; Richter and Lorenz, 2002; Steward and Schuman, 2003; Smith, 2004; Kindler et al., 2005). For these reasons, structural and functional determinants of dendritic RNA transport are of considerable interest to students of neuronal plasticity (Smith, 2004).
The number of RNAs that have been identified in dendrites has steadily increased over the past few years, and the total number of dendritic RNAs has been estimated at several hundred (Eberwine et al., 2001). Dendritic RNAs form a diverse and complex group. They include mRNAs that encode various types of synaptodendritic proteins (such as receptors, kinases, cytoskeletal components, etc.), as well as untranslated RNAs (Brosius and Tiedge, 2004), such as ribosomal RNAs (rRNAs), tRNAs, micro RNAs, and small brain-specific cytoplasmic (BC) RNAs (Kindler et al., 2005; Cao et al., 2006). A further layer of complexity is introduced by the fact that dendritic RNAs are nonhomogeneously distributed along the dendritic extent and differentially localized to their individual destination sites within the dendritic arborization (Paradies and Steward, 1997; Kindler et al., 2005). Another subset of neuronal RNAs, only partially overlapping with dendritic RNAs, is targeted to the axonal domain, in particular during periods of developmental or regenerative growth (for review see Giuditta et al., 2002).
The complexities of neuronal RNA transport raise important questions. On the most fundamental level, spatial codes that are used by localized neuronal RNAs to specify their respective subcellular destinations remain poorly understood. Dendritic targeting elements (DTEs), which are cis-acting elements that contain codes to direct neuronal RNAs to or along dendrites, have been identified in several dendritic RNAs (Kindler et al., 2005). Frequently, however, the functional description of DTEs has been rudimentary, and code-carrying RNA motifs have not yet been deciphered (Smith, 2004).
Considerable progress has been made in recent years in the elucidation of structure-function relationships in RNAs (for review see Noller, 2005). There has been growing awareness of the functional relevance of 3D RNA motifs, and the question is thus raised whether dendritic targeting codes may find expression in the architectural designs of such motifs. To address this question, we performed a functional dissection of cis-acting targeting elements in neuronal BC1 RNA (Muslimov et al., 1997). BC1 RNA is a small untranslated dendritic RNA that has been implicated in translational control mechanisms (Wang et al., 2002; Kondrashov et al., 2005; Wang et al., 2005). We report here that the 5′ stem-loop domain of BC1 RNA contains two DTEs; BC1 DTE1, which is responsible for somatic export, and BC1 DTE2 which is responsible for long-range distal dendritic targeting. Both BC1 DTEs are characterized by structural motifs—stem-bulge and GA kink-turn (KT) motifs, respectively—that feature distinct 3D conformations. Our data indicate that such motifs serve as spatial codes in dendritic RNA targeting.
Results
BC1 RNA was chosen for a functional dissection of dendritic targeting codes for the following two reasons: (1) the RNA is transported along the entire dendritic extent, including distal segments and tips (Muslimov et al., 1997, 1998), and (2) the secondary structure of BC1 RNA, including its targeting-competent 5′ domain, has been experimentally established by chemical and enzymatic probing (Rozhdestvensky et al., 2001).
BC1 RNA (152 nt) is naturally subdivided into the following three major structural domains: a 74-nt 5′ BC1 domain that forms a stable stem-loop structure, a central stretch of 22 uninterrupted A-residues, and a 3′ BC1 domain that consists of a punctuated A-rich region and a 3′-terminal stem-loop structure (Rozhdestvensky et al., 2001). For the purpose of this analysis, we focus on the targeting-determinant 5′ BC1 domain, as other BC1 domains do not appear to contribute to dendritic targeting competence (Muslimov et al., 1997).
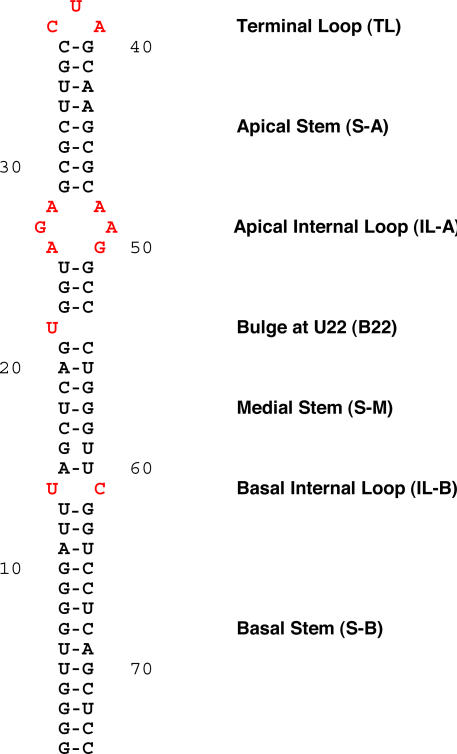
The experimentally established secondary structure of the 5′ BC1 domain (Rozhdestvensky et al., 2001) can be described as an A-form RNA helix with several nonhelical bulge and loop components (Fig. 1). Because RNA functionality is typically expressed by such nonhelical structural motifs (Lilley, 1995; Leontis and Westhof, 2003; Noller, 2005), we decided to examine each of these 5′ BC1 motifs for their contributions to targeting competence. Dendritic transport of the resulting BC1 derivatives was examined using a previously established microinjection protocol with neurons in primary culture (Muslimov et al., 1997; Shan et al., 2003). 35S-labeled RNAs were used throughout because in highly structured RNA motifs fluorescent and other side chains may easily perturb nucleotide–nucleotide or nucleotide–protein interactions that are required for architectural integrity or protein recognition. Dendritic targeting competence was calibrated against wild-type full-length BC1 RNA, which is transported along the entire dendritic extent (Fig. 2 A; Muslimov et al., 1997).
Figure 1.
Secondary structure of the 5′ BC1 domain. Structure was experimentally established by chemical and enzymatic probing (Rozhdestvensky et al., 2001). Helical elements are referred to as stems, and nonhelical elements are referred to as bulges or loops. Nonhelical motifs are shown in red.
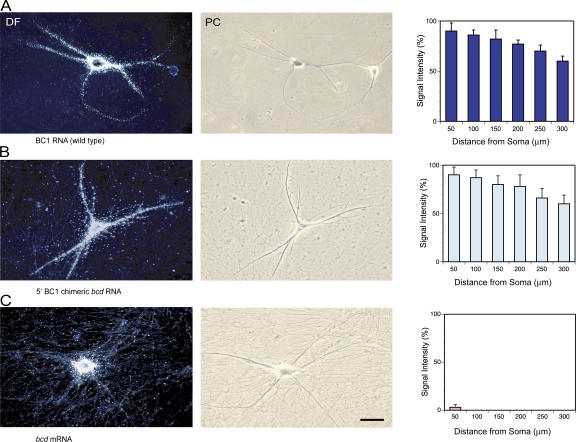
Figure 2.
Dendritic delivery of wild-type BC1 RNA and 5′ BC1 chimeric bcd RNA in sympathetic neurons in culture. (A) Full-length BC1 RNA was transported throughout the entire dendritic extent (Muslimov et al., 1997). The photomicrographs show dark-field (DF; left) and phase-contrast (PC; right) images of injected neurons. BC1 signal appears as white silver grains in DF images. (B) 5′ BC1 chimeric bcd RNA was distally delivered along dendrites, reaching dendritic tips. The 5′ BC1 domain was inserted 5′ of the bcd open reading frame. (C) bcd mRNA remained restricted to neuronal perikarya and was not detectable in proximal dendritic domains. Quantitative data (right) are presented as the mean ± the SEM of relative signal intensities along the dendritic extent. Bar, 50 μm.
We also ascertained whether the targeting elements that are contained within the 5′ BC1 domain are sufficient to specify distal dendritic delivery. To this end, we generated chimeric RNAs in which the 5′ stem-loop domain of BC1 RNA has been introduced into Drosophila melanogaster bcd mRNA. bcd mRNA is a transcript that is specific to dipteran flies and is not expressed in other eukaryotes (Wimmer et al., 2000). Microinjected into sympathetic neurons in culture, bcd mRNA remained restricted to neuronal somata and did not enter dendritic domains (Fig. 2 C). In contrast, chimeric bcd RNAs containing the 74-nt 5′ BC1 stem-loop domain were delivered along the entire dendritic extent (Fig. 2 B). The data thus indicate that in the system used, targeting elements contained within the 5′ BC1 domain are sufficient to direct distal dendritic delivery of a normally nondendritic RNA.
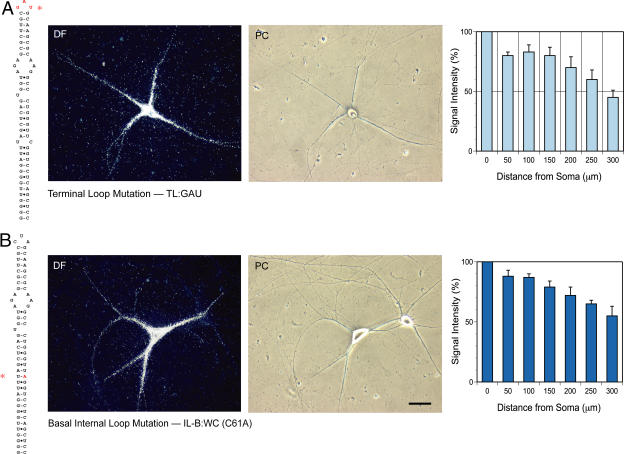
To identify targeting-competent motifs in the 5′ BC1 stem-loop domain, we worked in the apical-to-basal direction, beginning with the terminal CUA-loop motif. Terminal loop (TL) mutant full-length BC1 RNA, having the apical CUA triplet replaced with the corresponding GAU antisense sequence, still entered dendrites and was transported to distal dendritic segments (>300 μm), in a manner reminiscent of wild-type BC1 RNA (Figs. 2 and 3 A). Although it appeared that TL mutant BC1 RNA did not always reach extreme-distal dendritic regions and dendritic tips, statistical analysis revealed no substantial differences in the extent of dendritic labeling between TL mutant BC1 RNA and wild-type BC1 RNA. However, the possibility cannot be ruled out that the TL mutant exhibited reduced transport to distal-most dendritic segments, but that this reduction was not diagnosed in a statistically significant manner because of resolution limitations. The question of whether a short-range local targeting or anchoring code resides in the terminal loop motif must therefore remain open at this time.
Figure 3.
Derivatives of BC1 RNA that express undiminished dendritic targeting competence. Alterations made in the 5′ BC1 domain are shown in red and are indicated by an asterisk in the secondary structure representations (left). The photomicrographs show dark-field (DF; left) and phase-contrast (PC; right) images of injected neurons. (A) Terminal loop (TL) mutant BC1 RNA. In a sense–antisense mutation, the terminal loop triplet CUA was converted to GAU (TL:GAU). (B) Basal internal loop (IL-B) mutant BC1 RNA. C61 was exchanged with A to convert the loop to a standard WC pair (IL-B:WC). Histograms (right) show quantitative analyses (compare with Fig. 2 A). Statistical analysis revealed no significant differences in signal intensities between mutant and wild-type BC1 RNA at any point along the dendritic extent (e.g., one-way analysis of variance [ANOVA] for interval point 300 μm; P > 0.5). Quantitative data are presented as the mean ± the SEM of relative signal intensities along the dendritic extent. Bar, 50 μm.
Long-range dendritic transport: distal targeting
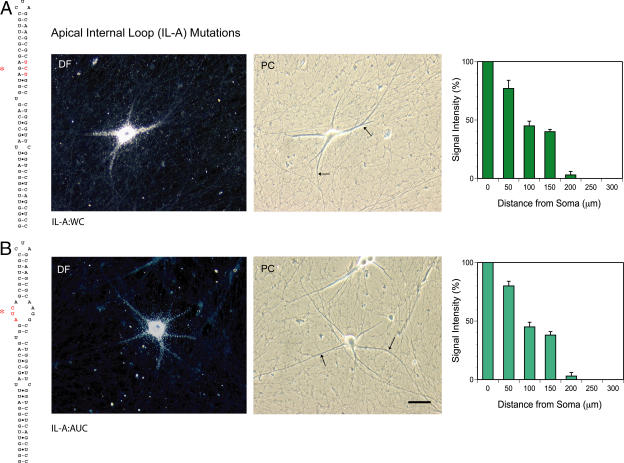
We next performed a functional dissection of the apical internal loop (IL-A; Fig. 1). This structure is separated from the terminal CUA-loop by eight canonical Watson–Crick (WC) base pairs (six of which are of the “hard” G-C type; Fig. 1). Inaccessibility to chemical probes (Rozhdestvensky et al., 2001) indicates helical structure of this apical stem. The apical internal loop itself has the hallmarks of a GA motif, containing four A and two G residues that, with the exception of G27, are accessible to chemical probes (Rozhdestvensky et al., 2001). GA motifs, which are core components of KT motifs that cause characteristic tight kinks in A-form RNA helices, feature noncanonical (non-WC) purine·purine base pairs (Vidovic et al., 2000; Klein et al., 2001; Rozhdestvensky et al., 2003). Does the GA internal loop motif in the apical 5′ BC1 domain act as a code to specify dendritic targeting?
To address this question, we first converted the GA apical internal loop into an A-form helical stem. Standard WC base pairs were introduced by exchanging three nucleotides in the 3′ strand of the loop (to generate mutant IL-A:WC; Fig. 4 A). This substitution resulted in a substantially reduced range in dendritic transport; IL-A:WC mutant full-length BC1 RNA advanced, on average, to only about half of the dendritic distance that was covered by wild-type BC1 RNA (Fig. 4 A). Thus, BC1 RNA with the GA apical internal loop replaced by a helical stem was still delivered to proximal dendrites, but it now failed to reach distal dendritic segments at 200 μm and beyond. Signal intensities in proximal-most dendritic segments (≤50 μm) did not differ substantially between apical internal loop mutant and wild-type BC1 RNA. Thus, it appears that IL-A:WC mutant BC1 RNA is exported normally from the soma, but is deficient in long-range transport along the dendritic extent.
Figure 4.
Long-range dendritic targeting codes: the GA apical internal loop (IL-A) motif. (A) Introduction of standard WC base pairs (IL-A:WC). (B) Conversion sense–antisense (IL-A:AUC). Both mutations resulted in reduced long-range dendritic targeting; somatic export remained unaffected, but dendritic transport was now restricted to proximal-medial segments. The photomicrographs show dark-field (DF; left) and phase-contrast (PC; right) images of injected neurons. Arrows in phase-contrast images demarcate the distal-most extent of dendritic labeling signal. Extended incubation times did not result in changes of the observed distribution patterns. Statistical analyses performed (comparison of both BC1 derivatives with wild-type BC1 RNA; Fig. 2) were one-way ANOVA for interval point 50 μm (measured from base of dendrite) (P > 0.5); one-way ANOVA for interval points 100, 150, and 200 μm (P < 0.001 in each case); and Scheffe's multiple comparison post hoc analysis for both derivatives at interval points 100, 150, and 200 μm (P < 0.001 in each case). Quantitative data are presented as the mean ± the SEM of relative signal intensities along the dendritic extent. Bar, 50 μm.
In a second approach, we mutated the GA apical internal loop as shown in Fig. 4 B, replacing a UAG triplet in the 5′ loop strand with an antisense AUC triplet (mutant IL-A:AUC). This sense–antisense conversion disrupts noncanonical G·A base pairs in the apical internal loop without replacing them with WC base pairs, thus, resulting in an unstructured open loop. In addition, we reasoned that the UAG triplet could potentially base-pair with the CUA triplet of the terminal loop (either inter- or intramolecularly), and that such triplet–triplet interaction might be required for distal targeting (Ferrandon et al., 1997). As with the IL-A:WC mutation, the UAG→AUC conversion abolished distal dendritic targeting, but did not affect somatic export (Fig. 4 B). This phenotype could not be reverted to wild type (i.e., “rescued”) by the simultaneous replacement of the terminal loop CUA triplet with an antisense GAU triplet, a compensatory exchange that would restore the capacity for triplet–triplet interactions between TL and IL-A (unpublished data). These results confirm that the GA apical internal loop motif is required for distal dendritic targeting; at the same time, they indicate that triplet–triplet interactions between terminal loop and apical internal loop are not relevant for dendritic BC1 targeting competence.
In summary, the combined results suggest that the GA internal loop motif in the apical 5′ BC1 domain carries a code to specify distal dendritic targeting, and that both strands of the motif are required for code expression.
Somatic export: proximal targeting
The medial and basal segments of the 5′ BC1 domain feature two nonhelical motifs: an unpaired U-residue at medial position 22 and a U14-C61 basal internal loop structure. U22, located within the medial helical stem (S-M) that connects the GA apical internal loop with the basal U14-C61 internal loop, is one of the most accessible nucleotides of the entire 5′ BC1 domain (Rozhdestvensky et al., 2001). Therefore, U22 is most likely not part of an A-form helical structure, but forms a bulge (B22) and may thus serve as a recognition motif (Hermann and Patel, 2000). Deletion of U22 resulted in a total loss of dendritic targeting competence (Fig. 5 A). Microinjected B22 mutant (ΔU) BC1 RNA did not enter dendrites to any substantial extent (except for a directly soma-adjacent segment of a few micrometers). These results show that bulged unpaired U22 is indispensable for proximal dendritic targeting (i.e., somatic export). It thus appears that B22 is a critical determinant of the 3D structure of the medial 5′ BC1 domain (see Discussion) and, as such, constitutes part of the somatic export code.
Figure 5.
Somatic export codes. (A) Bulge mutant (B22) BC1 RNA (ΔU). (B) Shortening of the basal stem (S-B). Both mutations resulted in a complete loss of somatic export competence, as microinjected RNAs now remained confined to neuronal perikarya. Prolonged incubation times did not alter the observed distribution patterns. The photomicrographs show dark-field (DF; left) and phase-contrast (PC; right) images of injected neurons. Statistical analyses performed (comparison with wild-type BC1 RNA; Fig. 2) were one-way ANOVA for interval point 50 μm (P < 0.001) and Scheffe's multiple comparison post hoc analysis (P < 0.001 for both mutants). Quantitative data are presented as the mean ± the SEM of relative signal intensities along the dendritic extent. Bar, 50 μm.
The U14-C61 internal loop is the basal-most motif in the 5′ BC1 domain. In this basal internal loop (IL-B), C61 is exposed but U14 is not (Rozhdestvensky et al., 2001), indicating noncanonical interactions between the two nucleotides. We generated a C61A mutant to replace the basal internal loop with a standard WC base pair, thus converting the basal 5′ BC1 domain into an uninterrupted helical stem devoid of any extruded nucleotide bases (mutant IL-B:WC). This exchange had no apparent effect on the dendritic targeting competence of the RNA; dendritic delivery of IL-B:WC mutant BC1 RNA was indistinguishable from wild-type BC1 RNA (Fig. 3 B). We conclude that the U14-C61 basal internal loop motif does not contribute to a dendritic targeting code.
The basal third of the 5′ BC1 domain is a helical stem that is entirely made up of WC base pairs, including standard WC pairs and several embedded G·U-type wobble WC pairs (Fig. 1). This 13-bp basal stem (S-B) forms an A-form double helix that lacks unpaired or bulged nucleotides (Rozhdestvensky et al., 2001). With the exception of the exocyclic guanosine amino group in G·U wobble pairs (see Discussion), discriminating features of A-form WC pairs are buried within the deep groove and are therefore not usually available for protein recognition (Hermann and Patel, 1999, 2000). Therefore, we examined whether the presence of the basal helix, per se, is relevant for dendritic targeting. We made two modifications to the basal stem: (1) S-B and IL-B were eliminated and replaced with two G-C base pairs, and (2) the 13-bp S-B helix was shortened to 6 bp by replacing the nine basal-most base pairs with two G-C pairs. These modifications produced an identical transport-deficient phenotype; both 5′ BC1 derivatives were unable to direct somatic export (Fig. 5 B; the shortened-stem derivative is shown). These results indicate that the basal stem of the 5′ BC1 domain is required for export from the soma.
We conclude that proximal dendritic targeting is specified by elements in the medial-basal region of the 5′ BC1 domain. These elements include bulged unpaired U22, a helix breaker (see Discussion) in the medial stem, and the G·U- rich basal helical stem. We presume that at least some of the G·U wobble WC pairs in the basal stem constitute part of the BC1 somatic export code (see Discussion). This hypothesis will be tested in future experiments.
Structural motifs in the 5′ BC1 domain
Our results implicate several 5′ BC1 motifs in dendritic targeting. RNA motifs such as bulges and internal loops typically introduce distinct 3D elements into otherwise straight helical stems, and we suggest that dendritic targeting codes are expressed in such 3D structural elements. The question is thus raised whether targeting-determinant 5′ BC1 motifs are, in fact, manifest as 3D elements, i.e., whether they can be diagnosed as deviations from the straight/linear trajectory that defines the axis of an A-form helical duplex. A powerful method to probe and resolve such deviations is native PAGE (Bhattacharyya et al., 1990; Lilley, 1995; Luebke and Tinoco, 1996).
This analysis is based on the fact that altered trajectories (i.e., kinks or bends) in helical stems result in reduced electrophoretic mobilities that can be visualized at high resolution (for review see Lilley, 1995). We hypothesized that the GA apical internal loop represents a KT motif. KTs introduce sharp kinks to the helical axis (Vidovic et al., 2000; Klein et al., 2001), and such kinks can be diagnosed by native PAGE because they result in considerably reduced electrophoretic mobilities (Goody et al., 2004). The presence of a KT motif is therefore predicted to retard the 5′ BC1 domain on native PAGE; in contrast, if the noncanonical base pairs in the apical internal loop do not introduce a kink (i.e., if they act similar to standard WC base pairs), no such retardation should be observed.
We compared the mobility of the wild-type 5′ BC1 domain with that of a mutant domain in which the noncanonical base pairs of the GA apical internal loop motif have been replaced with standard WC base pairs (IL-A:WC). We observed that the WC mutant 5′ BC1 domain displays a considerably increased mobility, relative to the wild-type 5′ domain (Fig. 6, compare lanes 1 and 3). The data indicate that a KT in the wild-type domain has been eliminated in the mutant domain, resulting in a now “straightened” helical stem with higher mobility. Thus, the results corroborate our hypothesis that a KT motif is introduced by the GA apical internal loop. Because conversion of the noncanonical base pairs in this motif to standard WC base pairs results in a loss of distal dendritic targeting competence (Fig. 4 A), the combined results suggest that a code specifying distal dendritic targeting is expressed by a KT internal loop motif residing in the apical 5′ BC1 domain. We will henceforth refer to this motif as KT BC1.
Figure 6.
Differential electrophoretic mobilities of 5′ BC1 derivatives on native PAGE. Conversion of the apical KT motif (IL-A) to a WC helical stem and elimination of the single-U bulge (B22) both resulted in substantially increased relative mobilities. Highest relative mobility was observed with the IL-A:WC/ΔU double mutant. See Fig. 1 for motif abbreviations.
We next examined the mobility of the B22 mutant (ΔU) that is deficient in somatic export competence (Fig. 5). Deletion of this single unpaired U-residue resulted in a considerable increase in electrophoretic mobility, a relative increase even higher than that produced by the WC mutant apical internal loop motif (Fig. 6, lanes 1 and 4). This result suggests that unpaired U22 acts as a helix breaker by causing a sharp turn in the A-form duplex. An IL-A/B22 double mutant (i.e., simultaneous elimination of U22 and conversion of the KT apical internal loop motif to standard WC base pairing) showed the highest relative mobility of all examined 5′ BC1 derivatives (Fig. 6, lanes 1 and 5), a result which is consistent with a straight, rodlike helical structure. This double mutant does not exhibit any dendritic targeting competence (unpublished data).
In contrast, conversion of the basal U14-C61 internal loop to a U14-A61 standard WC base pair resulted in only a moderate increase in electrophoretic mobility (Fig. 6, lanes 1 and 2). This result suggests that only a slight bend in the helical stem (as opposed to a sharp turn) is caused by the basal internal loop (IL-B). This loop is targeting-irrelevant (Fig. 3 B). Similarly, an antisense mutation in the CUA terminal loop (TL) does not substantially alter electrophoretic mobility (not depicted). This mutation also failed to affect proximal or distal dendritic targeting competence of BC1 RNA (Fig. 3 A). It thus appears that those motifs in the 5′ BC1 domain that make little, if any, contributions to its 3D conformation also contribute little to dendritic targeting competence.
It is noteworthy that those 5′ BC1 derivatives that contain bulged U22 produce, in addition to the major band, a distinct minor band of lower mobility (Fig. 6, lanes 1–3). In contrast, derivatives lacking U22 produce only one band, indicating uniform mobility (Fig. 6, lanes 4 and 5). We interpret these results to indicate that B22 causes the RNA backbone to undergo a turn that can occur in one of two distinct conformations, with B22 possibly acting as a hinge. This finding raises the question, to be addressed in future work, whether one, or both, of these conformers is targeting competent, and whether shifts in the dynamic equilibrium between the two conformers will modulate somatic export competence.
Binding of heterogeneous nuclear ribonucleoprotein (hnRNP) A2 to KT BC1
Our data suggest that the KT BC1, which is located in the apical 5′ BC1 domain, serves as a long-range targeting code. A search algorithm (see Materials and methods) identified similar KT motifs in the DTEs of several targeted neural RNAs (see Discussion). Therefore, we hypothesized that a trans-acting factor (i.e., a decoding protein) that recognizes such KT motifs would interact with those RNAs that use them as spatial codes. One of the KT motifs identified by our search algorithm is located in the 3′ UTR of myelin basic protein (MBP) mRNA, an mRNA that is distally targeted in oligodendrocytes and neurons (Ainger et al., 1993; Shan et al., 2003). This KT MBP motif (Fig. 9) overlaps with a previously described 21-nt element in the MBP 3′ UTR that was initially recognized as an RNA transport signal (Ainger et al., 1997). It was later identified as a targeting-determinant response element for hnRNP A2 (A2RE; Hoek et al., 1998; Munro et al., 1999; Shan et al., 2000, 2003). We therefore reasoned that hnRNP A2 may also interact with KT BC1.
Figure 9.
The BC1 KT motif (KT BC1). Two general classes of KT motifs have been described, the classic KT and the minimal KT (Vidovic et al., 2000; Klein et al., 2001; Hardin and Batey, 2004). KT BC1 belongs to a subtype of the classic KT that is exemplified by KT-58 in 23S rRNA. MBP, CaMKIIα, and PKMζ mRNA contain in their respective 3′ UTRs classic KT motifs that match consensus (Tiedge, 2006). R, purine; N, any nucleotide; blue, canonical G-C pairs of the C-stem; red, noncanonical G·A pairs of the NC-stem; orange, nominally unpaired internal loop nucleotides, may engage in stacking and trans–Sugar-Edge/Sugar-Edge interactions; yellow, extruded nucleotide. Motifs are aligned with their C-stems (blue, G-C pairs) to the left, NC-stems (red, G·A pairs) to the right of the asymmetric internal loop. In the consensus KT, the transitional position between the NC-stem and the resumption of A-form helical structure by G-C base pairs (referred to as N·N) may be occupied by an intervening base pair, by an unpaired residue, or not at all. KTs in small untranslated RNAs often carry a G·U wobble pair in this position (Rozhdestvensky et al., 2003; Hamma and Ferré-D'Amaré, 2004), although a sheared (trans–Hoogsteen/Sugar Edge) G·U pair is also allowed (Lescoute et al., 2005). Flanking G-C base pairs may occur in either the G-C or the C-G orientation. In the KT-58 subtype (including KT BC1), internal loop G and C nucleotides may also be part of an extended C-stem (Lescoute et al., 2005) and can therefore be rendered in either color coding.
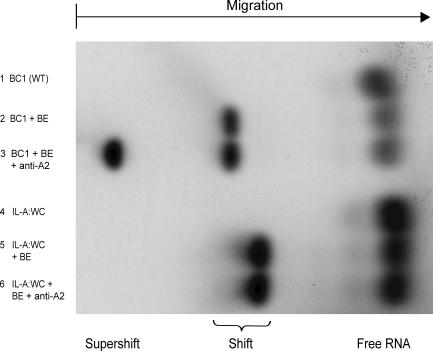
We decided to test this hypothesis by performing “supershift” electrophoretic mobility shift assays (EMSA). We used an antibody against hnRNP A2, the specificity of which has previously been documented (Ma et al., 2002), in supershift EMSA experiments with rat brain extracts, following established protocols (Muddashetty et al., 2002; Wang et al., 2002).
The results are shown in Fig. 7. Full-length BC1 RNA was shifted in brain extracts to lower relative mobility, as would be expected as a result of any BC1–protein interactions (Fig. 7, lane 2). The anti-A2 antibody produced a supershift, i.e., a further shift to even lower mobility as a result of antibody–A2–BC1 interactions (Fig. 7, lane 3). As a control, we used the IL-A:WC BC1 mutant, in which the apical KT motif has been replaced with an A-form helix by conversion to standard WC pairing through the exchange of three nucleotides in the 3′ strand of the motif. This WC mutant lacks the apical KT motif (i.e., is converted to a straightened helical stem; Fig. 6) and is long-range targeting incompetent (Fig. 4 A). No supershift in brain extract was observed with this apical WC mutant, indicating that it does not interact with hnRNP A2 (Fig. 7, lane 6). In addition, we examined the converse mutant (IL-A:AUC) in which the 5′ strand of the apical KT motif has been replaced with antisense, resulting in an open unstructured loop that is also long-range targeting incompetent (Fig. 4 B). The A2-supershift was likewise abolished (not depicted). The combined results thus indicate that the KT BC1 motif, with intact 5′ and 3′ strands, is indispensable for hnRNP A2 binding and distal dendritic targeting.
Figure 7.
Recognition of the KT BC1 motif by hnRNP A2. Wild-type BC1 RNA (lane 3) was supershifted by anti-A2 in brain extracts (BE). In contrast, the apical WC mutant (IL-A:WC) was not supershifted (lane 6; for structure, see Fig. 4 A). In addition, the regular shift (i.e., in the absence of anti-A2) produced in brain extract by the apical WC mutant was observed at higher relative mobility than the equivalent shift produced by wild-type BC1 RNA, suggesting that the apical WC mutant is interacting with fewer components in brain extract than is wild-type BC1 RNA. See Fig. 1 for motif abbreviations.
As a further control, we used B22 mutant (ΔU) BC1 RNA. U22, which forms a bulge in the vicinity of KT BC1, is required for somatic export competence (Fig. 5 A). Elimination of U22, although abolishing somatic export, had no apparent effect on the binding interaction with hnRNP A2 as the gel mobility pattern was not distinguishable from wild-type BC1 RNA (not depicted).
In summary, the data indicate that the interaction of hnRNP A2 with dendritic BC1 RNA is mediated by the KT motif structure in the 5′ BC1 domain. We conclude that BC1 RNA, which except for its long-range transport competence has little in common with MBP mRNA (i.e., in terms of nucleotide sequence, cellular function, and expressing cell types), is, by virtue of its KT motif, recognized by the same protein.
Discussion
3D architectural motifs serve as determinants of RNA function (Noller, 2005). We surmised that in dendritic RNAs, such motifs, when contained within cis-acting DTEs, may be carriers of spatial codes. To test this hypothesis, we functionally dissected the targeting-determinant 5′ domain of dendritic BC1 RNA. Our data (see Fig. 8 for synopsis) show that at least two spatial codes are expressed in the 5′ BC1 domain; a code to specify somatic export, contained within a cis-acting element that we call BC1 DTE1, and a code to specify long-range distal dendritic targeting, contained within BC1 DTE2. We identify a stem-bulge motif in the medial-basal region of the 5′ BC1 domain as a proximal DTE1 determinant, whereas a KT motif in the apical region serves as a distal DTE2 determinant.
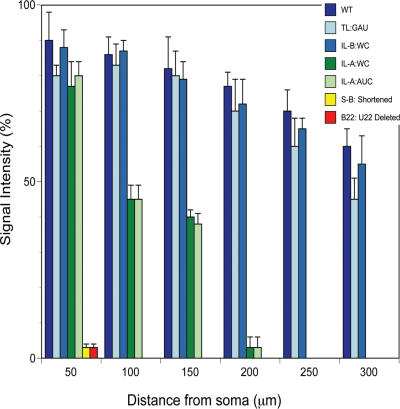
Figure 8.
Contributions of 5′ BC1 motifs to dendritic targeting competence. The histogram summarizes the quantitative analyses shown in Figs. 2–5 . (blue) Wild type and mutants that are indistinguishable from wild type. (green) Mutants that lack the long-range distal dendritic targeting code, but retain the somatic export code. (yellow/red) Mutants that lack the somatic export code. See Fig. 1 for motif abbreviations. Quantitative data are presented as the mean ± the SEM of relative signal intensities along the dendritic extent.
BC1 DTE1
Deletion of a single nucleotide, unpaired U22, abolishes somatic export. U22 forms a bulge and introduces a sharp turn into the medial helical stem (S-M) that connects the apical internal loop (IL-A) with the basal internal loop (IL-B). Structural analyses of ribosomal subunits have revealed that solvent-exposed unpaired U-residues are much more frequently observed than other unpaired nucleotides; in the context of a helical stem, they act as helix breakers (Nissen et al., 2001). Our data indicate that with a sharp turn at bulge 22, the medial-basal 5′ BC1 domain assumes the conformation of a stem-bulge motif, one that is a requisite determinant of the BC1 somatic export code.
In addition, we found that the presence of the basal helical stem is indispensable for somatic export. We consider the basal stem de facto contiguous with the medial stem, as the U14-C61 pair does not cause a substantial deviation from the helical axis. Because nucleotide bases in A-form RNA helices are not accessible through the deep groove, their identities are not usually recognizable features in helical stems (Hermann and Patel, 1999, 2000). However, the basal-medial stem contains an unusually high percentage of G·U wobble WC pairs. G·U pairs are known to provide RNA functionality in various biological contexts (Masquida and Westhof, 2000; Varani and McClain, 2000). Embedded in A-form helices, G·U wobble pairs provide features that can serve as protein recognition sites. Such features include the exocyclic guanosine amino group, which (unlike in standard WC pairs) is not hydrogen-bonded and is therefore accessible through the shallow groove. Owing to their conformational softness, G·U pairs also introduce flexibility to a helical stem, allowing for protein recognition by induced fit (Varani and McClain, 2000). It is therefore our hypothesis, which is to be tested in future experiments, that medial-basal G·U wobble pairs contribute to the BC1 somatic export code.
BC1 DTE2
Distal-targeting BC1 DTE2 acts in conjunction with proximal- targeting BC1 DTE1. These dendritic targeting codes therefore operate in modular fashion; i.e., the coordinated action of two types of codes is required to specify the final destination.
BC1 DTE2 is defined by an apical GA KT motif in the 5′ BC1 domain. KT BC1, which introduces a sharp kink in the RNA duplex, is long-range targeting-determinant; i.e., it encodes distal dendritic targeting. To our knowledge, this is the first time that a distinct architectural RNA motif has been identified as an intracellular spatial code. We hypothesize that KT motifs of this and similar types may serve as long-range targeting codes in other RNAs as well (for review see Tiedge, 2006).
KTs comprise a family of RNA motifs that have been described in diverse types of RNAs, including rRNAs, mRNAs, and especially small untranslated RNAs (Vidovic et al., 2000; Klein et al., 2001; Rozhdestvensky et al., 2003; Hamma and Ferré-D'Amaré, 2004). KT motifs are complex architectural RNA elements that feature double- and single-stranded components and act as recognition sites for various intermolecular interactions (Vidovic et al., 2000; Klein et al., 2001). They are characterized by the following hallmarks (Fig. 9).
Central to the KT motif is a core of tandem noncanonical base pairs, which are typically of the sheared G·A type (Vidovic et al., 2000) and form what is now referred to as the noncanonical (NC) stem (Klein et al., 2001; Lescoute et al., 2005). Sheared G·A pairs engage in A-to-G trans–Hoogsteen–Sugar-Edge interactions (Leontis and Westhof, 2001). The NC-stem, which also engages in A-minor interactions and cross-strand A-stacks (Vidovic et al., 2000; Klein et al., 2001), is flanked on one side by an A-form helix and on the other side by an extruded nucleotide (Fig. 9). In the classic KT motif, the extruded nucleotide is part of an internal loop, often asymmetric, that links the NC-stem with a canonical stem (the C-stem) of two or more standard WC (typically G-C) base pairs. In the minimal KT, the C-stem is lacking, but the unpaired extruded nucleotide is maintained (Fig. 9). The intramolecular interactions between nucleotides within the KT motif result in a sharp kink in the phosphodiester backbone (Vidovic et al., 2000; Klein et al., 2001). Identification of a KT consensus in a RNA is diagnostic of the presence of the KT motif structure (Klein et al., 2001).
The BC1 KT motif (KT BC1) belongs to a subtype of the classic KT (Fig. 9) that is exemplified by KT-58 in 23S rRNA (Klein et al., 2001; Lescoute et al., 2005). Because the geometry of NC-stem GA core is inverted, KT BC1 may assume the 3D structure of a reverse KT motif (Strobel et al., 2004). In this conformation, residue A28 (Fig. 9, yellow) assumes the strategic position of the extruded nucleotide. On the basis of previously established reactivities of nucleotide bases in KT motifs (Klein et al., 2001; Strobel et al., 2004; White et al., 2004), it is predicted that bases A28, A48, A49, and G50 in KT BC1 should be relatively accessible, whereas bases U25, A26, G27, and G51 should be more protected. This is in fact the pattern that was experimentally observed, with A28 being one of the most accessible bases in the entire 5′ BC1 domain (Rozhdestvensky et al., 2001).
It is noteworthy that in those positions where interspecies nucleotide substitutions have been observed in different rodent 5′ BC1 domains, these exchanges have been conservative. Such interspecies substitutions include G→A transitions (and vice versa) in the GA apical internal loop motif (Rozhdestvensky et al., 2001). Importantly, stem, loop, and bulge structures, as well as the apical KT motif, remain conserved in all cases. Similarly, dendritic BC200 RNA (Tiedge et al., 1993), which is the primate counterpart (analogue) of rodent BC1 RNA, contains a KT motif of the KT-58 subtype in its 5′ domain. Although DTEs have not yet been established in BC200 RNA, microinjected BC200 RNA is delivered to distal dendritic segments of sympathetic neurons in culture (unpublished data).
KT motifs as spatial codes
Our results suggest that KT BC1 is a structural RNA motif that serves as a distal dendritic targeting code. We asked whether similar motifs are contained in other distally targeted RNAs, and we used a KT search algorithm (see Materials and methods) to address this question.
This search algorithm identified a classic KT motif in the 3′ UTR of MBP mRNA. The putative KT MBP motif matches 100% with the classic KT consensus (Fig. 9). MBP mRNA has previously been shown to contain a targeting element (initially called RTS, for RNA transport signal, but later renamed A2RE, for hnRNP A2 response element) that is responsible for its transport along oligodendrocytes processes (Ainger et al., 1997; Munro et al., 1999; Shan et al., 2003). The 3′ strand of KT MBP turns out to represent A2RE11, a previously identified 11-nt targeting-determinant subelement of A2RE (Munro et al., 1999; Shan et al., 2003). KT MBP may thus represent a long-range targeting determinant in the 3′ UTR of MBP mRNA.
A2RE is recognized by the trans-acting factor hnRNP A2 (Munro et al., 1999; Shan et al., 2003; Smith, 2004). Point mutations in A2RE that abolish or substantially diminish hnRNP binding and RNA transport competence (Munro et al., 1999; Shan et al., 2003) all target nucleotides that are strategically important within the KT motif organization (for review see Tiedge, 2006). Therefore, we hypothesized that hnRNP A2 is a targeting trans-acting factor that engages a subtype of KT spatial codes. This hypothesis was tested with BC1 RNA, which—other than containing a KT motif and being distally targeted—shares little similarity with MBP mRNA. Our results show that hnRNP A2 binds to KT BC1 in the apical 5′ domain, and that both strands of the KT BC1 motif are required for this interaction. These results suggest that hnRNP A2 operates as a KT BC1 spatial decoder. It remains to be established whether other KT subtypes are recognized by hnRNP A2, and conversely, which A2 isoforms (Hatfield et al., 2002) interact with KT BC1 and other putative KT motifs. Also, although BC1 RNA and hnRNP A2 colocalize to the dendritic domain, it is unknown which A2 isoform colocalizes with BC1 RNA. In any case, however, our data raise the possibility that at least some KT-encoded RNA-targeting mechanisms use the A2 pathway.
hnRNP A2 belongs to the family of RNA-binding proteins that contain RNA recognition motifs (RRMs). Members of this large and highly diverse family use various different RNA recognition strategies (Maris et al., 2005). Recognition is often mediated by structural elements in which single-stranded (i.e., unpaired and extruded nucleotides) and non-WC components are embedded in double-stranded A-form helical context (Maris et al., 2005). For example, extruded uridine in KT-15 of 23S rRNA is engaged by a tight binding pocket that is formed by ribosomal proteins L7Ae and L15e (Klein et al., 2001; Moore et al., 2004), the latter an RRM-type protein with βαββ topology (Klein et al., 2004). Double-stranded context is often important for recognition; if removed, RRM–RNA binding affinities drop substantially (Maris et al., 2005).
KT motifs exist in a dynamic equilibrium between an open structure and the highly ordered and tightly kinked conformation (Dennis and Omer, 2005). In these and other architectural RNA motifs, transition from one conformational state to another can often be induced by protein binding through “induced fit” mechanisms (Williamson, 2000; Goody et al., 2004; Maris et al., 2005). At this time, we do not know which conformation KT BC1 will assume as a result of protein engagement. Similarly, A2RE11 alone binds to hnRNP A2 and is targeting competent (Munro et al., 1999; Shan et al., 2000), but the full KT motif has not yet been investigated. We hope that structural information will eventually provide answers to these questions.
Few targeting elements have been identified in dendritic RNAs (Kindler et al., 2005), and code-carrying architectural RNA motifs have not yet been reported in such DTEs. The mRNAs encoding the Ca2+/calmodulin-dependent kinase II α subunit (CaMKIIα; Burgin et al., 1990; Paradies and Steward, 1997; Blichenberg et al., 2001) and protein kinase M ζ (PKMζ) (Muslimov et al., 2004) have both been shown to be transported to distal dendritic domains. In CaMKIIα mRNA, a putative KT motif is located at nt 1,814–1,881 of the 3′ UTR, i.e., within a larger RNA segment that has previously been shown to be targeting-determinant (Blichenberg et al., 2001). PKMζ mRNA contains a putative KT motif within a long-range targeting-determinant 44-nt segment of its 3′ UTR (Muslimov et al., 2004). Putative KT CaMKIIα and KT PKMζ are both of the classic consensus type and are virtually identical to KT MBP (Fig. 9; Tiedge, 2006). In summary, it thus appears possible that KT motifs of the type shown in Fig. 9 serve as long-range targeting codes in various transported RNAs (Tiedge, 2006). We solicit further experimental work to corroborate or falsify this hypothesis.
Materials and methods
Plasmids and RNA preparation
Transcripts used in this work were generated from the following plasmids: (1) pBCX607 to generate wild-type BC1 RNA, as previously described (Cheng et al., 1996; Muslimov et al., 1997); (2) pBC1IL-A:WC to generate BC1 RNA with the GA apical internal loop motif replaced with standard WC base pairs; (3) pBC1ΔU22 to generate BC1 RNA lacking U22; (4) pBC1IL-A:WC/ΔU22 to generate BC1 RNA with the GA apical internal loop motif replaced with standard WC base pairs and U22 deleted; (5) pBC1IL-B:WC to generate BC1 RNA with the basal internal loop replaced with a standard WC base pair by a C61A transversion; (6) pKK541 to generate BC1 RNA with the terminal loop CUA triplet converted to antisense (TL:GAU); (7) pKK540 to generate BC1 RNA with the apical internal loop UAG triplet converted to antisense (IL-A:AUC); (8) pKK538 to generate BC1 RNA with both the terminal loop (TL) CUA triplet and the apical internal loop (IL-A) UAG triplet converted to antisense; (9) pKK542 to generate a 5′ BC1 derivative with the basal stem (S-B) shortened to 6 base pairs by replacing the 9 basal-most base pairs with two G-C pairs; (10) pKK543 to generate a 5′ BC1 derivative with the basal stem (S-B) and the basal internal loop (IL-B) eliminated and replaced with a two G-C base pairs; (11) pCMV-5′BC1-bcd to generate 5′ BC1 chimeric bcd RNA; and (12) pCMV-bcd to generate bcd mRNA (Muslimov et al., 1997). BC1 variants transcribed from pKK542 and pKK543 also contain nucleotides from vector pSL300 (Brosius, 1989); pSL300 nt 2191–2286 precede the respective GC-clamped BC1 segments, which are in turn followed by two nucleotides (AU) from the pSL300 EcoRV site.
All plasmids were verified by sequencing. Plasmids 9 and 10 were linearized with EcoRV, plasmids 11 and 12 with Asp718, and all other plasmids with DraI. 35S-labeled transcripts were generated using T3 or T7 RNA polymerase, as previously described (Muslimov et al., 2004). Excess unlabeled UTP was added to the reaction mixture to ensure that labeled transcripts were full-length, and the size and integrity of all transcripts were monitored as previously described (Muslimov et al., 1997).
Cell culture and microinjection
Primary cultures of sympathetic neurons at low density were generated from superior cervical ganglia of embryonic day 20–21 Sprague-Dawley rat embryos, as previously described (Higgins et al., 1991; Muslimov et al., 1997). Work with animals was approved by the SUNY Brooklyn Institutional Animal Care and Use Committee. Cells were plated on polylysine-treated glass coverslips, and basement membrane extract (Matrigel; BD Biosciences) was added at 100 μg/ml to induce dendritic growth.
RNAs were injected as previously described (Muslimov et al., 1997, 2004). In brief, RNAs were 35S radiolabeled at 3 × 106 cpm/μl and microinjected into the perikaryal regions of sympathetic neurons in culture at volumes of several femtoliters per pulse. 0.4% Lucifer yellow was coinjected for calibration purposes. Nuclear injections were occasionally performed to check if nuclear experience of a transcript was targeting-relevant. No such case was observed. Injection amounts were varied over a concentration range of one order of magnitude (corresponding to an estimated few thousand injected RNA molecules in the low-injection routine) to ensure that observed dendritic targeting patterns were concentration-independent (Muslimov et al., 1997). The stability of injected RNAs was monitored by PAGE (Muslimov et al., 1997). In addition, the means of integrated total signal intensities per injected cell were assessed for each transcript, using MetaMorph imaging software (Universal Imaging Corporation; see next section). This intensity (i.e., the mean of the sum of somatic and dendritic intensities) can be used as an indicator of intracellular RNA stability after injection and incubation. No substantial differences were observed between the various injected transcripts whenever equivalent level injection routines were used. These results indicate similar intracellular stabilities of those transcripts for the duration of the experiments.
Except for time course experiments, cells were incubated for 4 h at 35°C before fixation and emulsion autoradiography (Muslimov et al., 1997). Time course experiments were performed to ascertain whether any observed differences in the extent of dendritic delivery were the result of differential transport rates. We found this not to be the case. For example, for those BC1 derivatives that were observed not to exit the soma or not to proceed to distal dendrites, incubation times were varied. However, prolonged incubation did not result in increased extents of dendritic delivery.
Data acquisition and analysis
Neurons were photographed at room temperature on a microscope (Microphot-FXA; Nikon) using dark-field and phase-contrast optics. The following objectives were used (all Nikon): Plan Fluor 10×/0.30 NA, 160/0.17; PhC Plan 20×/0.50 NA, DL 160/0.17; Plan 20×/0.50 NA, DIC 160/0.17; and Ph3 DL Plan 40×/0.65 NA, 160/0.17. Digital images were acquired with a three–charge-coupled device camera (DKC-5000; Sony) or a cooled charge-coupled device camera (CoolSNAP HQ; Roper Scientific). RNA distribution profiles along dendrites were established as previously described (Muslimov et al., 2004). To quantify transport competence of microinjected RNAs, silver grain densities were measured along dendrites at 50-μm interval points, up to a maximal distance of 300 μm. Signal intensities at 0-μm interval points (base of dendrites) were assigned a relative value of 100%; average absolute signal intensities did not substantially differ at this interval point between wild-type and mutant RNAs. Statistical analysis was performed with at least 26 dendrites (from at least 14 neurons) for each RNA injected (i.e., wild-type BC1 RNA and each of the BC1 derivatives).
MetaMorph software was used for quantitative analysis of signal intensities along dendrites (Iacoangeli et al., 2004). Grayscale images were thresholded to distinguish silver grains, and the area above threshold (number of pixels) was measured for each interval point. A thresholded area of identical dimensions was also measured in a nearby region of the same coverslip that was not populated by neurons. This value was considered background. A dendritic labeling signal was recorded if it exceeded background levels by a factor of three or more (Muslimov et al., 1997). Photoshop and Illustrator software (both Adobe) was used to generate final composite illustrations.
Statistical analyses were performed using SPSS software (SPSS, Inc.). A modified version of DNAMAN software (Lynnon Corporation) was used to interrogate RNAs for KT motifs.
Native PAGE and EMSA
Native PAGE was performed in 8% polyacrylamide gels (with a 19:1 ratio of acrylamide/bisacrylamide) in 90 mM Tris-borate, pH 8.3, in the presence of 15 mM MgCl2 at room temperature for 12 h at 15 V (Bhattacharyya et al., 1990; Goody et al., 2004). Supershift experiments were performed as previously described (Muddashetty et al., 2002; Wang et al., 2002). Anti-A2 antibodies (Ma et al., 2002) were provided by R. Smith (University of Queensland, Brisbane, Australia).
Acknowledgments
We thank Dr. Yue Huang (Lynnon Corporation, Canada) for help in developing and using algorithms for the identification of KT motif secondary structure conformations, Drs. Hans von Gizycki and Jeremy Weedon (State University of New York Brooklyn Scientific Computing Center) for statistical consultation, and Dr. Dennis Higgins for advice and discussion.
This paper is dedicated to the memory of Dennis Higgins, colleague, mentor, friend.
This work was supported, in part, by the New York City Council Speaker's Fund for Biomedical Research (I.A. Muslimov), Deutsche Forschungsgemeinschaft grant BR 754/2 (J. Brosius), Nationales Genomforschungsnetz grant NGFN 0313358A (J. Brosius), and National Institutes of Health grants HD043428 and NS046769 (H. Tiedge).
Abbreviations used in this paper: ANOVA, analysis of variance; CaMKIIα, Ca2+/calmodulin-dependent kinase II α subunit; DTE, dendritic targeting element; EMSA, electrophoretic mobility shift assay; hnRNP, heterogeneous nuclear ribonucleoprotein; KT, kink-turn; MBP, myelin basic protein; PKMζ, protein kinase M ζ; RRM, RNA recognition motif; rRNA, ribosomal RNA; WC, Watson–Crick.
References
- Ainger, K., D. Avossa, F. Morgan, S.J. Hill, C. Barry, E. Barbarese, and J.H. Carson. 1993. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J. Cell Biol. 123:431–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainger, K., D. Avossa, A.S. Diana, C. Barry, E. Barbarese, and J.H. Carson. 1997. Transport and localization elements in myelin basic protein mRNA. J. Cell Biol. 138:1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya, A., A.I. Murchie, and D.M. Lilley. 1990. RNA bulges and the helical periodicity of double-stranded RNA. Nature. 343:484–487. [DOI] [PubMed] [Google Scholar]
- Blichenberg, A., M. Rehbein, R. Müller, C.C. Garner, D. Richter, and S. Kindler. 2001. Identification of a cis-acting dendritic targeting element in the mRNA encoding the alpha subunit of Ca2+/calmodulin-dependent protein kinase II. Eur. J. Neurosci. 13:1881–1888. [DOI] [PubMed] [Google Scholar]
- Brosius, J. 1989. Superpolylinkers in cloning and expression vectors. DNA. 8:759–777. [DOI] [PubMed] [Google Scholar]
- Brosius, J., and H. Tiedge. 2004. RNomenclature. RNA Biol. 1:81–83. [DOI] [PubMed] [Google Scholar]
- Burgin, K.E., M.N. Waxham, S. Rickling, S.A. Westgate, W.C. Mobley, and P.T. Kelly. 1990. In situ hybridization histochemistry of Ca2+/calmodulin-dependent protein kinase in developing rat brain. J. Neurosci. 10:1788–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao, X., G. Yeo, A.R. Muotri, T. Kuwabara, and F.H. Gage. 2006. Noncoding RNAs in the mammalian central nervous system. Annu. Rev. Neurosci. 29:77–103. [DOI] [PubMed] [Google Scholar]
- Cheng, J.-G., H. Tiedge, and J. Brosius. 1996. Identification and characterization of BC1 RNP particles. DNA Cell Biol. 15:549–559. [DOI] [PubMed] [Google Scholar]
- Dennis, P.P., and A. Omer. 2005. Small non-coding RNAs in Archaea. Curr. Opin. Microbiol. 8:685–694. [DOI] [PubMed] [Google Scholar]
- Eberwine, J., K. Miyashiro, J.E. Kacharmina, and C. Job. 2001. Local translation of classes of mRNAs that are targeted to neuronal dendrites. Proc. Natl. Acad. Sci. USA. 98:7080–7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon, D., I. Koch, E. Westhof, and C. Nüsslein-Volhard. 1997. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 16:1751– 1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuditta, A., B.B. Kaplan, J. van Minnen, J. Alvarez, and E. Koenig. 2002. Axonal and presynaptic protein synthesis: new insights into the biology of the neuron. Trends Neurosci. 25:400–404. [DOI] [PubMed] [Google Scholar]
- Goody, T.A., S.E. Melcher, D.G. Norman, and D.M. Lilley. 2004. The kink-turn motif in RNA is dimorphic, and metal ion-dependent. RNA. 10:254–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamma, T., and A.R. Ferré-D'Amaré. 2004. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 A resolution. Structure. 12:893–903. [DOI] [PubMed] [Google Scholar]
- Hardin, J.W., and R.T. Batey. 2004. Curse of the hairpin loop. Structure. 12:731–732. [DOI] [PubMed] [Google Scholar]
- Hatfield, J.T., J.A. Rothnagel, and R. Smith. 2002. Characterization of the mouse hnRNP A2/B1/B0 gene and identification of processed pseudogenes. Gene. 295:33–42. [DOI] [PubMed] [Google Scholar]
- Hermann, T., and D.J. Patel. 1999. Stitching together RNA tertiary architectures. J. Mol. Biol. 294:829–849. [DOI] [PubMed] [Google Scholar]
- Hermann, T., and D.J. Patel. 2000. RNA bulges as architectural and recognition motifs. Structure. 8:R47–R54. [DOI] [PubMed] [Google Scholar]
- Higgins, D., P.J. Lein, D.J. Osterhout, and M.I. Johnson. 1991. Tissue culture of mammalian autonomic neurons. In Culturing Nerve Cells. G. Banker and K. Goslin, editors. MIT Press, Cambridge. 177–205.
- Hoek, K.S., G.J. Kidd, J.H. Carson, and R. Smith. 1998. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 37:7021–7029. [DOI] [PubMed] [Google Scholar]
- Iacoangeli, A., Y. Lin, E.J. Morley, I.A. Muslimov, R. Bianchi, J. Reilly, J. Weedon, R. Diallo, W. Böcker, and H. Tiedge. 2004. BC200 RNA in invasive and preinvasive breast cancer. Carcinogenesis. 25:2125–2133. [DOI] [PubMed] [Google Scholar]
- Job, C., and J. Eberwine. 2001. Localization and translation of mRNA in dendrites and axons. Nat. Rev. Neurosci. 2:889–898. [DOI] [PubMed] [Google Scholar]
- Kindler, S., H. Wang, D. Richter, and H. Tiedge. 2005. RNA transport and local control of translation. Annu. Rev. Cell Dev. Biol. 21:223–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, D.J., P.B. Moore, and T.A. Steitz. 2004. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 340:141–177. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., T.M. Schmeing, P.B. Moore, and T.A. Steitz. 2001. The kink-turn: a new RNA secondary structure motif. EMBO J. 20:4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov, A.V., M. Kiefmann, K. Ebnet, T. Khanam, R.S. Muddashetty, and J. Brosius. 2005. Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP). J. Mol. Biol. 353:88–103. [DOI] [PubMed] [Google Scholar]
- Leontis, N.B., and E. Westhof. 2001. Geometric nomenclature and classification of RNA base pairs. RNA. 7:499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis, N.B., and E. Westhof. 2003. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 13:300–308. [DOI] [PubMed] [Google Scholar]
- Lescoute, A., N.B. Leontis, C. Massire, and E. Westhof. 2005. Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res. 33:2395–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley, D.M. 1995. Kinking of DNA and RNA by base bulges. Proc. Natl. Acad. Sci. USA. 92:7140–7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luebke, K.J., and I. Tinoco Jr. 1996. Sequence effects on RNA bulge-induced helix bending and a conserved five-nucleotide bulge from the group I introns. Biochemistry. 35:11677–11684. [DOI] [PubMed] [Google Scholar]
- Ma, A.S., K. Moran-Jones, J. Shan, T.P. Munro, M.J. Snee, K.S. Hoek, and R. Smith. 2002. Heterogeneous nuclear ribonucleoprotein A3, a novel RNA trafficking response element-binding protein. J. Biol. Chem. 277:18010–18020. [DOI] [PubMed] [Google Scholar]
- Maris, C., C. Dominguez, and F.H. Allain. 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 272:2118–2131. [DOI] [PubMed] [Google Scholar]
- Masquida, B., and E. Westhof. 2000. On the wobble GoU and related pairs. RNA. 6:9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, T., Y. Zhang, M.O. Fenley, and H. Li. 2004. Molecular basis of box C/D RNA-protein interactions; cocrystal structure of archaeal L7Ae and a box C/D RNA. Structure. 12:807–818. [DOI] [PubMed] [Google Scholar]
- Muddashetty, R., T. Khanam, A. Kondrashov, M. Bundman, A. Iacoangeli, J. Kremerskothen, K. Duning, A. Barnekow, A. Hüttenhofer, H. Tiedge, and J. Brosius. 2002. Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 321:433–445. [DOI] [PubMed] [Google Scholar]
- Munro, T.P., R.J. Magee, G.J. Kidd, J.H. Carson, E. Barbarese, L.M. Smith, and R. Smith. 1999. Mutational analysis of a heterogeneous nuclear ribonucleoprotein A2 response element for RNA trafficking. J. Biol. Chem. 274:34389–34395. [DOI] [PubMed] [Google Scholar]
- Muslimov, I.A., E. Santi, P. Homel, S. Perini, D. Higgins, and H. Tiedge. 1997. RNA transport in dendrites: a cis-acting targeting element is contained within neuronal BC1 RNA. J. Neurosci. 17:4722–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov, I.A., G. Banker, J. Brosius, and H. Tiedge. 1998. Activity-dependent regulation of dendritic BC1 RNA in hippocampal neurons in culture. J. Cell Biol. 141:1601–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslimov, I.A., V. Nimmrich, A.I. Hernandez, A. Tcherepanov, T.C. Sacktor, and H. Tiedge. 2004. Dendritic transport and localization of protein kinase Mζ mRNA: implications for molecular memory consolidation. J. Biol. Chem. 279:52613–52622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen, P., J.A. Ippolito, N. Ban, P.B. Moore, and T.A. Steitz. 2001. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. USA. 98:4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller, H.F. 2005. RNA structure: reading the ribosome. Science. 309:1508–1514. [DOI] [PubMed] [Google Scholar]
- Paradies, M.A., and O. Steward. 1997. Multiple subcellular mRNA distribution patterns in neurons: a nonisotopic in situ hybridization analysis. J. Neurobiol. 33:473–493. [DOI] [PubMed] [Google Scholar]
- Richter, D. 2001. Cell Polarity and Subcellular RNA Localization. Springer, Berlin. 221 pp.
- Richter, J.D., and L.J. Lorenz. 2002. Selective translation of mRNAs at synapses. Curr. Opin. Neurobiol. 12:300–304. [DOI] [PubMed] [Google Scholar]
- Rozhdestvensky, T.S., A. Kopylov, J. Brosius, and A. Hüttenhofer. 2001. Neuronal BC1 RNA structure: evolutionary conversion of a tRNAAla domain into an extended stem-loop structure. RNA. 7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhdestvensky, T.S., T.H. Tang, I.V. Tchirkova, J. Brosius, J.P. Bachellerie, and A. Hüttenhofer. 2003. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 31:869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan, J., K. Moran-Jones, T.P. Munro, G.J. Kidd, D.J. Winzor, K.S. Hoek, and R. Smith. 2000. Binding of an RNA trafficking response element to heterogeneous nuclear ribonucleoproteins A1 and A2. J. Biol. Chem. 275:38286–38295. [DOI] [PubMed] [Google Scholar]
- Shan, J., T.P. Munro, E. Barbarese, J.H. Carson, and R. Smith. 2003. A molecular mechanism for mRNA trafficking in neuronal dendrites. J. Neurosci. 23:8859–8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, R. 2004. Moving molecules: mRNA trafficking in mammalian oligodendrocytes and neurons. Neuroscientist. 10:495–500. [DOI] [PubMed] [Google Scholar]
- Steward, O., and E.M. Schuman. 2003. Compartmentalized synthesis and degradation of proteins in neurons. Neuron. 40:347–359. [DOI] [PubMed] [Google Scholar]
- Strobel, S.A., P.L. Adams, M.R. Stahley, and J. Wang. 2004. RNA kink turns to the left and to the right. RNA. 10:1852–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge, H. 2006. K-turn motifs in spatial RNA coding. RNA Biol. In press. [DOI] [PMC free article] [PubMed]
- Tiedge, H., W. Chen, and J. Brosius. 1993. Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J. Neurosci. 13:2382–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani, G., and W.H. McClain. 2000. The G·U wobble base pair: a fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 1:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidovic, I., S. Nottrott, K. Hartmuth, R. Lührmann, and R. Ficner. 2000. Crystal structure of the spliceosomal 15.5kD protein bound to a U4 snRNA fragment. Mol. Cell. 6:1331–1342. [DOI] [PubMed] [Google Scholar]
- Wang, H., A. Iacoangeli, S. Popp, I.A. Muslimov, H. Imataka, N. Sonenberg, I.B. Lomakin, and H. Tiedge. 2002. Dendritic BC1 RNA: functional role in regulation of translation initiation. J. Neurosci. 22:10232–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., A. Iacoangeli, D. Lin, K. Williams, R.B. Denman, C.U.T. Hellen, and H. Tiedge. 2005. Dendritic BC1 RNA in translational control mechanisms. J. Cell Biol. 171:811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells, D.G., J.D. Richter, and J.R. Fallon. 2000. Molecular mechanisms for activity-regulated protein synthesis in the synapto-dendritic compartment. Curr. Opin. Neurobiol. 10:132–137. [DOI] [PubMed] [Google Scholar]
- White, S.A., M. Hoeger, J.J. Schweppe, A. Shillingford, V. Shipilov, and J. Zarutskie. 2004. Internal loop mutations in the ribosomal protein L30 binding site of the yeast L30 RNA transcript. RNA. 10:369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson, J.R. 2000. Induced fit in RNA-protein recognition. Nat. Struct. Biol. 7:834–837. [DOI] [PubMed] [Google Scholar]
- Wimmer, E.A., A. Carleton, P. Harjes, T. Turner, and C. Desplan. 2000. Bicoid-independent formation of thoracic segments in Drosophila. Science. 287:2476–2479. [DOI] [PubMed] [Google Scholar]