Abstract
The impact of chronic inflammation on the expression of human α-defensins 5 and 6 (HD-5, HD-6), β-defensins 1 and 2 (hBD-1, hBD-2) and lysozyme in epithelial cells of small and large intestine was investigated. Intestinal specimens from 16 patients with ulcerative colitis (UC), 14 patients with Crohn's disease (CD) and 40 controls with no history of inflammatory bowel disease were studied. mRNA expression levels of the five defence molecules were determined in freshly isolated epithelial cells by real-time quantitative RT-PCR. Specific copy standards were used allowing comparison between the expression levels of the different defensins. HD-5 and lysozyme protein expression was also studied by immunohistochemistry. Colonic epithelial cells from patients with UC displayed a significant increase of hBD-2, HD-5, HD-6 and lysozyme mRNA as compared to epithelial cells in controls. Lysozyme mRNA was expressed at very high average copy numbers followed by HD-5, HD-6, hBD-1 and hBD-2 mRNA. HD-5 and lysozyme protein was demonstrated in metaplastic Paneth-like cells in UC colon. There was no correlation between hBD-2 mRNA levels and HD-5 or HD-6 mRNA levels in colon epithelial cells of UC patients. Colonic epithelial cells of Crohn's colitis patients showed increased mRNA levels of HD-5 and lysozyme mRNA whereas ileal epithelial cells of Crohn's patients with ileo-caecal inflammation did not. Chronic inflammation in colon results in induction of hBD-2 and α-defensins and increased lysozyme expression. hBD-1 expression levels in colon remain unchanged in colitis. The high antimicrobial activity of epithelial cells in chronic colitis may be a consequence of changes in the epithelial lining, permitting adherence of both pathogenic bacteria and commensals directly to the epithelial cell surface.
Keywords: α-defensin, β-defensin, inflammatory bowel disease, intestinal epithelial cell, lysozyme
INTRODUCTION
Host defence relies on both innate and adaptive immune mechanisms. Innate immunity is the first line of defence, instrumental in keeping invading microorganisms at bay during the time it takes to mount an effective adaptive immune response. Innate immune components also participate in activating antigen-specific adaptive immune responses. It includes a number of constitutively expressed and inducible humoral factors such as antimicrobial peptides/proteins, e.g. the defensins, the cathelicidins and lysozyme [1–4]. Two major groups of defensins are found in mammals, α-defensins and β-defensins. They differ in pairing of the three intrachain disuphide bonds but have similar tertiary structures showing triple-stranded antiparallel β-sheets. In humans, six α- and four β-defensins have so far been identified. Only one cathelicidin, hCAP18, is produced in humans. It is associated preferentially with granules of neutrophils. Of the six human α-defensins, four (also denoted human neutrophil peptides 1–4) are predominantly produced and stored in granules of phagocytes while human α-defensin 5 and 6 (HD-5, HD-6) are expressed in Paneth cell granules [5–7]. Paneth cells are of epithelial cell lineage and are situated at the bottom of the crypts in the small intestine. HD-5 is also found in female reproductive tissues [8,9]. Mucosal epithelial cells of various tissues including the respiratory, digestive, urinary and reproductive systems as well as keratinocytes produce β-defensins [10–15]. Lysozyme is expressed in normal human intestine [16–18] and human β-defensin 1 (hBD-1) appears to be expressed constitutively by most epithelial cells in both small and large intestine [14,15]. hBD-2 mRNA is generally not expressed in normal human intestine [14,15] but bacteria and proinflammatory cytokines can induce its expression in gastrointestinal epithelium [14,19], as well as in lung epithelium [20,21], skin [11] and oral mucosa [22]. hBD-2 functions as a NF-κB target gene in epithelial cells [14,23,24]. hBD-3 and hBD-4 were discovered most recently and were shown to be expressed in some parts of the gastrointestinal tract [25,26].
Comparatively little is known about the expression of epithelial defensins and lysozyme in chronic inflammatory bowel disease (IBD). hBD-2 was shown to be expressed in colonic epithelium of patients with UC by immunohistochemistry [14], and using the same technique Cunliffe and associates reported recently that HD-5 is expressed by metaplastic Paneth cells in the colon of UC and Crohn's colitis patients [27]. To throw light on the role of defensins in IBD we undertook a quantitative analysis of the mRNA expression levels of defensins and lysozyme in epithelial cells of the small and large intestine of patients with UC and CD and in controls with no history of IBD. HD-5 and lysozyme expression was confirmed at the protein level by immunohistochemistry.
MATERIALS AND METHODS
Patients
Specimens were obtained from patients suffering from UC (n = 16), CD (n = 14) or other benign conditions and cancer (n = 40) by surgical resection.
UC samples were from 11 male and five female patients 23–72 years old (mean ± s.d.: 40 ± 14 years). Two UC patients had no medication the last 4 weeks prior to sampling, six received corticosteroids alone, six received corticosteroids in combination with 5-aminosalicylic acid (5-ASA) compounds, one received corticosteroids in combination with azathioprine and 5-ASA compounds, whereas one patient received 5-ASA compounds only. Eight specimens were from patients with substantially active disease, six from patients with moderately active disease and two from patients with inactive disease, as judged by clinical symptoms and routine histopathology.
CD samples were from four male and 10 female patients 18–56 years old (34 ± 14 years). Nine specimens were from colon, five with ileo-caecal localization and four with colonic localization of active disease. Eight specimens were from small intestine, seven with ileal and one with jejunal localization of disease. Three CD patients provided both ileal and colonic samples. Two CD patients had no medication during the last 4 weeks prior to sampling, eight received corticosteroids alone or in combination with 5-ASA or azathioprine, three received azathioprine in combination with 5-ASA, whereas one patient received azathioprine alone.
Control colon specimens were from 11 male and 12 female patients (68 ± 10 years) with colorectal cancer (n = 18) and non-inflammatory benign conditions (n = 5). Control ileum specimens were from six male and three female patients (69 ± 15 years) with the following diagnoses: Meckel's diverticulitis (n = 1), caecal tubular adenoma (n = 2), gastrointestinal cancer (n = 6). Control jejunum was from two male and six female patients (64 ± 15 years) with gastric and pancreatic cancer (n = 5), gastric ulcer (n = 1) or noninflammatory benign conditions (n = 2). All patients received a single intravenous dose of antibiotics 2 h prior to surgery according to preoperative standard procedure. None of the control patients had been subjected to radio- or chemotherapy, long-standing antibiotic medication or steroid treatment. The control samples were taken distant to macroscopically detectable lesions. This study was approved by the ethical committee at the Medical and Odontological Faculty of Umeå University Hospital and the patients gave their informed consent.
Isolation of intestinal epithelial cells
Epithelial cells were isolated from fresh intestinal specimens, as described earlier [28,29]. Briefly, the mucosal layer was cut into small pieces and treated with 0·1 mm (small intestine) or 0·2 mm (large intestine) dithiotreitol (DTT) with shaking to free cells from the tissue. The supernatant containing epithelial cells from the luminal/villous compartment (referred to as l/v-IEC) and intra-epithelial lymphocytes was collected. Remaining tissue pieces were treated subsequently with 72·5 U/ml of collagenase type IV (Worthington, Freehold, NJ, USA) in heat inactivated human AB+ serum with vigorous shaking at 37°C for 30 min. The cell suspension was then passed through a stainless steel sieve, resulting in a cell fraction containing epithelial cells mainly from the crypt compartment (c-IEC), lamina propria leucocytes and stromal cells. Both cell fractions were then washed in Tris-buffered Hanks's balanced salt solution (TH) containing 0·2% human serum albumin (HSA) and suspended in 66·7% Percoll (Pharmacia, Uppsala, Sweden) overlaid with a gradient of 50%, 44% and 20% Percoll and centrifuged. l/v-IEC and c-IEC were recovered at the interface between 44% and 20% Percoll. The isolated epithelial cells were subsequently depleted of leucocytes by treatment with paramagnetic beads charged with anti-CD45 MoAbs (Dynabeads M-450, Dynal, Oslo, Norway) or anti-CD11b MoAb OKM-1 (American Tissue Culture Collection, Rockville, MD, USA). Magnetic beads were added at a ratio of 20 : 1 and cells were incubated at 4°C for 30 min with slow rotation. The suspension was then treated three times by magnet and unbound cells were counted, washed in RNase-free phosphate buffered saline (PBS) and frozen at −80°C for RNA extraction.
Alternatively, a total epithelial cell population was prepared by a method modified from Ishii et al. [30]. Briefly, the mucosa was dissected from the muscle layer, cut into small pieces and incubated in PBS supplemented with 1·5 mm ethylenediamine-tetraacetic acid (EDTA), 2·6/5·2 mm DTT (small intestine/large intestine), fungizone, penicillin and streptomycin at 37°C for 15 min without agitation. After centrifugation the tissue pieces were suspended in the same solution without DTT, and incubated for 20 min at 37°C with vigorous shaking, followed by vortexing for 2 min. Tissue pieces were then allowed to sediment and the supernatant containing released epithelium was collected. Fresh medium was added subsequently to the tissue pieces and the procedure was repeated. The epithelium was finally treated with collagenase type IV in TH containing 0·4% HSA, fungizone, penicillin and streptomycin for 1 h at 37°C with gentle agitation. After washing the cells were subjected to Percoll gradient centrifugation and depleted of leucocytes as described above.
Cell lines
Four established adherent human colon carcinoma cell lines were used. Moderately differentiated HT-29 cells and well-differentiated LS174T cells were grown in Parker199 medium (SBL Vaccin, Stockholm, Sweden) supplemented with 8% fetal calf serum (FCS) and penicillin–streptomycin. Well-differentiated T-84 cells were grown in Dulbecco/Vogt modified Eagle's minimal essential medium: F12-HAM (ratio 1 : 1), supplemented with 15 mm HEPES buffer, 5% FCS, penicillin and streptomycin. Well-differentiated HCT-8 cells were grown in Parker199 : F12-HAM (ratio 1 : 1), 8% FCS, penicillin, and streptomycin. Cells were maintained at 37°C in humidified air with 5% CO2. Cell cultures were trypsinized weekly by incubation with 1% trypsin and 0·5% EDTA in PBS (pH 7·3) for 5 min at 37°C.
Cytokines, bacteria and lipopolysaccharide (LPS)
Recombinant human interleukin-1β (rhIL-1β) and tumour necrosis factor-α (rhTNF-α) were obtained from Endogen (Woburn, MA, USA), and interferon-γ (rhIFN-γ) from Promega (Madison, WI, USA). Bacterial LPS from Esherichia coli O55: B5 was obtained from Sigma. Live bacteria used for stimulation of epithelial cells were E. coli D21, Bacillus megaterium Bm11, and Micrococcus luteus (kindly provided by Professor Hans G. Boman, MTC, Karolinska Institute, Solna, Sweden) and Salmonella typhimurium LT2 (kindly provided by Professor Glenn Björk, Department of Microbiology, Umeå University, Umeå, Sweden).
Stimulation of colon carcinoma cell lines
Colon carcinoma cells were trypsinized and washed in the appropriate complete tissue culture medium; 1·5 × 106 cells in 3 ml were seeded in six-well tissue culture plates and grown for 48 h to confluence. Cells were stimulated at 37°C with 3 or 10 µg/ml of LPS, 100 ng/ml of rhIL-1β, 10 ng/ml of rhTNF-α or 200 U/ml of rhIFN-γ, respectively. Cells were also stimulated for 1 h with 1 × 108 live bacteria from overnight cultures without the addition of antibiotics. The tumour cells were then washed thoroughly in medium containing 50 µg/ml of gentamycin in order to kill any remaining extracellular bacteria and incubated further in media containing gentamycin for an additional 5 h. The cells were then harvested by trypsinization, washed in RNase-free PBS and frozen at −80°C for RNA extraction.
Immunoflow cytometry
To determine the purity of the isolated IEC fractions, 0·1 × 106 cells were stained with phycoerythrin conjugated anti-CD45 MoAbs (Dakopatts, Glostrup, Denmark) and flourescein-isothiocyanate conjugated anti-epithelial antigen MoAb BerEp4 (Dakopatts) in TH containing 0·2% HSA, 0·02% NaN3, penicillin and streptomycin. Conjugated irrelevant concentration and isotype-matched MoAb (Dakopatts) served as controls. Cells were analysed on a FACScan flowcytometer (Becton-Dickinson, Mountain View, CA, USA) using the cellquest program.
RNA extraction and real time quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR)
Total RNA was extracted by the acid guanidine–phenol–chloroform method [31]. Precipitated RNA was dissolved in RNase free water containing 1000 U/ml of the RNase inhibitor rRNasin (Promega, Madison, WI, USA) and stored at −80°C.
Real-time qRT-PCR assays for quantitative determination of HD-5, HD-6, hBD-1, hBD-2 and lysozyme mRNA were constructed using the TaqMan EZ® technology (Applied Biosystems). Primers and probes are listed in Table 1. Primers were chosen to hybridize to different exons and a reporter dye-labelled probe was placed over an exon border within the amplicon. Emission from released reporter dye was monitored by the ABI Prism® 7700 Sequence Detection System (Applied Biosystems). All assays gave a linear relation between log concentrations of standard RNA and PCR cycles over a range of at least 5 logs. For each system a specific RNA copy standard was prepared (see below). Determinations were carried out in triplicates and expressed as copies of mRNA/µl using the external copy standard. The concentration of 18S rRNA (Applied Biosystems) was determined in each sample and the result expressed as mRNA copies of defensin/lysozyme per unit of 18S rRNA.
Table 1.
Sequences of primers and probes used in real-time quantitative RT-PCR
| 5′-primer | 3′-primer | Probe | Amplicon length | |
|---|---|---|---|---|
| hBD-1 | 5′-CTGCTGTTTACTCTCTGCTTACTTTT-3′ | 5′-CCTCCACTGCTGACGCA-3′ | 5′-AGAAAGTTACCACCTGAGGCCATCTCA-3′ | 107 bp |
| hBD-2 | 5′-CTCGTTCCTCTTCATATTCCTGA-3′ | 5′-CTAGGGCAAAAGACTGGATGAC-3′ | 5′-CCTATACCACCAAAAACACCTGGAAGAGG-3′ | 111 bp |
| HD-5 | 5′-GACAACCAGGACCTTGCTATCT-3′ | 5′-ACGGGTAGCACAACGGC-3′ | 5′-CTTAGAACCTCAGGTTCTCAGGCAAGAGC-3′ | 114 bp |
| HD-6 | 5′-AGAGGATGCAAGCTCAAGTCTTA-3′ | 5′-AGTGCAGGTCCCATAGGAATAT-3′ | 5′-CTTTGGGCTCAACAAGGGCTTTCACT-3′ | 106 bp |
| lysozyme | 5′-AAAACCCCAGGAGCAGTTAAT-3′ | 5′-CAACCCTCTTTGCACAAGCT-3′ | 5′-CCTGCAGTGCTTTGCTGCAAGATAA-3′ | 94 bp |
Cloning and sequencing
RT-PCR products were subjected to gel electrophoresis in 1·5% SeaKem®LM agarose (FMC BioProducts, Rockland, ME, USA), fragments were cut out and purified with QiaexII® Gel extraction kit (Qiagen, Germany) and then ligated into dT-treated EcoRV pBluescript (SK + or KS +). Competent E. coli XL1blueMRF′ was transformed with plasmids and grown on plates containing 100 µg/ml ampicillin, 12·5 µg/ml tetracycline, 20 µg/ml X-gal and 40 µg/ml IPTG. Transformants were checked for insert of the expected size on agarose gel after restriction cleavage of plasmids using XbaI/XhoI (Life Technologies). DNA, 2–3 µg, was used for cycle-sequencing using T7 or Rev primers and Thermo Sequenase fluorescent labelled primer cycle sequencing kit with 7-deaza-dGTP (Amersham/Pharmacia Biotech, UK). Cycle-sequenced products were analysed by ALFexpress (Pharmacia Biotech).
RNA copy standard preparation
Total RNA from normal jejunal epithelial cells (HD-5, HD-6), peripheral blood granulocytes (lysozyme) and thymus (hBD-1 and hBD-2) were used as the starting materials for copy standard preparations and optimization of the assays (see above). The primers used for RT-PCR are given in Table 2. The PCR products, which include the respective sequences amplified in quantitative RT-PCR, were cloned, sequenced and used as templates for in vitro transcription with T7 polymerase/RiboProbe® In Vitro Transcription Systems (Promega). Linearized DNA, 3–7 µg, was used in large-scale synthesis reactions carried out at 37°C for 2–3 h. The reaction products were then treated with 1 U/µg of RNase-free DNase (Promega) for 30–40 min at 37°C, followed by extractions with phenol : chloroform : isoamylalcohol (25 : 24 : 1) and chloroform : isoamylalcohol (24 : 1). RNA was precipitated with 2·5 volumes of 99·5% ethanol and 0·5 volumes of 7·5 m ammonium acetate at −70°C for at least 1 h. DNase treatment was repeated at least twice. Finally, the copy standards were checked by RT-PCR and PCR to evaluate the content of DNA which proved to be less than 0·2% for all of them. Concentration of the transcripts was calculated on the basis of the O.D.260 value, the molecular weight of the transcript and Avogadro's number. The standards were finally diluted to 108 copies/µl.
Table 2.
Primers used in RT-PCR for cloning of cDNA and construction of copy standards
| 5′-primer | 3′-primer | Amplicon length | |
|---|---|---|---|
| hBD-1 | 5′-AGTGTTGCCTGCCAGTCGCC-3′ | 5′- TCTGCGTCATTTCTTCTGGTCACTC −3′ | 257 bp |
| hBD-2 | 5′-AGCCATCAGCCATGAGGGTCTT-3′ | 5′-CTGATGAGGGAGCCCTTTCTGAAT-3′ | 261 bp |
| HD-5 | 5′-CCCAGCCATGAGGACCATC-3′ | 5′-TCTATCTAGGAAGCTCAGCGACAG-3′ | 306 bp |
| HD-6 | 5′-AGCGACCCTAGCCATGAGAACC-3′ | 5′-ACAAAGTTGATGGCAATGTATGGGA-3′ | 406 bp |
| lysozyme | 5′-GAACTCTGAAAAGATTGGGAATGGA-3′ | 5′-ACAACCTTGAACATACTGACGGACA-3′ | 356 bp |
Immunohistochemistry
Fresh tissue samples were rinsed with Hanks's balanced salt solution, snap frozen in isopentane precooled in liquid N2 and stored at −80°C. Seven µm sections were air-dried and fixed in 4% paraformaldehyde in PBS (pH 7·4) for 15 min and then rinsed in cold 0·02 m PBS (pH 7·2) containing 0·005% saponin (PBS/saponin). Free aldehyde groups were blocked with 0·1 m glycine in PBS/saponin for 10 min at room temperature, rinsed and incubated with PBS, 0·2% bovine serum albumin (BSA) and 0·05% saponin (PBS/BSA/saponin) for 45 min at room temperature. Sections were incubated with antilysozyme MoAb BTI (Paesel + Lorei, Hanau, Germany) or anti-HD-5 polyclonal rabbit antiserum (kindly provided by Professor Tom Ganz, UCLA, Los Angeles, USA) diluted in PBS/BSA/saponin for 1 h at room temperature. Irrelevant mouse IgG1 MoAb (Dakopatts) and preimmune rabbit serum served as negative controls, respectively. For antilysozyme staining the endogenous avidin activity was blocked with biotin blocking system (Dakopatts). Endogenous peroxidase activity was blocked by incubation in PBS containing 0·03% H2O2 and 2 mm NaN3 at 37°C for 1 h, followed by incubation in PBS/BSA/saponin for 30 min at room temperature. Lysozyme sections were incubated with biotinylated F(ab′)2-fragments of sheep antimouse Ig (Amersham) for 1 h at room temperature followed by 30 min incubation with ABC complex/HRP-solution (Dakopatts). HD-5 sections were incubated with HRP conjugated F(ab′)2-fragments of donkey antirabbit IgG (Amersham) for 1 h at room temperature. Sections were developed with 0·05% 3,3′-diaminobenzidine tetrahydrochloride and 0·03% H2O2 in 0·05 m Tris HCl buffer (pH 7·6) and counterstained with methyl-green. Two antisera against hBD-2 were tested in immunohistochemistry; BD-2 ab9871 from Abcam, UK and HBD21-S from Alpha Diagnostic International, USA.
Statistical analyses
The significance of differences in antimicrobial peptide mRNA expression levels between groups was evaluated by two-tailed Mann–Whitney's rank sum test. Two-tailed Wilcoxon's assigned rank sum test was used when comparing mRNA expression levels in paired samples of c-IEC and l/v-IEC. Analyses of correlation between mRNA levels of different antimicrobial peptides were performed using the Spearman's rank correlation test.
RESULTS
Intestinal epithelial cell populations
A cell isolation procedure developed for the isolation of epithelial cells and leucocytes from the same mucosal sample was used. Here epithelial cells localized in the villi of small intestine and at the free luminal surface of colon are enriched in the ‘l/v’-fraction. Epithelial cells in the crypts are preferentially recovered in the ‘c’-fraction. The relative contribution to the total number of epithelial cells in the ‘l/v’- and ‘c’-fractions was 1 : 4 in small intestine and 1 : 3 in large intestine with no significant difference between UC colon and normal colon. As epithelial cells are separated from leucocytes by Percoll gradient fractionation followed by removal of the residual leucocytes by magnetic beads charged with anti-CD45 MoAb and anti-CD11b MoAb, both epithelial cell fractions were free of leucocytes (<1% positive cells by immunoflow cytometry with anti-CD45 MoAb). The percentage of BerEp+ cells was >95%.
Epithelial cells were also isolated by detaching intact crypts and crypt epithelial cells from the basal membrane using EDTA as described by Ishii et al. [30]. Analysis of five colon samples prepared in this way revealed that the median number of mRNA copies/18S rRNA unit for the four defensins and lysozyme was almost identical to the corresponding median values obtained for the ‘c’-fraction (data not shown).
Differential expression of mRNA for antimicrobial peptides and lysozyme in epithelial cells from normal human small and large intestine
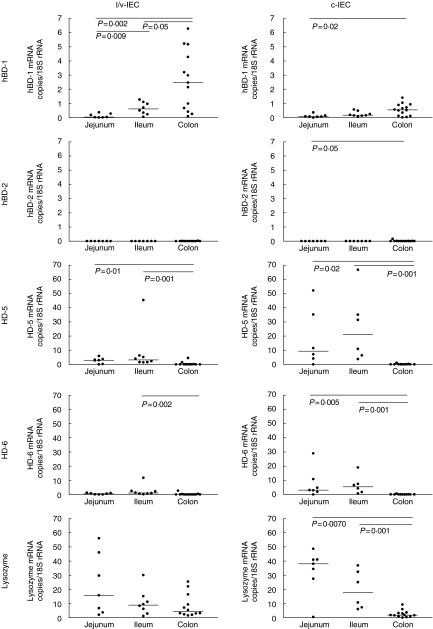
The number of mRNA copies for hBD-1, hBD-2, HD-5, HD-6 and lysozyme per 18S rRNA unit was determined in freshly isolated luminal/villous (l/v-IEC) and cryptal (c-IEC) epithelial cells from apparently normal human jejunum, ileum and colon by real-time qRT-PCR. The results are shown in Fig. 1. We found that 0·74 units of 18S rRNA correspond to one epithelial cell (mean value of 105 determinations). No significant differences were seen between 18S rRNA levels in IEC isolated from small and large intestine or IEC from UC colon compared to control colon. Thus, the values of mRNA copies/18S rRNA unit are close to the average number of mRNA copies/cell.
Fig 1.
Expression levels of defensin and lysozyme mRNAs as determined by qRT-PCR in c- and l/v-IECs freshly isolated from jejunum, ileum and colon of patients with no history of intestinal inflammation. Statistically significant differences are indicated. Horizontal bars indicate median values.
hBD-1 mRNA was expressed throughout the intestine with the highest expression levels in colonic epithelial cells and clearly lower levels in ileal and jejunal epithelial cells. In contrast, hBD-2 mRNA was undetectable or expressed at very low levels in epithelial cells of both small and large intestine. HD-5 and HD-6 mRNA showed a very similar expression pattern to each other with no expression in colon and high expression levels in both jejunum and ileum. It can also be seen that the ‘c’-fraction contained between four and 11 times higher concentrations of HD-5 and HD-6 mRNA than the ‘l/v’-fraction. This result is consistent with the fact that Paneth cells are the major source of the α-defensins. Paneth cells are restricted normally to the bottom of the crypts in small intestine and are consequently expected to preferentially accumulate in the ‘c’-fraction of ileal and jejunal samples. Lysozyme mRNA was expressed at higher levels in small intestine than in large intestine. The expression pattern of lysozyme in the two cell fractions was more complex. While lysozyme is approximately 2 times more abundant in the ‘c’-fraction than in the ‘l/v’-fraction in small intestine the reverse is true for colon. The difference was statistically significant between ‘c’- and ‘l/v’-fractions from the same sample in colon (P = 0·02, n= 11) but did not reach statistical significance in small intestine (P > 0·05, n= 11). The occurrence of lysozyme mRNA in the two fractions is due to lysozyme being expressed in several different cell types (see below).
It can be noted that the number of mRNA copies/18S rRNA unit for HD-5 and HD-6 in small intestine are comparable, about 5–25 (‘c’-fraction containing the majority of the cells; Fig. 1). Because Paneth cells will constitute only about 10% of all the epithelial cells the average number of mRNA copies for these defensins in Paneth cells would be ∼ 50–250 copies/cell. These values put these mRNAs in the intermediate abundance class [32]. Normal colonic epithelial cells expressed an average of 1–3 copies of hBD-1 mRNA and ∼ 3 copies of lysozyme mRNA/18S rRNA unit (Fig. 1).
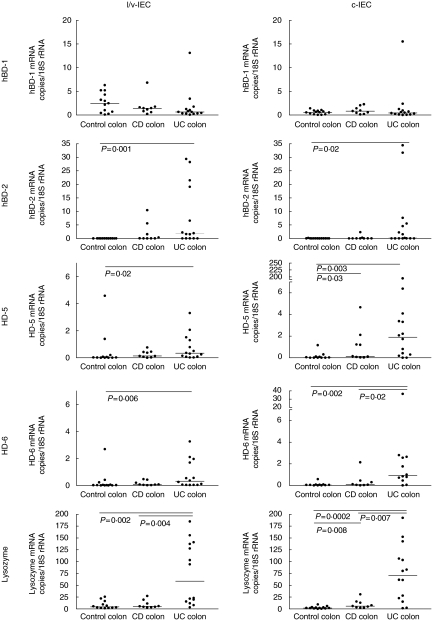
hBD-2, α-defensins and lysozyme mRNA levels are increased in colonic epithelial cells of UC patients
The mRNA levels of the five antimicrobial components were determined subsequently in colon epithelial cells from UC and CD patients and compared to the levels in epithelial cells of control colon (Fig. 2). The colon samples from UC patients were all from diseased tissue while four samples from the CD patients were from diseased colon (Crohn's colitis) and five colon CD samples were from patients with active disease restricted to the ileo-caecal region. As can be seen, the mRNA levels of hBD-1 were not significantly different between IBD patients and controls irrespective of whether the ‘l/v’- or ‘c’- fractions were compared. In contrast, hBD-2 mRNA expression was clearly induced in UC colon. The median copy number of hBD-2 mRNA/18S rRNA unit was increased to 0·2–1·8 compared to < 0·01 in control colon. HD-5 and HD-6 mRNA levels in UC colon were also elevated significantly compared to controls with median copies/18S rRNA unit values similar to those of hBD-2 mRNA, i.e. 0·3–1·9. It should be noted that normal colon does not express these defensins (Fig. 2). Finally, lysozyme mRNA was also expressed at highly elevated levels in UC colon with a 10-fold increase compared to control colon (Fig. 2; median copies/18S rRNA values 60–70).
Fig 2.
Expression levels of defensin and lysozyme mRNAs as determined by qRT-PCR in c- and l/v-IECs freshly isolated from UC-, CD- and control colon. Statistically significant differences are indicated. Horizontal bars indicate median values.
The colon samples from the CD patients displayed an interesting mRNA expression pattern. The three highest values for hBD-2, HD-5, HD-6 and lysozyme mRNA in the CD colon group were all from patients with Crohn's colitis. The fourth Crohn's colitis sample had a low value similar to those of CD patients with ileo-caecal localization of active disease.
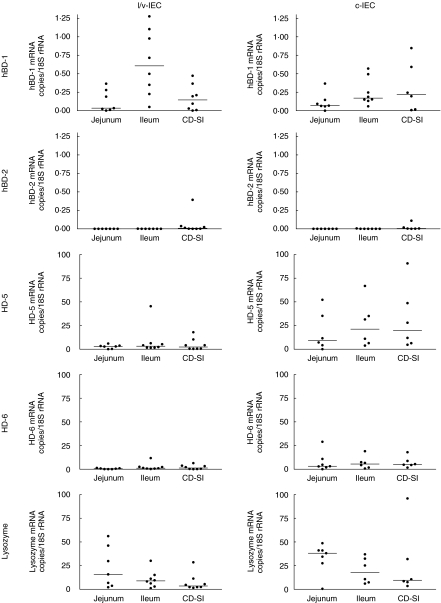
mRNA levels for defensins and lysozyme are generally not increased in diseased small intestine epithelial cells from CD patients.
Figure 3 shows the results from analysis of epithelial cells from inflamed small intestinal mucosa of CD patients compared to control ileum and jejunum. As can be seen, there are no statistically significant differences between normal and diseased ileum with respect to mRNA for any of the five antimicrobial components.
Fig 3.
Expression levels of defensin and lysozyme mRNAs as determined by qRT-PCR in c-IECs and l/v-IECs freshly isolated from CD small intestine (CD-SI) and control ileum and jejunum. Horizontal bars indicate median values.
mRNA levels for antimicrobial peptides and lysozyme in relation to inflammatory status
IEC of UC patients were analysed for correlation between hBD-1, hBD-2, HD-5, HD-6 and lysozyme mRNA expression levels and disease activity. Eight specimens were from patients with active disease, six from patients with moderately active disease and two from patients with inactive disease. mRNA expression levels were assigned to either of three groups: high, intermediate and low. Patients with active disease generally had high hBD-2, HD-5, HD-6 and lysozyme mRNA levels while patients with inactive disease had low levels. However, the difference did not reach statistical significance due to the low number of patients with inactive disease.
Lack of correlation between mRNA levels for hBD-2 on one hand and HD-5, HD-6 and lysozyme on the other hand in epithelial cells from UC colon
The mRNA levels for hBD-2, HD-5, HD-6 and lysozyme in individual colon c-IEC samples from UC patients were compared with each other. HD-5, HD-6 and lysozyme mRNA levels were correlated, while the hBD-2 mRNA levels were not correlated with the levels of HD-5, HD-6, or lysozyme (Table 3).
Table 3.
Correlation between defensin and lysozyme mRNA levels in c-IEC of UC colon
| mRNA species compared | P* | r |
|---|---|---|
| hBD-2/HD-5 | NS | – |
| hBD-2/HD-6 | NS | – |
| hBD-2/lysozyme | NS | – |
| HD-5/HD-6 | 0·002 | 0·8 |
| HD-5/lysozyme | 0·04 | 0·5 |
| HD-6/lysozyme | 0·001 | 0·8 |
Correlation between indicated defensins or lysozyme mRNA copies/18S rRNA in freshly isolated c-IEC of UC colon was analysed by Spearman's test.
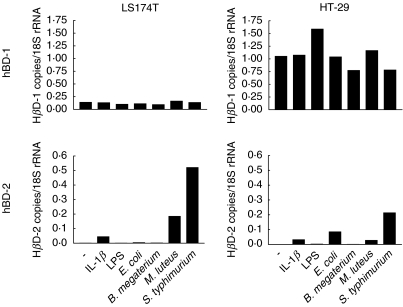
hBD-2 mRNA is induced by live bacteria and IL-1β
Colon carcinoma cells were stimulated in vitro with live E. coli, Bacillus megaterium, Micrococcus luteus or Salmonella typhimurium or with E. coli 055 LPS or the cytokine IL-1β for 1 h (Fig. 4). Live S typhimurium, E. coli and M. luteus bacteria and the cytokine IL-1β induced hBD-2 mRNA while B. megaterium was inactive as was purified LPS. hBD-1 mRNA was expressed in the cell lines. As expected, none of the agents increased hBD-1 mRNA expression (Fig. 4). It can also be seen that S. typhimurium induced hBD-2 mRNA to levels comparable to those of hBD-1 mRNA. IFN-γ (4, 24, 96 h of incubation), TNF-α (24 h of incubation) or LPS (1, 4, 24 and 96 h of incubation) did not induce hBD-2 mRNA in any of the four colon carcinoma cell lines studied (data not shown).
Fig 4.
Stimulation of confluent cultures of HT-29 and LS174T for 1 h with 10 µg/ml of LPS, 100 ng/ml of rhIL-1β, 1 × 108 E. coli D21, 1 × 108 B. megaterium Bm11, 1 × 108 M. luteus, or 1 × 108 S. typhimurium LT2, followed by removal of bacteria and continued culturing for 5 h in fresh medium containing 50 µg/ml gentamycin to kill remaining extracellular bacteria. Total RNA was extracted and the expression levels of defensin mRNAs were determined by qRT-PCR with copy standards.
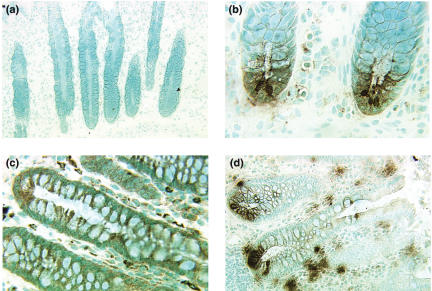
HD-5 and lysozyme protein are expressed in cryptal cells in UC colon
The ability of the anti-HD-5 antibody to specifically stain Paneth cells was evaluated on tissue sections of normal human small intestine. An intense staining of the Paneth cells at the bottom of the crypts was seen (data not shown). These cells were also intensely stained by the antilysozyme MoAb. Figure 5 shows representative results of immunohistochemical staining of UC colon and control colon. No HD-5 positive cells are seen in normal human colon (Fig. 5a). However, colon ascendens, colon transversum and colon sigmoideum of UC patients contained groups of HD-5-positive cells (13/18 samples displayed positively stained cells) located in the lower parts of the crypts (Fig. 5b). These cells were also positively stained with antibodies against lysozyme (8/13 samples displayed positively stained cells; Fig. 5d). Anti-lysozyme MoAb also stained goblet cells in normal colon as well as lamina propria macrophages (Fig. 5c). Two antisera to hBD-2 were tested on tissue sections of UC colon, but no positively stained cells were detected.
Fig 5.
Immunohistochemical staining for detection of HD-5 and lysozyme expressing cells in UC and control colon. (a) Control colon stained with anti-HD-5 antiserum. No stained cells could be detected. (b) UC colon stained with anti-HD-5 antiserum. Positive cells are seen at different locations in the crypt, but most commonly towards the crypt base. (c) Control colon stained with antilysozyme MoAb. Stained goblet cells as well as lamina propria macrophages can be seen. (d) UC colon stained with antilysozyme MoAb. Positive cells are seen at different locations in the crypt, but most commonly towards the crypt base. Original magnification: 55 × (a, d), and 220 × (b,c).
DISCUSSION
The aim of this study was to investigate whether the expression levels of α- and β-defensins and lysozyme in epithelial cells of human intestine are affected by chronic inflammation, as seen in UC and CD. For this purpose we used purified epithelial cells depleted of intraepithelial lymphocytes and lamina propria leucocytes by density fractionation followed by treatment with magnetic beads charged with anti-CD45 MoAb and anti-CD11b MoAb. Less than 1% of the cells in the epithelial cell fraction were leucocytes. This was considered important, as murine intraepithelial lymphocytes were shown to express cryptidins 1 and 3, i.e. α-defensins expressed by Paneth cells in the small intestine of mice, and β-defensins [33]. As yet it has not been investigated whether human intraepithelial lymphocytes can express defensins. The expression pattern along the normal human intestine (proximal to distal) as well as between intestinal epithelial cells of different states of differentiation (l/v-IEC versus c-IEC) were analysed and compared with the corresponding cell type in the chronically inflamed intestine of patients suffering from UC or CD. As expected, the two α-defensins HD-5 and HD-6 as well as lysozyme mRNAs were mainly expressed by small intestinal c-IEC in controls. The clear difference in expression levels for HD-5 and HD−6 in c-IEC and l/v-IEC from small intestine reassures that the isolation procedure that we use really separates the cells of the luminal compartment and the cells from the crypt into different populations. Lysozyme, being produced by other cell types than Paneth cells as well, notably goblet cells, was also detected in the l/v-IEC fraction in all three intestinal regions. The β-defensin hBD-1 was detected along the entire normal intestine, but was expressed more abundantly in colon with a tendency for higher expression levels in the more mature epithelial cells in the l/v-compartment. In contrast, hBD-2 was detected only rarely in normal IECs. This is in line with the findings of Frye et al. [15], who found no evidence for hBD-2 in normal human intestine using crude extracts of whole biopsies and a semiquantitative RT-PCR assay.
The most striking result of the study was the statistically significant increase in hBD-2, HD-5, HD-6 and lysozyme mRNA expression levels in colonic IECs of UC patients. This increase was observed in both l/v-IECs and c-IECs. Patients with inactive disease had low levels of defensin mRNA. However, because almost all patients had active or moderately active disease (14/16), no firm conclusion can be drawn about the relationship between disease activity and defensin/lysozyme expression levels. Interestingly, expression levels of HD-5, HD-6 and to some degree, lysozyme, correlated with each other in individual UC colon samples while the levels of hBD-2 did not correlate with the HD-5 and HD-6 levels. This indicates that different mechanisms are responsible for their expression. Immunohistochemical staining of UC colon with anti-HD-5 antibodies and antilysozyme MoAb revealed the presence of Paneth-like cells in the crypts. Groups of positive cells were located not only in the base of crypts but also higher up. In addition, antilysozyme MoAb stained goblet cells. Our result is in complete agreement with a recent study by Cunliffe et al. [27], who studied the expression of HD-5 in UC colon by immunohistochemistry. It is known that the chronic intestinal inflammation in UC can lead to differentiation to metaplastic Paneth cells [34]. We conclude that metaplastic Paneth cells in colon of UC patients produce HD-5, HD-6 and lysozyme and that the expression levels of HD-5 and HD-6 in these cells are similar (this study and [27]).
hBD-2 mRNA was clearly up-regulated in UC colon while the mRNA level of hBD-1 was essentially unchanged. It is interesting to note that the expression level of hBD-2 mRNA in colon epithelial cells from UC patients reached the level at which hBD-1 normally is expressed i.e. ∼ 1 copy/cell. Thus, a significant additional antimicrobial defence is induced in the colon epithelium as a consequence of the disease process. It can also be noted that the l/v-IEC of UC colon had higher expression levels of hBD-2 mRNA than the c-IEC indicating that the mature epithelial cells facing the gut lumen produce the highest levels of hBD-2. We were unsuccessful in demonstrating hBD-2 protein using two commercially available antihBD-2 antisera in immunohistochemistry. We do not think that this is due to lack of the protein, but rather to the fact that the antibodies react poorly with the native defensins. O'Neil et al. [14] has shown recently that hBD-2 mRNA will result in a functional protein which can be stained with another anti-hBD-2 antibody in colonic epithelium. It has been shown previously that live enteric bacteria can up-regulate the levels of hBD-2 mRNA and protein in colon carcinoma cell lines [14]. Their results are confirmed and extended with quantitative measurements of mRNA levels in this study (Fig. 4). The consistency between the studies proves the utility and reliability of the qRT-PCR assays as tools to detect changes in mRNA levels in human cells. Furthermore, our results on the induction of hBD-1 and -2 in adenocarcinoma cell lines support the view that hBD-2 is induced in colon by IL-1β and bacteria but not by IFN-γ, TNF-α or LPS. This finding is compatible with the idea that hBD-2 is induced in epithelial cells through contact with enteric bacteria. The hBD-2 gene contains several NF-κB-motifs in its proximal promoter region [23,24] and induction of hBD-2 in colon carcinoma cell lines by IL-1β and live enteroinvasive bacteria is NF-κB dependent [14]. Thus, it is likely that the up-regulation of hBD-2 in the colon epithelium of UC patients is indeed NF-κB dependent. NF-κB plays an important role in the regulation of several genes involved in the inflammatory process [35–38].
CD patients displayed an interesting difference with respect to expression of defensins and lysozyme in small and large intestine. When small intestinal samples from patients with the disease located to the small intestine were analysed we found no difference in mRNA expression levels for any of the five antimicrobial components analysed compared to controls (Fig. 3). However, the three highest values of hBD-2, HD-5, HD-6 and lysozyme were from patients with active Crohn's colitis (Fig. 2). Because the ‘microbial load’ is much higher in the large intestine than in the small intestine, this finding supports the notion that up-regulation of hBD-2 is caused by components of the microbial flora. Whether it is a direct effect of the interaction between live microorganisms and the epithelium or an indirect effect from secretion of proinflammatory cytokines as IL-1β from immune cells and/or epithelial cells cannot be decided from these experiments. Toll-like receptors expressed on intestinal epithelial cells [39–42] are likely to be involved if the effect is direct. In IBD, alterations in glycosylation of mucosal glycoproteins may result in weakening of the mucosal barrier [43] which leads to closer contact of the luminal flora (endogenous) with the mucosal layer and higher numbers of bacteria within the mucus layer [44]. Whether the disease process weakens the apical glycocalyx/mucin layer of colon epithelium making the apical [45] and/or basolateral surface accessible to intestinal bacteria or whether a pathogenic microorganism with ability to penetrate the mucosa is responsible for the observed induction of hBD-2 is unknown.
The expression of high levels of α- and β-defensins in colon epithelial cells of IBD patients may help not only in protecting the mucosa by killing microorganisms; defensins may also act as chemotactic factors for immune cells [1], leading to enhancement of adaptive immune responses. In this case we must assume that defensins are also released baso-laterally. Defensins can exert cytotoxic effects on host epithelial cells [46] and induce apical conductance in Cl− secretory epithelia [47]. The marked lymphocyte infiltration, in particular of T cells in UC colon [48], may be caused at least partly by excessive defensin production in the diseased intestine. Similarly, excessive defensin production may contribute to the diarrhoea affecting UC patients.
Acknowledgments
This project was supported by grants from the ‘Network for Inflammation Research’ funded by the Swedish Foundation for Strategic Research (S.H.), the Swedish Research Council, Medicine (Å.D., 72X-14060 and S.H., 06X-09945), the Swedish Natural Science Research Council (M.-L.H., B650-19981072) and the Medical Faculty of Umeå University (S.H.). The skilful technical assistance of Marianne Sjöstedt is gratefully acknowledged. We thank Dr Åke Öberg and colleges for surgical samples, Professor Tom Ganz (UCLA, Los Angeles, USA) for kindly providing the anti-HD-5 antiserum and Professors Hans G. Boman and Vladimir Baranov for stimulating and enlightening discussions.
References
- 1.Yang D, Chertov O, Oppenheim JJ. The role of mammalian antimicrobial peptides and proteins in awakening of innate host defenses and adaptive immunity. Cell Mol Life Sci. 2001;58:978–89. doi: 10.1007/PL00000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boman HG. Innate immunity and the normal microflora. Immunol Rev. 2000;173:5–16. doi: 10.1034/j.1600-065x.2000.917301.x. [DOI] [PubMed] [Google Scholar]
- 3.Schroder JM. Epithelial antimicrobial peptides: innate local host response elements. Cell Mol Life Sci. 1999;56:32–46. doi: 10.1007/s000180050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehrer RI, Ganz T. Antimicrobial peptides in mammalian and insect host defence. Curr Opin Immunol. 1999;11:23–7. doi: 10.1016/s0952-7915(99)80005-3. [DOI] [PubMed] [Google Scholar]
- 5.Jones DE, Bevins CL. Paneth cells of the human small intestine express an antimicrobial peptide gene. J Biol Chem. 1992;267:23216–25. [PubMed] [Google Scholar]
- 6.Jones DE, Bevins CL. Defensin-6 mRNA in human Paneth cells: implications for antimicrobial peptides in host defense of the human bowel. FEBS Lett. 1993;315:187–92. doi: 10.1016/0014-5793(93)81160-2. [DOI] [PubMed] [Google Scholar]
- 7.Porter EM, Liu L, Oren A, et al. Localization of human intestinal defensin 5 in Paneth cell granules. Infect Immun. 1997;65:2389–95. doi: 10.1128/iai.65.6.2389-2395.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quayle AJ, Porter EM, Nussbaum AA, et al. Gene expression, immunolocalization, and secretion of human defensin-5 in human female reproductive tract. Am J Pathol. 1998;152:1247–58. [PMC free article] [PubMed] [Google Scholar]
- 9.Svinarich DM, Wolf NA, Gomez R, et al. Detection of human defensin 5 in reproductive tissues. Am J Obstet Gynecol. 1997;176:470–5. doi: 10.1016/s0002-9378(97)70517-9. [DOI] [PubMed] [Google Scholar]
- 10.Bensch KW, Raida M, Magert HJ, et al. hBD-1: a novel β-defensin from human plasma. FEBS Lett. 1995;368:331–5. doi: 10.1016/0014-5793(95)00687-5. [DOI] [PubMed] [Google Scholar]
- 11.Harder J, Bartels J, Christophers E, et al. A peptide antibiotic from human skin. Nature. 1997;387:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 12.Bals R, Wang X, Wu Z, et al. Human β-defensin 2 is a salt-sensitive peptide antibiotic expressed in human lung. J Clin Invest. 1998;102:874–80. doi: 10.1172/JCI2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valore EV, Park CH, Quayle AJ, et al. Human β-defensin-1: an antimicrobial peptide of urogenital tissues. J Clin Invest. 1998;101:1633–42. doi: 10.1172/JCI1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Neil DA, Porter EM, Elewaut D, et al. Expression and regulation of the human β-defensins hBD-1 and hBD-2 in intestinal epithelium. J Immunol. 1999;163:6718–24. [PubMed] [Google Scholar]
- 15.Frye M, Bargon J, Lembcke B, et al. Differential expression of human alpha- and beta-defensins mRNA in gastrointestinal epithelia. Eur J Clin Invest. 2000;30:695–701. doi: 10.1046/j.1365-2362.2000.00696.x. [DOI] [PubMed] [Google Scholar]
- 16.Geyer G. Lysozyme in Paneth cell secretions. Acta Histochem. 1973;45:126–32. [PubMed] [Google Scholar]
- 17.Ghoos Y, Vantrappen G. The cytochemical localization of lysozyme in Paneth cell granules. Histochem J. 1971;3:175–8. doi: 10.1007/BF01002560. [DOI] [PubMed] [Google Scholar]
- 18.Mason DY, Taylor CR. The distribution of muramidase (lysozyme) in human tissues. J Clin Pathol. 1975;28:124–32. doi: 10.1136/jcp.28.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Neil DA, Cole SP, Martin-Porter E, et al. Regulation of human β-defensins by gastric epithelial cells in response to infection with Helicobacter pylori or stimulation with interleukin-1. Infect Immun. 2000;68:5412–5. doi: 10.1128/iai.68.9.5412-5415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Becker MN, Diamond G, Verghese MW, et al. CD14-dependent lipopolysaccharide-induced β-defensin-2 expression in human tracheobronchial epithelium. J Biol Chem. 2000;275:29731–6. doi: 10.1074/jbc.M000184200. [DOI] [PubMed] [Google Scholar]
- 21.Singh PK, Jia HP, Wiles K, et al. Production of β-defensins by human airway epithelia. Proc Natl Acad Sci USA. 1998;95:14961–6. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathews M, Jia HP, Guthmiller JM, et al. Production of β-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect Immun. 1999;67:2740–5. doi: 10.1128/iai.67.6.2740-2745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu L, Wang L, Jia HP, et al. Structure and mapping of the human β-defensin HBD-2 gene and its expression at sites of inflammation. Gene. 1998;222:237–44. doi: 10.1016/s0378-1119(98)00480-6. [DOI] [PubMed] [Google Scholar]
- 24.Harder J, Meyer-Hoffert U, Teran LM, et al. Mucoid Pseudomonas aeruginosa, TNF-α, and IL-1β, but not IL-6, induce human β-defensin-2 in respiratory epithelia. Am J Respir Cell Mol Biol. 2000;22:714–21. doi: 10.1165/ajrcmb.22.6.4023. [DOI] [PubMed] [Google Scholar]
- 25.Harder J, Bartels J, Christophers E, et al. Isolation and characterization of human β-defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276:5707–13. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 26.Garcia JR, Krause A, Schulz S, et al. Human β-defensin 4: a novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J. 2001;15:1819–21. [PubMed] [Google Scholar]
- 27.Cunliffe RN, Rose FR, Keyte J, et al. Human defensin 5 is stored in precursor form in normal Paneth cells and is expressed by some villous epithelial cells and by metaplastic Paneth cells in the colon in inflammatory bowel disease. Gut. 2001;48:176–85. doi: 10.1136/gut.48.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lundqvist C, Hammarström ML, Athlin L, et al. Isolation of functionally active intraepithelial lymphocytes and enterocytes from human small and large intestine. J Immunol Meth. 1992;152:253–63. doi: 10.1016/0022-1759(92)90147-l. [DOI] [PubMed] [Google Scholar]
- 29.Lundqvist C, Melgar S, Yeung MMW, et al. Intraepithelial lymphocytes in human gut have lytic potential and a cytokine profile that suggest T helper 1 and cytotoxic functions. J Immunol. 1996;157:1926–34. [PubMed] [Google Scholar]
- 30.Ishii S, Steele G, Jr, Ford R, et al. Normal colonic epithelium adheres to carcinoembryonic antigen and type IV collagen. Gastroenterology. 1994;106:1242–50. doi: 10.1016/0016-5085(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 31.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 32.Alberts B. Molecular biology of the cell. 3. New York: Garland Publications; 1994. [Google Scholar]
- 33.Shires J, Theodoridis E, Hayday AC. Biological insights into TCRγδ+ and TCRαβ+ intraepithelial lymphocytes provided by serial analysis of gene expression (SAGE) Immunity. 2001;15:419–34. doi: 10.1016/s1074-7613(01)00192-3. [DOI] [PubMed] [Google Scholar]
- 34.Symonds DA. Paneth cell metaplasia in diseases of the colon and rectum. Arch Pathol. 1974;97:343–7. [PubMed] [Google Scholar]
- 35.Lenardo MJ, Baltimore D. NF-κB: a pleiotropic mediator of inducible and tissue-specific gene control. Cell. 1989;58:227–9. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- 36.Tak PP, Firestein GS. NF-κB: a key role in inflammatory diseases. J Clin Invest. 2001;107:7–11. doi: 10.1172/JCI11830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naumann M. Nuclear factor-κ B activation and innate immune response in microbial pathogen infection. Biochem Pharmacol. 2000;60:1109–14. doi: 10.1016/s0006-2952(00)00390-7. [DOI] [PubMed] [Google Scholar]
- 38.Jobin C, Sartor RB. The I-κ B/NF-κ B system: a key determinant of mucosal inflammation and protection. Am J Physiol Cell Physiol. 2000;278:C451–62. doi: 10.1152/ajpcell.2000.278.3.C451. [DOI] [PubMed] [Google Scholar]
- 39.Cario E, Rosenberg IM, Brandwein SL, et al. Lipopolysaccharide activates distinct signaling pathways in intestinal epithelial cell lines expressing Toll-like receptors. J Immunol. 2000;164:966–72. doi: 10.4049/jimmunol.164.2.966. [DOI] [PubMed] [Google Scholar]
- 40.Fusunyan RD, Nanthakumar NN, Baldeon ME, et al. Evidence for an innate immune response in the immature human intestine: toll-like receptors on fetal enterocytes. Pediatr Res. 2001;49:589–93. doi: 10.1203/00006450-200104000-00023. [DOI] [PubMed] [Google Scholar]
- 41.Sierro F, Dubois B, Coste A, et al. Flagellin stimulation of intestinal epithelial cells triggers CCL20-mediated migration of dendritic cells. Proc Natl Acad Sci USA. 2001;98:13722–7. doi: 10.1073/pnas.241308598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cario E, Podolsky DK. Differential alteration in intestinal epithelial cell expression of toll-like receptor 3 (TLR3) and TLR4 in inflammatory bowel disease. Infect Immun. 2000;68:7010–7. doi: 10.1128/iai.68.12.7010-7017.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rhodes JM. Unifying hypothesis for inflammatory bowel disease and associated colon cancer: sticking the pieces together with sugar. Lancet. 1996;347:40–4. doi: 10.1016/s0140-6736(96)91563-9. [DOI] [PubMed] [Google Scholar]
- 44.Schultsz C, Van Den Berg FM, Ten Kate FW, et al. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology. 1999;117:1089–97. doi: 10.1016/s0016-5085(99)70393-8. [DOI] [PubMed] [Google Scholar]
- 45.Hammarström S, Baranov V. Is there a role for CEA in innate immunity in the colon? Trends Microbiol. 2001;9:119–25. doi: 10.1016/s0966-842x(01)01952-7. [DOI] [PubMed] [Google Scholar]
- 46.Lehrer RI, Lichtenstein AK, Ganz T. Defensins: antimicrobial and cytotoxic peptides of mammalian cells. Ann Rev Immunol. 1993;11:105–28. doi: 10.1146/annurev.iy.11.040193.000541. [DOI] [PubMed] [Google Scholar]
- 47.Merlin D, Yue G, Lencer WI, et al. Cryptdin-3 induces novel apical conductance (s) in Cl− secretory, including cystic fibrosis, epithelia. Am J Physiol Cell Physiol. 2001;280:C296–302. doi: 10.1152/ajpcell.2001.280.2.C296. [DOI] [PubMed] [Google Scholar]
- 48.Yeung MMW, Melgar S, Baranov V, et al. Characterisation of mucosal lymphoid aggregates in ulcerative colitis: immune cell phenotype and TcR-γδ expression. Gut. 2000;47:215–27. doi: 10.1136/gut.47.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]