Abstract
Combinatorial antibody libraries were constructed from the spleen of a patient with concomitant systemic lupus erythematosus and idiopathic thrombocytopenia. Following selection of the libraries with DNA, a panel of 15 anti-DNA Fabs was isolated. Sequence analysis of these antibodies coupled with measurements of their affinities for ss- and dsDNA were used to investigate the role of somatic mutation in affinity maturation of the anti-DNA response. Examination of the germline genes used by these Fabs supports previous studies that suggest there is no restriction of the gene usage in the anti-DNA response. However, data are presented indicating that VH3 genes and the A27 Vκ paired with the Jκ1 may be over-expressed in the anti-DNA repertoire. Analysis of the role of somatic mutation in increasing affinity for DNA indicates that affinity maturation has occurred and suggests that the CDR1 and CDR2 of the heavy chain are of importance in this process.
Keywords: human spleen lupus autoantibodies autoimmunity
INTRODUCTION
Systemic lupus erythematosus is characterized by high affinity antibodies to double-stranded DNA (dsDNA). The occurrence of such antibodies has been correlated with both disease flares and renal involvement [1–3]. However, the role of antigenic drive and the relative importance of somatic mutation in the production and pathogenicity of these high affinity antibodies are still unclear. Several studies have shown that human anti-DNA antibodies are somatically mutated, with a strong bias toward replacement mutations in the CDRs and increasing rates of mutation correlating with the switch from IgM to IgG [4–6]. This has been interpreted as evidence of affinity maturation [5,6]. However, increases in somatic mutation are not always apparent in such instances, e.g. Mannheimer-Lory and co-workers found no difference in the mutation rate of IgG and IgM anti-DNA antibodies [7]. The role of replacement mutations in increasing antibody affinity for DNA has also proved equivocal, with some studies indicating that mutations are important, whereas others find little correlation between affinity for dsDNA and mutation rate [8–11]. Certainly, it appears that some high-affinity anti-DNA antibodies can be encoded by genes that are essentially germ-line [8] and it is likely that the particular rearrangements of V, D and J segments in SLE patients determine the affinities of these antibodies [11]. The role of basic amino acids such as arginines and lysines in the CDR3s of such antibodies has been highlighted [8,9]. There are, however, other examples of anti-DNA antibodies where somatic mutations do appear to contribute to the affinity for dsDNA [10,11]. Thus, overall, it appears that there is evidence of somatic mutation, focused on the CDRs, in a proportion of anti-dsDNA antibodies from SLE patients. It is still not fully clear, however, if this is due to affinity maturation mediated by DNA or if the somatic mutations observed are incidental to affinity maturation in response to another antigen.
To resolve these issues, it is necessary to examine many individual anti-DNA antibodies and to try to correlate their affinity and specificity with germline gene usage and incidence of somatic mutation. The relative lack of human monoclonal anti-DNA antibodies (particularly of the IgG class) from SLE patients presents a problem here. Conventional techniques for generating human monoclonal antibodies tend to be inefficient and, in most cases, peripheral blood lymphocytes that contain relatively few IgG-producing B cells have been used [12]. An alternative is to generate human anti-DNA antibodies from SLE patients using repertoire cloning techniques, where DNA is used to select antibodies from a combinatorial library representing the heavy and light chain genes expressed by the patients' B cells. This method of sampling the human antibody response substantially increases the number of variable regions available for analysis [8,11].
This report seeks to add to the data on human IgG anti-DNA antibodies. We have used repertoire cloning techniques to construct a combinatorial library from the splenic lymphocytes of a patient with SLE and concomitant thrombocytopenia. By selecting against dsDNA, 15 IgG Fabs were isolated. We have analysed the sequences and affinities of these antibodies for ss- and dsDNA and in particular have examined the role of somatic mutation in increasing affinity for DNA.
MATERIALS AND METHODS
MRNA isolation and patient details
The spleen was taken from a 20-year-old-male with active SLE and concurrent idiopathic thrombocytopenia. Arthritis began at the age of five and he was found to be ANA positive. He was anti-Ro antibody positive at the age of 6. For the last 10 years frequent mouth ulcers were noted and for the last eight years he had had episodes of thrombocytopaenia. He had been treated with oral prednisolone and danazol. The thrombocytopaenia was usually steroid responsive although the platelet drop could be quite profound, reaching 9 × 109/l, for example. At the time of the splenectomy the platelet count was 322 × 109 per litre. His dsDNA antibody titre was 352 IU/ml. ANF was homogeneous at 1/320, and C3 and C4 levels were normal. He was also anti-Ro antibody positive but anti-La and anti-RNP antibody negative.
Total RNA was isolated from homogenized splenic tissue using an Ultraspec™ RNA isolation kit (Biotex Laboratories, Edmonton, Canada).
Library construction
Initial library construction used the pComB3HSS phage display vector (a gift from the Scripps Research Institute, La Jolla, USA) and was carried out as described previously [13–17]. After reverse transcription of 5 µg of RNA (SuperscriptII™, Gibco, Paisley, UK) the γ1 Fd region and κ light chains were amplified by PCR using C region and a previously described panel of VH and Vκ first framework specific primers [17,18] and additional VH and Vκ primers described in the Cold Spring Harbour Laboratory Course Manual, namely: VH2f, (5′-CAGGTGCAGCTACTCGAGTCG GG-3′); VH4f, (5′-CAGGTGCAGCTGCTCGAGTCGGG-3′); VH6a, (5′-CAGGTACAGCTCGAGCAGTCAGG-3′); VH6f, (5′-CAGGTACAGCTGCTCGAGTCAGGTCCA-3′); Vκ1s, (5′-GA CATCGAGCTCACCCAGTCTCCA-3′); Vκ2a, (5′-GATATTGA GCTCACTCAGTCTCCA-3′); Vκ3b, (5′-GAAATTGAGCTCA CG(G/A)CAGTCTCCA-3′). The heavy chain Fd and light chain PCR products were gel-purified, extracted (Wizard® PCR Preps DNA Purification System, Promega, Southampton, UK) and reamplified using extension primers with a 5′-poly (GA) tail to increase restriction enzyme digestion and cloning efficiency [16]. The light- and heavy chain PCR fragments were cut with SacI/XbaI and SpeI/XhoI, respectively (GibcoBRL, Paisley, UK) and ligated sequentially into pre-cut pComB3HSS vector [15,17]. After electroporation into Escherichia coli, XL-1Blue a light chain library of 3·3 × 107 independent clones was generated, followed by a combinatorial (Fd × κ) library of 1 × 107 independent primary clones.
Library panning for dsDNA binding phage
The library was panned against biotinylated plasmid dsDNA [19]. Briefly, 8-well ELISA strips (Maxisorb, Nunc, Roskilde, Denmark) coated with streptavidin were incubated overnight at 4°C with biotinylated dsDNA (0·1 mg/ml), washed with distilled water and blocked with 3% (w/v) BSA in PBS. Successive rounds of panning using freshly prepared phage were then performed as described [15] using increased washing stringency (1 ×, 3 ×, 6 ×, 10 ×) with TBS 0·05% Tween 20 to remove unbound phage at each stage.
Production of soluble Fabs
Soluble Fab fragments were generated using DNA isolated from a number of independent clones after the final round of panning, as previously described [15]. Bacterial lysates/supernates were then analysed for the presence of anti-dsDNA Fabs by ELISA. Additionally, two complementary oligomers coding for hexa-histidine (His) tag (5′-CTAGCCACCATCATCACCACCATT-3′; 5′-CTAGAATGGTGGTGATGATGGTGG-3′) were ligated into the COOH-terminal of the light-chain DNA, via the XbaI site, to allow purification of the fragments on a nickel agarose affinity column (Qiagen, Crawley, UK).
Enzyme-linked immunosorbent assay (ELISA)
Soluble Fabs were screened for dsDNA binding by ELISA. Microtitre wells were coated with dsDNA as described above, blocked with TBS containing 3% bovine serum albumin (BSA) for 1 h at 37°C and incubated with bacterial lysate/purified Fab for 1 h at 37°C. After three washes with TBS 0·05% Tween 20, wells were incubated at 37°C for 1 h with Fab-specific, HRP-labelled goat anti-human IgG (Sigma, Poole, UK) diluted 1 : 1000 in TBS-1% BSA. After washing as above, plates were developed with ABTS substrate tablets (Sigma) according to the manufacturer's instructions.
Determination of binding kinetics by surface plasmon resonance
Purified anti-dsDNA Fabs were analysed on the BIAcore system (BIAcore 2000, Biacore, Uppsala, Sweden). Biotinylated pure double-stranded plasmid DNA or single-stranded random oligonucleotide DNA was coupled to a streptavidin-coated sensor chip, yielding a change in response units (RU) of 1935. All determinations were performed at 25°C in 10 mm HEPES, 150 mm NaCl, 2 mm CaCl2, 0·05% BIAcore surfactant P20, pH 7·4 (HBS-M), with a constant flow rate of 5 µl/min and an injected sample volume of 30 µl for all Fabs at a concentration of 10−7 m. The calculation of the off- and on-rates was performed with the BIAevaluation Program 2·0 (Pharmacia Biosensor, Uppsala, Sweden). Biacore was used to measure affinity rather than ELISA as this provides more detailed information to compare the relative affinities of the antibodies isolated.
Fab sequence determination
Nucleic acid sequencing of selected clones was carried out on a 373-A automated DNA sequencer (Applied Biosystems, Warrington, UK) using a Taq fluorescent dideoxy terminator cycle sequencing kit (Applied Biosystems), as described previously [17]. The V, J and D gene usage was determined using the VBASE program [20].
Statistics
Comparisons between group means were carried out using the student's t-test. Correlation was carried out by Pearson Correlation coefficients.
RESULTS
Variable region gene usage
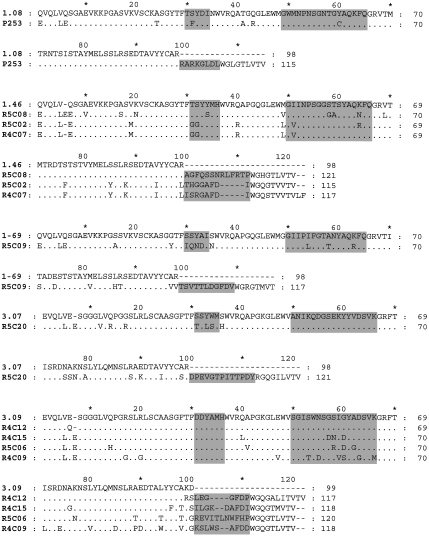
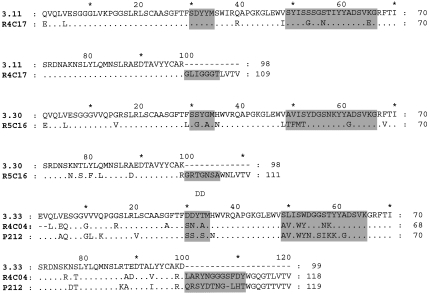
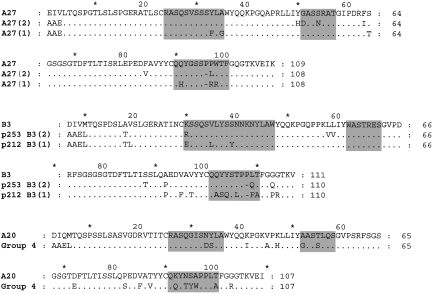
After four rounds of panning, DNA from selected phage was sequenced and used to generate soluble Fab fragments. Fabs giving OD405 readings of> 0·5 against dsDNA as bacterial lysates/supernates in ELISA were selected for further analysis and purification. Table 1 shows the variable region genes used by 15 anti-DNA Fabs. Figures 1 and 2 show the deduced amino acid sequences compared with those of the appropriate germline gene. Five of the nine VH, and two of the three VL, germline genes used have previously been described in association with anti-DNA antibodies [21]. J and D region germline genes have been assigned, although in many cases the CDR3 of the heavy chain appears to have arisen by a complex process and the accurate prediction of the germline genes used was not always possible.
Table 1.
Variable region genes used in 15 anti-DNA antibodies and the combined R/S ratio for all CDRS except CDR3 of the heavy chain. The variable region genes marked in italics have previously been described in IgG anti-DNA antibodies [12] Variable region genes were assigned using VBASE [21] in the case of VH, VL, and JL. D region genes and JH genes were assigned by a combination of VBASE and manual comparison
| Antibody code | VH family | VH Locus | D Locus | JH Locus | VL Locus* | JL Locus | Total R/S ratio for the CDRS of the heavy and light chain combined |
|---|---|---|---|---|---|---|---|
| R5C08 | 1 | 1–46 | D3-22 | JH5B | A20 | Jκ4 | 2·0 |
| R4C17 | 3 | 3–11 | D-5–24 | JH4A | A27(2) | Jκ1 | 6·0 |
| R4C09 | 3 | 3–9 | D4-11/4/17 | JH5B | A27(1) | Jκ1 | 3·5 |
| R4C12 | 3 | 3–9 | D6-? | JH5B | A27(1) | Jκ1 | 2·0 |
| R5C02 | 1 | 1–46 | D3-22 | JH3B | A27(1) | Jκ1 | 1·4 |
| P253 | 1 | 1–8 | D1-1/D1-20 | JH5B | B3(2) | Jκ4 | 7·0 |
| R5C20 | 3 | 3–7 | A20 | Jκ4 | 2·7 | ||
| R4C07 | 1 | 1–46 | D3-22 | JH3B | A27(1) | Jκ1 | 1·5 |
| P212 | 3 | 3–33 | D3-3 | JH3B | B3(1) | Jκ4 | 4·3 |
| R4C15 | 3 | 3–9 | D7-27 | JH3B | A27(1) | Jκ1 | 2·3 |
| R4C03 | 3 | 3–48 | D3-16 | JH4B | A20 | Jκ4 | 2·4 |
| R5C06 | 3 | 3–9 | D4-23 | JH5B | A27(2) | Jκ1 | 4·3 |
| R4C04 | 3 | 3–33 | D3-3 | JH4A | A27(2) | Jκ1 | 4·0 |
| R5C16 | 3 | 3–30 | D7-27 | JH6a/b/c | A20 | Jκ4 | 3·8 |
| R5C09 | 1 | 1–69 | D4-17 | JH3A | A20 | Jκ4 | 4·2 |
| Mean | 3·4 |
Numbers in parenthesis indicate the light chain somatic variant used.
Fig. 1.
A comparison of the deduced VDJ region amino acid sequences of the heavy chains of 15 anti-DNA antibodies with those of the corresponding VH germline genes. The CDRs are highlighted. The DNA sequences have been submitted to EMBL and are listed under the following accession numbers: R4C12 AJ306669, R4C15 AJ306670, R4C17 AJ306671, R4C03 AJ306672, R4C04 AJ306673, R4C07 AJ306674, R4C09 AJ306675, R5C20 AJ306676, R5C02 AJ306677, R5C06 AJ306678, R5C08 AJ306679, R5C09 AJ306680, R5C16 AJ307663, P212 AJ306681, P253 AJ306682.
Fig. 2.
A comparison of the deduced VJ region amino acid sequences of the light chains used by 15 anti-DNA antibodies with those of the corresponding VLVJ germline genes. The CDRs are highlighted. The DNA sequences have been submitted to EMBL and are listed under the following accession numbers: A27(1) AJ307664, A27(2) AJ307665, B3(1) AJ307666, B3(2) AJ307667, Group 4 AJ307668.
Replacement to silent ratio and mutation rates
The mean replacement to silent ratio (R/S) for all the CDRs combined [Table 1], the CDRs 1 and 2 of the heavy chain combined [Table 2] and the CDRs 1, 2 and 3 of the light chain combined [Table 3] were greater than 3. The mean total mutation rate for the heavy chain was 7·58% (range 2·16–17·56: median 7·41) and the mean percentage of the heavy chain mutations that were transitions was 49·89% (range 28·1–77·8%: median 45·5%). The mean total mutation rate for the light chain was 8·68% (range 3·35–22·15: median 4·37) and the mean percentage of the light chain mutations that were transitions was 49·4% (range 27·3–81·8%: median 47·1%).
Table 2.
The replacement to silent mutation ratios of the heavy chains for the 15 anti-DNA Fabs. Figures in italics are those for which there are no silent mutations
| Antibody code | R/S ratio in CDR 1 and 2 | R/S ratio in CDR 1 | R/S ratio in CDR 2 | R/S ratio in the framework region | Transition mutation rate |
|---|---|---|---|---|---|
| R5C08 | 1·3 | 1·0 | 1·5 | 2·5 | 42·9 |
| R4C17 | 3·0 | 0·0 | 3·0 | 3 | 72·7 |
| R4C09 | 3·0 | 0·0 | 3·0 | 0·7 | 35·7 |
| R4C12 | 0·0 | 0·0 | 0·0 | 2·5 | 77·8 |
| R5C02 | 1·0 | 1·0 | 0·7 | 4·5 | 66·7 |
| P253 | 3·0 | 2·0 | 1·0 | 5 | 45·5 |
| R5C20 | 2·5 | 2·5 | 0·0 | 2 | 38·5 |
| R4C07 | 1·0 | 1·0 | 0·5 | 3·6 | 65·5 |
| P212 | 3·0 | 2·0 | 1·0 | 3 | 47·8 |
| R4C15 | 3·0 | 0·0 | 3·0 | 3 | 53·3 |
| R4C03 | 3·0 | 4·0 | 2·0 | 3 | 37·3 |
| R5C06 | 5·0 | 0·0 | 5·0 | 8 | 57·9 |
| R4C04 | 7·0 | 2·0 | 5·0 | 2·75 | 33·3 |
| R5C16 | 8·0 | 4·0 | 4·0 | 3·75 | 30·8 |
| R5C09 | 9·0 | 6·0 | 3·0 | 1·625 | 28·1 |
| Mean | 3·4 | 1·7* | 2·2 | 3·2 | 48·9 |
Probability value, P > 0·003 when compared with combined R/S ratio for CDR1 and 2; not significant when compared with the R/S ratio for CDR2.
Table 3.
The replacement to silent mutation ratios and homologies with germline genes of the light chains for the 15 anti-DNA Fabs. Figures in italics are those for which there are no silent mutations
| VL | Jκ | Combined R/S ratio for CDRs 1, 2 and 3 | R/S ratio for CDR1 | R/S ratio CRD2 | R/S ratio CRD3 | R/S ratio in the framework region | Transition mutation rate |
|---|---|---|---|---|---|---|---|
| A27(1)1 | Jκ1 | 2 | 1 | 0 | 3 | 0·5 | 27·3 |
| A27(2)2 | Jκ1 | 3 | 0 | 2 | 1 | 2 | 81·8 |
| A20 | Jκ4 | 2·75 | 3 | 2 | 2 | 1·7 | 50·0 |
| B3(1)3 | Jκ4 | 3·5 | 3 | 0 | 3·67 | 3·8 | 41·0 |
| B3(2)4 | Jκ4 | 4 | 2 | 0 | 2 | 4·0 | 47·1 |
| Mean | 3·05 | 1·8 | 0·8* | 2·33 | 2·4 | 49·4 |
Probability value, P > 0·005 when compared with total ratio of the CDRs and> 0·03 when compared with the CDR3, but not significant when compared with the CDR1·
Somatic variant of A27/Jκ1 light chain used by antibodies R4C07, R4C09, R4C12, R4C15 and R5C02.
Somatic variant of A27/Jκ1 light chain used by antibodies R4C04, R4C17 and R5C06.
Somatic variant of B3/Jκ4 light chain used by antibody P212.
Somatic variant of B3/Jκ4 light chain used by antibody P253. The sequence differences between these light chains are described in Fig. 2.
The R/S ratios for each CDR of the heavy chain for each antibody and their means are shown in Table 2 and for both CDR1 and CDR2 the mean was below 3. Table 4 shows the distribution of replacement to silent ratios for the heavy chains categorized according to the type of light chain used. Clear differences are seen between the groups, in that for antibodies that use A27 [1] light chain there is no evidence of increased R/S ratios in either CDR1 and 2 of the heavy chain. This contrasts with antibodies that use A27 [2] light chain which show a marked focus of mutations in the CDR2 of the heavy chain compared with the CDR1 of the heavy chain. This pattern is also reflected in those antibodies that use B3, although the R/S ratio for the CDR2 of the heavy chain is not as high. In those antibodies that use A20 light chain, high R/S ratios are seen in both CDR 1 and 2 of the heavy chain. The R/S ratio for each CDR of the light chain for each antibody and their means are shown in Table 3 and for CDR1, 2 and 3 the mean was below 3.
Table 4.
A comparison of the mean replacement to silent ratios of the heavy chains paired with specific light chains. Antibodies using the different somatic variants of the A27/Jκ1 light chain are treated as two separate groups. As only two antibodies used the somatic variants of the B3/Jκ4 light chain, these they are treated together. Mean ± s.e.m.
| Number of antibodies | R/S ratio for heavy chain CDRs 1 and 2 | R/S ratio for CDR1 | R/S ratio for CDR2 | |
|---|---|---|---|---|
| A27 (1) | 6 | 2·83 ± 0·27 | 1·33 ± 0·19 | 1·69 ± 0·12 |
| A27 (2) | 3 | ″ 5 ± 0·33 | 0·67 ± 0·19 | 4·33 ± 0·19 |
| B3 | 2 | 2·97 ± 0* | 0·53 ± 0* | 2·88 ± 0* |
| A20 | 4 | 4·55 ± 0·37 | 3·50 ± 0·19 | 2·10 ± 0·15 |
The numbers of mutations in both heavy chains was identical. For an explanation of A27 (1) and A27 (2) see the legend to Table 3.
The effect of replacement to silent ratio on antibody affinity
The Ka and Kd for the binding of all antibodies to ssDNA and dsDNA is shown in Table 5. Significant correlations were found between the combined R/S ratio for the CDR1 and 2 of the heavy chain and the Ka values for ssDNA and dsDNA (correlation coefficients = 0·8, P = 0·0003 and 0·86, P = 0·0001, respectively) and the Kd against ssDNA (correlation coefficient = −0·71 P = 0·003). By contrast, there was no significant correlation between the R/S ratio of the framework regions, when compared with Ka (correlation coefficients of − 0·03 for ssDNA and 0·04 for dsDNA) or Kd (correlation coefficients of − 0·18 for ssDNA and 0·18 for dsDNA). This strong correlation between affinity and somatic mutations is due to increased numbers of replacement mutations rather than decreasing numbers of silent mutations in the combined CDR1 and 2 of the heavy chain (correlation coefficients of 0·63 P = 0·01 for ssDNA and 0·74 P = 0·002 for dsDNA when compared for ka, and − 0·73 P = 0·002 for ssDNA when compared for kDa) (data not shown). No such correlation was seen when the light chain was studied.
Table 5.
The affinity constants Kaand Kd rates for 15 anti-DNA antibodies as determined by BIAcore analysis
| Antibody code | ssKa × 108 | ssKd × 10−7 | dsKa × 108 | dsKd × 10−7 |
|---|---|---|---|---|
| R5C08 | 0·30 | 0·34 | 0·05 | 2·16 |
| R4C17 | 0·56 | 0·18 | 0·08 | 1·18 |
| R4C09 | 0·16 | 0·64 | 0·17 | 0·60 |
| R4C12 | 0·10 | 1·04 | 0·19 | 0·54 |
| R5C02 | 0·16 | 0·62 | 0·19 | 0·53 |
| P253 | 0·19 | 0·52 | 0·23 | 0·44 |
| R5C20 | 0·15 | 0·68 | 0·23 | 0·43 |
| R4C07 | 0·19 | 0·52 | 0·26 | 0·39 |
| P212 | 0·18 | 0·56 | 0·27 | 0·38 |
| R4C15 | 0·19 | 0·53 | 0·27 | 0·37 |
| R4C03 | 0·25 | 0·39 | 0·41 | 0·24 |
| R5C06 | 0·34 | 0·29 | 0·42 | 0·24 |
| R4C04 | 0·28 | 0·36 | 0·48 | 0·21 |
| R5C16 | 0·58 | 0·17 | 0·62 | 0·16 |
| R5C09 | 0·81 | 0·12 | 0·83 | 0·12 |
DISCUSSION
In this study, we have used repertoire cloning to select 15 anti-DNA IgG antibodies from the splenic lymphocytes of a patient with SLE. It is well documented that these techniques can be used to isolate high affinity antibodies with the same antigen binding specificity as the donor serum [16,22]. Nevertheless, a persistent criticism of these methods is that, as the heavy and light chain genes are randomly combined, the pairings observed in the antibodies selected are not necessarily those that occur in vivo. Whilst this is acknowledged, it should be noted that chain-shuffling experiments on antibodies isolated from libraries indicate that the heavy and light chain pairings required to generate anti-dsDNA specificities are relatively non-promiscuous [11,23]. The technique provides an efficient means of generating human antibodies and, coupled with an analysis of their binding properties, provides valuable information on the elements of the antibody response that may be used in the expression of IgG anti-DNA antibodies.
Analyses of variable gene use have, in this and other studies [21], failed to find any preference in the germline genes used. However there is a preponderance of VH3 genes in the IgG anti-DNA Fabs described here and by Rahman et al. [21] (64% of 42 antibodies and Fabs), compared with current estimates for all used antibody genes (approximately 40% of 1200) [24]. The number of human IgG anti-DNA antibodies that have been analysed, however, is still relatively small and clearly the nature of the immunoglobulin repertoire in SLE patients and its difference from normal individuals requires further study.
From the studies summarized in [21] and those reported here, a total of eight light chains that use the Vκ gene A27 have now been described in IgG anti-DNA antibodies/Fabs. Of the seven in which the J region can be attributed, six are paired with Jκ1. Studies on the antibody repertoires of normal and SLE patients indicate that Jκ1 occurs in 20–30% of all antibodies [25,26]. This would suggest a preferential pairing of A27 and Jκ1 in anti-DNA antibodies, an observation that requires further investigation.
Although it has been suggested that antigen driven somatic mutation is important in the development of human anti-DNA antibodies, very few studies have been able to correlate the accumulation of mutations with changes in affinity (summarized in [5,6]). Studies that use site-directed mutagenesis to revert anti-DNA antibodies to their germline equivalents show contradictory data with regard to a role for somatic mutation in determining affinity [9,10] Here, the replacement to silent ratios of greater than 3 observed in the combined CDRs of 10 of the 15 heavy chains and three of the light chains strongly indicate that an antigen driven process is occurring. This is further supported by the mean purine to pyrimidine transition rate of 48·9% for the heavy chain and 49·4% for the light chain even though somatic hypermutation is inherently biased in favour of purine to pyrimidine transitions, with a rate of 66% expected by random [27]. Taken together, these findings imply that affinity maturation in response to antigen has occurred in the development of these antibodies. It has been shown that, in antibodies generally, the CDR1 of the heavy chain has a higher inherent replacement to silent ratio than CDR2 [28]. Here, however, the mean replacement to silent mutation ratio in the CDR1 of eight of the 15 H chains is lower than in CDR2 [Table 2]. It is possible that antigen selection determines the lower replacement to silent ratio in the CDR1 of the heavy chain, although further studies are needed.
Analysis of affinity measurements shows a strong correlation between the R/S ratio in the CDRs 1 and 2 of the heavy chain and Ka for both ss- and dsDNA and for Kd for ssDNA. This contrasts with a lack of correlation when the R/S ratio of the heavy chain framework regions are compared with both Ka and Kd. Whilst it is unlikely that all of these mutated residues are involved in DNA binding, the increasing level of mutation in the heavy chain CDRs with increasing affinity of the antibodies is quite striking. However, as only two of the antibodies (R5C20 and R4C07) appear to be clonally related, it is difficult in this study to determine the contribution of the CDR3 of the heavy chain, or of individual amino acids to affinity. A difference between the related antibodies is the presence of an arginine in place of a glycine in the CDR3 of the heavy chain of the slightly higher affinity R4C07 [Fig. 1]. Arginine and lysine have been reported to be important in determining the affinity of anti-DNA antibodies [8,9]. In general, however, whilst this panel of antibodies contains several basic amino acids there appears to be no direct relationship between affinity and the number of basic amino acids. Clearly, future site-directed mutagenesis studies will be important in determining the role of individual amino acids in DNA specificity.
When heavy chains are classified according to the light chain used, then it appears that somatic mutation may be more important in some groups than others. Notably there is a much higher R/S ratio in the heavy chain of antibodies that use the A27 [2] light chain when compared with those that use A27 [1], suggesting that the light chain plays a more prominent role in the latter group. This pattern suggests that mutations in each CDR can play a significant role in determining the specificity and affinity of anti-DNA antibodies, depending on their context within the structure of the combining site. Modelling of the Fab:DNA interaction and 3-dimensional structural studies on the Fabs would obviously provide further information here.
In conclusion the germline genes used by this panel of antibodies show characteristics consistent with other human anti-DNA antibodies and studies on variable gene usage in patients with SLE. Evidence is also provided that the high replacement to silent ratio observed here and by others is significantly correlated with increasing affinity. However, the role of individual mutations in determining affinity for DNA must be verified, e.g. by structural studies or mutagenesis. The correlation between mutation and increasing Ka for dsDNA indicates that affinity maturation of the response to dsDNA has occurred. This panel of anti-bodies provides an excellent resource to investigate antigen: antibody interactions in anti-DNA antibodies. The libraries will also provide a resource to study the antibody response to other autoantigens, such as platelets; these studies are currently in progress.
Acknowledgments
This project was supported by a grant from the Arthritis Research Campaign.
References
- 1.Ehrenstein M, Longhurst C, Isenberg DA. Production and analysis of IgG monoclonal anti-DNA antibodies from systemic lupus erythematosus (SLE) patients. Clin Exp Immunol. 1993;92:39–45. doi: 10.1111/j.1365-2249.1993.tb05945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.ter Borg EJ, Horst G, Hummel EJ, Limburg PC, Kallenberg CG. Measurement of increases in anti-double-stranded DNA antibody levels as a predictor of disease exacerbation in systemic lupus erythematosus. A long-term, prospective study. Arthritis Rheum. 1990;33:634–43. doi: 10.1002/art.1780330505. [DOI] [PubMed] [Google Scholar]
- 3.Swaak AJ, Aarden LA, Statius van Eps LW, Feltkamp TE. Anti-dsDNA and complement profiles as prognostic guides in systemic lupus erythematosus. Arthritis Rheum. 1979;22:226–35. doi: 10.1002/art.1780220304. [DOI] [PubMed] [Google Scholar]
- 4.Winkler TH, Fehr H, Kalden JR. Analysis of immunoglobulin variable region genes from human IgG anti-DNA hybridomas. Eur J Immunol. 1992;22:1719–28. doi: 10.1002/eji.1830220709. [DOI] [PubMed] [Google Scholar]
- 5.Isenberg DA, Ehrenstein MR, Longhurst C, Kalsi JK. The origin, sequence, structure, and consequences of developing anti-DNA antibodies. A human perspective. Arthritis Rheum. 1994;37:169–80. doi: 10.1002/art.1780370204. [DOI] [PubMed] [Google Scholar]
- 6.Isenberg DA, Ravirajan CT, Rahman A, Kalsi J. The role of antibodies to DNA in systemic lupus erythematosus – a review and introduction to an international workshop on DNA antibodies held in London, May 1996. Lupus. 1997;6:290–304. doi: 10.1177/096120339700600316. [DOI] [PubMed] [Google Scholar]
- 7.Manheimer-Lory A, Katz JB, Pillinger M, Ghossein C, Smith A, Diamond B. Molecular characteristics of antibodies bearing an anti-DNA-associated idiotype. J Exp Med. 1991;174:1639–52. doi: 10.1084/jem.174.6.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbas SM, Ditzel HJ, Salonen EM, Yang WP, Silverman GJ, Burton DR. Human autoantibody recognition of DNA. Proc Natl Acad Sci USA. 1995;92:2529–33. doi: 10.1073/pnas.92.7.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, Schettino EW, Padlan EA, Ikematsu H, Casali P. Structure-function analysis of a lupus anti-DNA autoantibody: central role heavy chain complementarity-determining region 3 Arg binding double- and single-stranded DNA. Eur J Immunol. 2000;30:2015–26. doi: 10.1002/1521-4141(200007)30:7<2015::AID-IMMU2015>3.0.CO;2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahman A, Haley J, Radway-Bright E, Nagl S, Low DG, Latchman DS, Isenberg DA. The importance somatic mutations V (lambda) gene 2a2 human monoclonal anti-DNA antibodies. J Mol Biol. 2001;307:149–60. doi: 10.1006/jmbi.2000.4491. [DOI] [PubMed] [Google Scholar]
- 11.Roben P, Barbas SM, Sandoval L, Lecerf JM, Stollar BD, Solomon A, Silverman GJ. Repertoire cloning of lupus anti-DNA autoantibodies. J Clin Invest. 1996;98:2827–37. doi: 10.1172/JCI119111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watts RA, Winska-Wiloch H, Muller S, Isenberg DA. Analysis of factors affecting human hybridoma production. Hum Antibodies Hybridomas. 1990;1:160–5. [PubMed] [Google Scholar]
- 13.Barbas CF, Lerner RA. Combinatorial immunoglobulin libraries on the surface of phage (Phabs): rapid selection of antigen-specific Fabs. Methods: a Companion to Meth Enzymol. 1991;2:119–24. [Google Scholar]
- 14.Persson MA, Caothien RH, Burton DR. Generation of diverse high-affinity human monoclonal antibodies by repertoire cloning. Proc Natl Acad Sci USA. 1991;88:2432–6. doi: 10.1073/pnas.88.6.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burton DR, Barbas CF, 3rd, Persson MA, Koenig S, Chanock RM, Lerner RA. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc Natl Acad Sci USA. 1991;88:10134–7. doi: 10.1073/pnas.88.22.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williamson RA, Burioni R, Sanna PP, Partridge LJ, Barbas CF, 3rd, Burton DR. Human monoclonal antibodies against a plethora of viral pathogens from single combinatorial libraries. Proc Natl Acad Sci USA. 1993;90:4141–5. doi: 10.1073/pnas.90.9.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clayton R, Cooke ID, Partridge LJ, Moore HD. A combinatorial phage display library for the generation of specific Fab fragments recognizing human spermatozoa and inhibiting fertilizing capacity in vitro. Biol Reprod. 1998;59:1180–6. doi: 10.1095/biolreprod59.5.1180. [DOI] [PubMed] [Google Scholar]
- 18.Silverman GJ, Roben P, Bouvet JP, Sasano M. Superantigen properties of a human sialoprotein involved in gut-associated immunity. J Clin Invest. 1995;96:417–26. doi: 10.1172/JCI118051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emlen W, Jarusiripipat P, Burdick G. A new ELISA for the detection of double-stranded DNA antibodies. J Immunol Meth. 1990;132:91–101. doi: 10.1016/0022-1759(90)90402-h. [DOI] [PubMed] [Google Scholar]
- 20.Cook GP, Tomlinson IM. The human immunoglobulin VH repertoire. Immunol Today. 1995;16:237–42. doi: 10.1016/0167-5699(95)80166-9. [DOI] [PubMed] [Google Scholar]
- 21.Rahman A, Latchman DS, Isenberg DA. Immunoglobulin variable region sequences of human monoclonal anti-DNA antibodies. Semin Arthritis Rheum. 1998;28:141–54. doi: 10.1016/s0049-0172(98)80031-0. [DOI] [PubMed] [Google Scholar]
- 22.Hexham JM, Partridge LJ, Furmaniak J, Petersen VB, Colls JC, Pegg C, Rees Smith B, Burton DR. Cloning and characterisation of TPO autoantibodies using combinatorial phage display libraries. Autoimmunity. 1994;17:167–79. doi: 10.3109/08916939409010651. [DOI] [PubMed] [Google Scholar]
- 23.Calcutt MJ, Kremer MT, Giblin MF, Quinn TP, Deutscher SL. Isolation and characterization of nucleic acid-binding antibody fragments from autoimmune mice-derived bacteriophage display libraries. Gene. 1993;137:77–83. doi: 10.1016/0378-1119(93)90254-z. [DOI] [PubMed] [Google Scholar]
- 24.Knappik A, Ge L, Honegger A, Pack P, Fischer M, Wellnhofer G, Hoess A, Wolle J, et al. Fully synthetic human combinatorial antibody libraries (HuCAL) based on modular consensus frameworks and Cdrs randomized with trinucleotides. J Mol Biol. 2000;296:57–86. doi: 10.1006/jmbi.1999.3444. [DOI] [PubMed] [Google Scholar]
- 25.Foster SJ, Brezinschek HP, Brezinschek RI, Lipsky PE. Molecular mechanisms and selective influences that shape the kappa gene repertoire of Igm+ B cells. J Clin Invest. 1997;99:1614–27. doi: 10.1172/JCI119324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorner T, Foster SJ, Farner NL, Lipsky PE. Immunoglobulin kappa chain receptor editing in systemic lupus erythematosus. J Clin Invest. 1998;102:688–94. doi: 10.1172/JCI3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuberger MS, Milstein C. Somatic hypermutation. Curr Opin Immunol. 1995;7:248–54. doi: 10.1016/0952-7915(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 28.Chang B, Casali P. The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol Today. 1994;15:367–73. doi: 10.1016/0167-5699(94)90175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]