Abstract
X-linked hyper-IgM syndrome (HIGM1) (MIM#308230), is a severe primary immunodeficiency caused by mutations in the gene coding for CD40 ligand (CD40L or CD154), a member of the tumour necrosis factor (TNF) superfamily. The interaction of this protein with its ligand, CD40, mediates crucial processes in the immune response. The variety of defects that have been described in HIGM1 patients range from a complete lack of CD40L protein expression to missense mutations that interfere with its interaction with CD40L. In this study we describe three families – a total of seven HIGM1 patients and carriers, presenting a spectrum of severity in clinical evolution. In two of these families, patient DNA samples were available for genetic studies. In the third, carrier detection was performed on female family members. The results of immunological studies – the different patterns of CD40L expression and binding capacity as measured by flow cytometry – and molecular diagnosis are presented. Three novel mutations were identified: an intron mutation that partially interferes with the splicing process (intron 3, position + 5 G/T); a missense mutation (Ser222 Phe) located in the molecular region which interacts with the receptor and which abrogates binding capacity; and a 14 base pair deletion leading to a frameshift and a premature truncated mutation (del I 171 X 195). An attempt to correlate protein expression and function of the CD40L mutants with clinical disease evolution is described.
Keywords: CD40 ligand, molecular diagnosis, receptor–ligand interaction, X-linked hyper-IgM syndrome
INTRODUCTION
The X-linked hyper-IgM syndrome (HIGM1) is a primary immunodeficiency characterized by recurrent infections, associated with very low or absent IgG, IgA and IgE levels but normal or elevated serum IgM levels [1,2].
This syndrome is caused by mutations in the gene coding for CD40 ligand (CD40L or CD154) [3–5]. The genomic sequence for this molecule spans over 13 kb of the Xq26.3–q27.1 region and is composed of five coding exons and four introns. CD40L is a type II transmembrane protein expressed mainly by activated CD4+ T cells [6,7] and is a member of the tumour necrosis factor (TNF) superfamily of cytokines. It is composed of 261 amino acids and three functional domains: the intracellular (22 amino acids), the transmembrane (24 amino acids) and the extracellular (215 amino acids) domain. The fourth and fifth exons encode for the C-terminal region, named TNF homology (TNFH), which characterized this family of proteins. In addition to the cell-associated full-length 39 kDa protein, a soluble form of the ligand, able to associate in heterotrimeric complexes with the full-length variant, has been described. As suggested first by structural model predictions and confirmed later by the X-ray crystal structure description, CD40L adopts the typical three-dimensional structure of the TNF family, a sandwich of two β sheets with a jelly-roll or Greek key topology, and it binds its receptor as a symmetric homotrimer with three binding sites, each one created by the interaction of two monomers [8–10].
Its natural receptor, CD40, is a member of the TNF receptor superfamily expressed on a variety of cells including B cells, macrophages/monocytes, dendritic cells, vascular endothelial cells and epithelial cells [11]. The CD40–CD40L interaction plays an important role in B-cell development, growth and differentiation, and it rescues B cells from apoptosis at some stages of differentiation [4]. This interaction is also essential for isotype switching and memory B-cell generation [12].
Patients with HIGM1 have recurrent bacterial and opportunistic infections, starting in the first year of life. Pneumocystis carinii pneumonia is a common presenting infection. These patients may develop chronic hepatitis (progressing frequently to cirrhosis), sclerosing cholangitis, and display a susceptibility to liver and biliary tract tumours (rarely observed in other primary immunodeficiencies) [13]. Chronic diarrhoea is often associated with Cryptosporidium infection, which may also contribute to the high frequency of sclerosing cholangitis [13,14]. Parvovirus-induced aplastic anaemia is another characteristic manifestation in some cases [15]. Neutropenia is observed in about 50% of the patients, and is either chronic or cyclic and often associated with oral ulcers. Anaemia, thrombocytopenia, neurological alterations and autoimmune manifestations have also been documented in HIGM1 [14].
Mutations in the activation-induced cytidine deaminase (AID) gene involved in the isotype switching process [16,17] and in the CD40 gene [18] have been identified as causing autosomal recessive forms of hyper-IgM (named HIGM2 and HIGM3, MIM#605258 and MIM#606843, respectively). It has been reported that mutations in the NF-κB essential modifier (NEMO) result in X-linked hyper-IgM syndrome with hypohydrotic ectodermal dysplasia (XHM-ED MIM#300291) [19,20].
In this study we describe three unrelated HIGM1 families, analyse the CD40L expression and the binding capacity of mutant proteins by flow cytometry, characterize the mutation of the CD40L gene in each family and attempt to correlate genotype with phenotype.
MATERIALS AND METHODS
Patients
Family 1
The first patient from this family, born in 1972, was studied at age 3 years due to a history of recurrent otitis and bronchitis, two incidences of pneumonia and frequent diarrhoea since age 6 months; their brother died of bronchopneumonia at age 4 months. The immunological evaluation of the patient revealed an IgG level of 45 mg/dl, an IgA level of less than 5 mg/dl and an IgM level of 350 mg/dl. T and B cell phenotype and lymphocyte proliferation in response to mitogens were normal. The patient was diagnosed with HIGM1 and began receiving intramuscular immunoglobulin (IMIG) at age 3 years. At age 12 years he started on intravenous immunoglobulin (IVIG) replacement (400 mg/kg every 3 weeks). He had frequent Herpes simplex infections and several incidences of pneumonia, developing bronchiectasis. Several times he developed very intense neutropenia (5–10 neutrophils/µl). At age 22 he presented with chronic hepatitis caused by HCV infection and was hospitalized with diarrhoea caused by Salmonella B9. One year later he was diagnosed with leishmaniasis. At age 25 he was again hospitalized with generalized oedemas, hepatic insufficiency and leishmaniasis, and he died of massive haemorrhage.
The second patient, born in 1975, was evaluated at age 4 months because the family's medical history was suggestive of immunodeficiency. He had suffered minor infections. The levels of serum IgG, IgA and IgM were 54, 9·5 and 250 mg/dl, respectively. T and B cell phenotype and lymphocyte proliferation were normal. At age 8 months he was diagnosed with HIGM1 and treatment with IMIG was initiated. At age 6 years he had two incidences of pneumonia and 1 year later, due to intestinal tuberculosis, he underwent an intestinal resection. At age 9 years, he started on IVIG at a dosage of 300 mg/kg every 3 weeks. Since then, he has had a history of recurrent upper respiratory tract infections, sinusitis and diarrhoea. Bronchiectasis was detected. A sister in the family was also studied to determine her putative carrier status.
Family 2
The patient from family 2, born in 1981, was evaluated at age 8 years for a history of recurrent otitis, three incidences of pneumonia and several upper respiratory tract infections. The family history was negative for immune deficiency. Immunological studies showed normal T and B lymphocyte phenotype and normal proliferation. Serum immunoglobulin levels were as follows: IgG 34·7 mg/dl, IgA 7·7 mg/dl and IgM 185 mg/dl. He was diagnosed as HIGM1 and started on treatment with IVIG (300 mg/kg every 15 days). Otherwise, he has remained well, experiencing only upper respiratory tract infections, recurrent otitis and conjunctivitis.
Family 3
The patient, born in 1972, was apparently well but was studied at age 6 months because of a family history suggesting X-linked immunodeficiency disease. Two brothers had died of pneumonia (one of them by Pneumocystis carinii pneumonia) at 11 months and 7 months of age, respectively. The immunological evaluation revealed an IgG level of 95 mg/dl, an undetectable IgA level and an IgM level of 135 mg/dl. He was diagnosed with hypogammaglobulinaemia and treatment with IMIG was started. He had several bacterial and fungal infections and at age 2 he contracted viral meningoencephalitis and died. His sister was clinically well but presented transient hypogammaglobulinaemia during infancy.
Flow cytometry studies
The study protocol was approved by the Hospital's Ethic Commission and blood samples were drawn after obtaining written informed consent from the family members.
The assay was performed on peripheral blood mononuclear cells (PBMCs) activated with PMA (Sigma, St Louis, MO, USA, 5 ng/ml) and ionomycin (Calbiochem, La Jolla, CA, USA, 500 ng/ml) after 6 h of incubation at 37°C and in an atmosphere containing 5% CO2. Expression of CD40L was tested on cells stained with anti-CD3 PerCP, anti-CD8 FITC (Becton Dickinson, San Jose, CA, USA) and anti-CD40L PE (PharMingen, San Diego, CA, USA) and processed as described previously [21]. Because exposure of T lymphocytes to phorbol esters such as PMA induces down-regulation of CD4 expression, CD4+ positive cells were inferred from CD8 negativity.
For the CD40 protein binding assay, the cells were resuspended in staining buffer (PBS containing 1% BSA and 0·2% sodium azide) and stained with CD40-Fcµ fusion protein (generated by Dr P. Lane, Basel) for 1 h at 4°C. Cells were washed once with staining buffer and incubated for 30 min at 4°C with PE-conjugated rabbit antihuman IgM (Dako, Glostrup, Denmark). The cells were again washed with staining medium, and fusion protein binding was determined on activated lymphocytes by flow cytometry. Anti-CD69 staining was performed as a control for T cell activation. The CD69+ cells were the ones that increased in size after activation, so the characteristic of greater size was also considered to be an activation criteria.
Polymerase chain reaction (PCR)
Each of the CD40L exons and adjacent intronic regions were P CR amplified, using the oligonucleotide primers described by Seyama et al. [22]. The reactions were carried out in a 50µl volume containing 1 µg of genomic DNA, 250 µm of each dNTP, 50 pmol of each primer, 1× reaction buffer (50 mm KCl, 10 mm Tris-HCl, 1·5 mm MgCl2) and 1·25 U Taq DNA polymerase (PE Applied Biosystems, Foster City, CA, USA). The samples were first denatured at 94°C for 2 min, and then subjected to 30 cycles of: denaturation at 93°C for 1 min, annealing at 55°C for 1 min and extension at 72°C for 2 min in a GeneAmp System 9600 (PE Applied Biosystems Thermocycler) [22].
Single-strand conformation polymorphism (SSCP)
SSCP analysis was performed as described previously [23]. The PCR product that exhibited a shifted mobility was sequenced to determine the type and exact mutation site.
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated from activated PBMCs (6 h incubation with PMA at 5 ng/ml and ionomycin at 500 ng/ml) according to the manufacturer's instructions (QIAamp RNA, Qiagen, Hilden, Germany). cDNA was prepared by reverse transcription from patients’ mRNA using reverse transcriptase and PCR kit Supercript (Gibco BRL/Life Technologies) and the oligonucleotide primers as described by Splawski et al. [24].
Sequence analysis
PCR products from patients with visible band shifts on SSCP were purified on Quiaquik columns (Quiagen) and sequenced on an automated ABI PRISM™ 310 Genetic Analyser (PE Applied Biosystems) using the same primers as used in the PCR reaction and the BigDye™ Terminator Cycle Sequencing Kit (PE Applied Biosystems). Detected mutations were confirmed by opposite direction sequencing and compared with the reported coding sequence (NCBI GenBank: X67878).
RESULTS
CD40L cell surface expression and CD40-Fcµ binding by flow cytometry
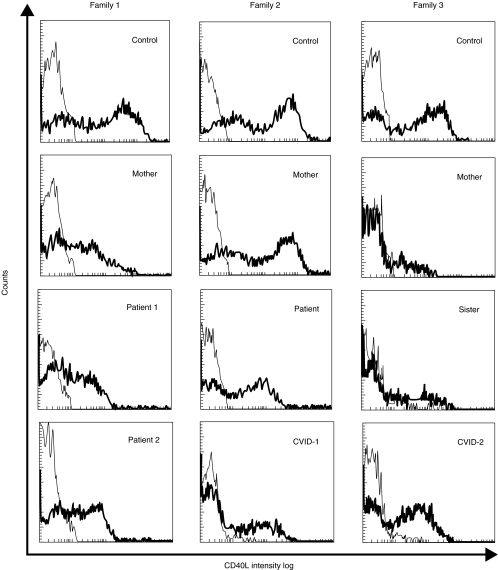
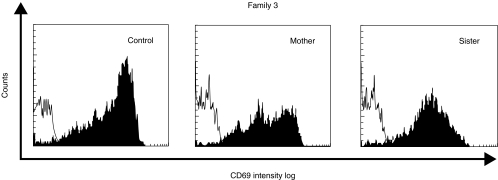
The three families included in this study presented different patterns of CD40L expression, as shown in Fig. 1 and summarized in Table 1. In the first family, both patients and their mother presented a normal percentage of CD4+ cells expressing CD40L. However, the fluorescence intensity was decreased in comparison with a normal control studied in parallel. In the sister, the fluorescence intensity and the percentage of CD4+CD40L+ cells were normal (Table 1). The patient from family 2 showed a slightly decreased percentage of CD4+CD40L+ cells and a lower fluorescence intensity, compared with a normal control. The mother's cells had normal expression of CD40L. Both mother and sister from family 3 showed a very low percentage of CD4+CD40L+ cells and a decreased fluorescence intensity versus a normal control studied in the same assay, in spite of the correct stimulation of cells reflected by CD69 expression within the normal range (Fig. 2).
Fig. 1.
Flow cytometric study of CD40L expression on activated lymphocytes after stimulation with PMA and ionomycin for 6 h. The thin line represents the binding of the isotype control monoclonal antibody to activated lymphocytes and the thick line represents the CD40L expression in this cell population.
Table 1.
Percentage and mean fluorescence intensity of CD40L expression on CD8− subpopulation in the three families; for the two-colour analysis a gate was set on activated lymphocytes
| CD4+CD40L+ (%) | Arithmetic mean | Geometric mean | |
|---|---|---|---|
| Family 1 | |||
| Mother | 49 | 96 | 40·7 |
| Patient 1 | 40 | 45 | 37 |
| Patient 2 | 49 | 53 | 35 |
| Sister | 66 | 407 | 177 |
| Healthy control | 50 | 212 | 119 |
| Family 2 | |||
| Mother | 71 | 409 | 186 |
| Patient | 52 | 119 | 53 |
| Healthy control | 73 | 424 | 200 |
| Family 3 | |||
| Mother | 10 | 32·6 | 26 |
| Sister | 16 | 67 | 39·6 |
| Healthy control | 59 | 127 | 82 |
Fig. 2.
Flow cytometric study of CD69 expression on activated lymphocytes after stimulation with PMA and ionomycin for 6 h in female carriers of family 3. The thin line represents the binding of the isotype control monoclonal antibody to activated lymphocytes and the black area represents the CD69 expression in this cell population.
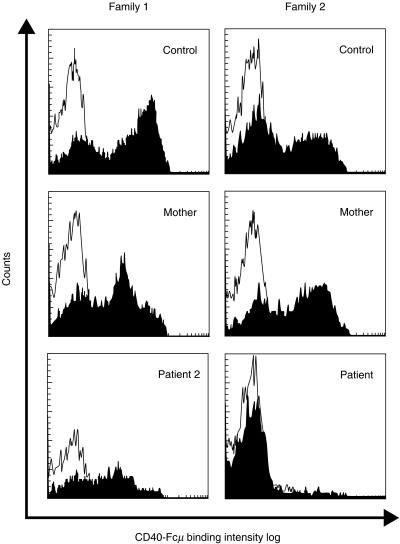
The CD40-Fcµ binding assay (Fig. 3) showed a very low staining intensity in patient 2 from family 1. His mother presented two positive populations: one with the same intensity as a normal control and the other with the low intensity of the patient's cells, in accordance with her carrier status. The patient from family 2 presented a complete absence of CD40-Fcµ binding (Fig. 3), in spite of correct cell activation demonstrated by the high level of CD69 expression (data not shown).
Fig. 3.
Flow cytometric study of CD40-Fcµ binding on activated lymphocytes after stimulation with PMA and ionomycin for 6 h in families 1 and 2. The thin line represents the binding of the isotype control monoclonal antibody to activated lymphocytes and the black area represents the CD40-Fcµ binding in this cell population.
Mutations of the CD40L gene
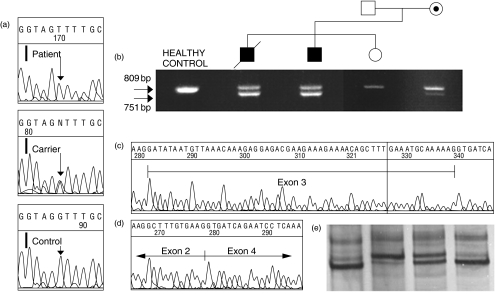
In family 1, SSCP analysis in both patients detected a shift in the exon 3 PCR product. Both the shifted and the normal band were observed in the mother's PCR product, in agreement with her carrier status. No altered bands were observed in the sister (Fig. 4e).
Fig. 4.
Family 1 molecular study. (a) Comparison of intron 3 sequence from the first patient, the mother and a healthy control. (b) Agarose gel showing the RT-PCR products obtained from different members. (c) Automated sequencing result obtained from the big products shown in b. (d) Exon 3 skipping in the sequence from the small band in both patients and the mother. (e) From left to right, SSCP analysis of non-carrier sister, one of the two patients, the carrier mother and a healthy control.
Sequencing of this region demonstrated a G/T transversion at the + 5 position of the intron 3 5′ splice site (Fig. 4a). In order to study the consequence of this mutation in the splicing process, RT-PCR was performed. Both patients presented an extra band with a smaller size than the expected product. This band, although fainter, was also present in the mother (Fig. 4b). While the expected product was identical with the wild-type sequence (Fig. 4c), the entire sequence of exon 3 was skipped in the small one (Fig. 4d). This deletion would cause a shift in the reading frame and the synthesis of a prematurely terminated polypeptide that lacks the TNF homology domain (I 107 X).
In family 2, the exon 5 PCR product of the patient presented an altered migration (data not shown). Sequencing of this exon revealed a C to T transition at the 686 position of the cDNA sequence. This mutation results in the substitution of the wild-type serine with phenylalanine (Ser222 Phe) in the extracellular domain of the protein.
In family 3, DNA samples from the deceased patients were not available. Thus, molecular diagnosis was performed on the genomic DNA from the putative carriers. All the exons were PCR amplified and the products were compared to those obtained from a healthy control DNA by SSCP. A shifted band was detected only in the exon 5 product (data not shown). Automated sequencing revealed the presence of two overlapping sequences: the wild-type, and another containing a 14-bp deletion starting at the 532 position of the cDNA sequence. The deletion in the mutated sequence leads to a frameshift and the appearance of a premature stop codon (del I 171 X 195). These results indicated that the mother and the sister were carriers of a mutation in the CD40L gene, suggesting that the clinical phenotype of the deceased brothers was a consequence of this severe mutation.
DISCUSSION
Low expression of CD40L has been detected in other immunodeficiencies apart from HIGM1, such as common variable immunodeficiency (CVID) [25–27]. Our data from a series of 39 CVID patients are in agreement with these results (Cambronero et al., manuscript submitted). To exclude the possibility of misdiagnosis in these patients, a CD40L gene study was performed on five male patients from this series who had low levels of IgG and IgA, normal or elevated IgM, and decreased CD40L expression (two of which are shown in Fig. 1). CD40L mutations were not found in any of these CVID patients, suggesting that the low level of CD40L expression was a secondary defect.
In the group of HIGM1 patients, several molecular mechanisms have been proposed to explain the functional consequences of the mutations. To date, 88 different defects throughout the gene have been collected in the database ‘CD40Lbase’ created for this disease [28].
The type of mutations range from large deletions that completely remove the coding region, so leading to an absence of protein expression, to missense mutations permitting the synthesis of a protein product but severely compromising the binding of the ligand receptor.
The three mutations identified in our patients have not been described previously and reflect the variety of molecular mechanisms that trigger the appearance of this immunodeficiency.
In family 1 the replacement of the wild-type guanine by thymine severely affects the efficiency of mRNA maturation. The appearance of the expected product in the RT-PCR suggests that some transcripts accomplish the maturation process correctly. Another mutation at the same position (G to A) has been described, affecting the splicing process in the same manner [29]. According to the classical studies that established the score for variations in the splice sites sequences [30], both mutations would provoke a similar deviation from the consensus sequence at this site.
In both patients from family 1, CD40 ligand expression can be detected by flow cytometry on activated CD4+ T lymphocytes. The intensity of expression is greatly diminished in comparison with a healthy control, however. The translation of the exon 3-skipped mRNA would lead to the production of a polypeptide lacking a great proportion of the extracellular domain, including the characteristic TNFH domain. The correctly spliced mRNA would be translated to generate wild-type molecules that would associate in functional homotrimers [31–33]. Recently, Su and coworkers [34] demonstrated by a co-transfection experiment in COS cells that CD40L mutants lacking the TNFH domain are still able to form a complex with the wild-type molecule. However, this association compromises the ability of the complex to mature and be expressed on the cell surface do, however, and therefore the probabilities of assembling a functional homotrimer are greatly decreased. The low number of CD40L molecules presented at the cell surface maintain ligand binding capacity. This is inferred by the similar staining intensity obtained with anti-CD40L and CD40-Fcµ. However, this low level of expression of normal CD40L is insufficient to reach the necessary threshold for correct B cell stimulation.
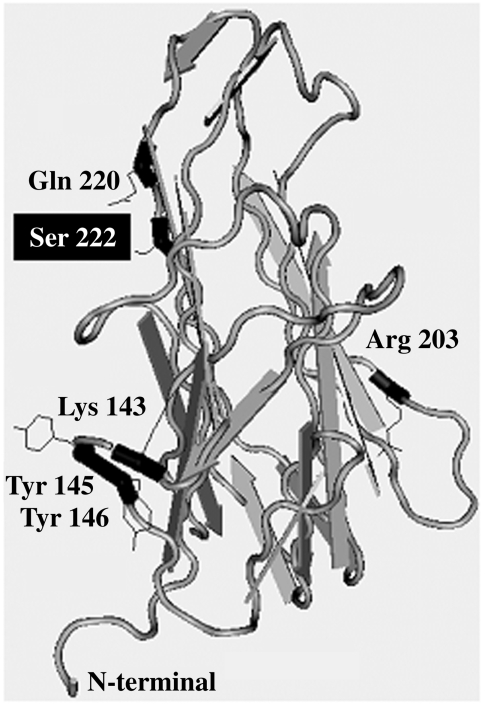
The family 2 patient presents a missense mutation (Ser222 Phe) in the TNF homologous domain. The majority of missense mutations collected in the ‘CD40Lbase’ are located in this region. Molecular models and an X-ray structure of the CD40L TNF homology domain, in conjunction with site-directed mutagenesis, have been used by others to identify residues involved in the binding of the ligand with its receptor. Five residues in CD40L (Lys143, Tyr145, Tyr146, Arg203 and Gln220), located along the groove that two monomers create after dimerization, have been identified as contributing to the interaction with CD40 [35]. Ser222 is located in an exposed position in the same beta strand as Gln220 and the side chain of Ser222 points to the interface of the dimer (Fig. 5) [8,36].
Fig. 5.
1ALY, crystal structure of human CD40L [8]. Only one monomer is shown. The residues involved in polar interactions with CD40 and the mutated Ser222 in family 2 are shown in black using the National Center for Biotechnology Information (NCBI) program Cn3D version 4·0 [36].
The ligand–receptor union has been described to be stabilized by polar interactions between CD40 and CD40L residues [37]. The substitution of a polar and hydrophilic amino acid (Ser) in this crucial position by an apolar and hydrophobic one (Phe) could severely disturb the overall structure of this part of the homotrimer. Bajorath et al. [38] classified missense mutations based on the effect on monomer structure. Class I mutations affect the internal core of the monomer, class II affect trimer formation and class III affect CD40 binding either directly or indirectly. The Ser222 Phe mutation could be classified as type III: CD40L-positive staining is detected in activated cells, but the binding capacity of this mutant protein is abolished completely, as demonstrated by the CD40-Fcµ binding assay. Thus, this alteration acts functionally as a null mutation. Receptor binding could be compromised either directly by the Ser222 Phe substitution itself or by the substitution affecting the nearby located Gln220.
Family 3 patients presented the most severe clinical phenotype. The profound defect in protein expression that could be inferred from the mutation in these patients may explain this observation. It is also noticeable in the mother and the sister that only a very small proportion of activated T cells expressed CD40L as assessed by flow cytometry. This molecule is not necessary for CD4+ T cell development. Hence, one could expect approximately half of the cells in both carriers to randomly inactivate the X-chromosome that harbours the mutant gene and therefore show normal levels of CD40L expression, and the other half to inactivate the X-chromosome of the wild-type gene, thereby resulting in complete lack of expression. However, it has been demonstrated that 5–20% of apparently normal women without X-linked disease carrier status may present non-random X-inactivation [39]. Deviations from non-random inactivation can lead to disease expression in X-linked hyper-IgM heterozygous carriers [40]. A pattern of X-chromosome inactivation skewed towards the normal chromosome is the most plausible explanation for the very low expression of CD40L in otherwise correctly activated CD4+ T cells presented by the female carrier members of family 3. Both are asymptomatic at present and display a competent immune system but, interestingly, the sister had transient hypogammaglobulinaemia until she was 2 years old. Both carriers are being monitored regularly at our clinic. These results suggest that a small number of T cells expressing CD40L are enough to conserve a functional immune response.
The similar phenotype caused by the different gene defects and the fact that low CD40L expression is not exclusive to this syndrome make molecular studies mandatory for correct diagnosis, especially taking into account that bone marrow precursor reconstitution in the first months of life has been revealed as a new therapeutical approach for HIGM1 with excellent results [41]. The family 2 patient has presented so far the mildest phenotype, which may suggest that the missense mutation is associated with a better prognosis. However, the CD40-Fcµ binding assay was completely negative, and severe manifestations may appear in the future. This underscores the importance of this assay before establishing a genotype-phenotype correlation in patients with decreased CD40L expression.
Acknowledgments
We thank Priscila Acebes Sanz, Milagros Arribas Escaso, Amparo Barranco Herrero, Josefa Borrega Guillén, Ana Carazo Álvarez, Begoña Castro Rodríguez and Josefina Moruno González for the technical assistance in the development of this work. R. Cambronero was supported by a grant from Instituto de Salud Carlos III (99/4223).
REFERENCES
- 1.Conley ME, Notarangelo LD, Etzioni A. Diagnostic criteria for primary immunodeficiencies. Clin Immunol. 1999;93:190–7. doi: 10.1006/clim.1999.4799. [DOI] [PubMed] [Google Scholar]
- 2.IUIS Scientific Group. Primary immunodeficiency diseases. Clin Exp Immunol. 1999;118(Suppl. 1):1–28. doi: 10.1046/j.1365-2249.1999.00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aruffo A, Farrington M, Hollenbaugh D, et al. The CD40 ligand, gp39, is defective in activated T cells from patients with X-linked hyper-IgM syndrome. Cell. 1993;72:291–300. doi: 10.1016/0092-8674(93)90668-g. [DOI] [PubMed] [Google Scholar]
- 4.Ramesh N, Fuleihan R, Geha R. Molecular pathology of X-linked immunoglobulin deficiency with normal or elevated IgM (HIGMX-1) Immunol Rev. 1994;138:87–104. doi: 10.1111/j.1600-065x.1994.tb00848.x. [DOI] [PubMed] [Google Scholar]
- 5.Kroczek RA, Graf D, Brugnoni D, et al. Defective expression of CD40 ligand on T cells causes ‘X-linked immunodeficiency with hyper-IgM (HIGM1)’. Immunol Rev. 1994;138:39–59. doi: 10.1111/j.1600-065x.1994.tb00846.x. [DOI] [PubMed] [Google Scholar]
- 6.Armitage RJ, Fanslow WC, Strockbine L, et al. Molecular and biological characterization of a murine ligand for CD40. Nature. 1992;357:80–2. doi: 10.1038/357080a0. [DOI] [PubMed] [Google Scholar]
- 7.Hollenbaugh D, Grosmaire LS, Kullas CD, et al. The human T cell antigen gp39, a member of the THF gene family, is a ligand for the CD40 receptor: expression of a soluble form of gp39 with B cell co-stimulatory activity. EMBO J. 1992;11:4313–21. doi: 10.1002/j.1460-2075.1992.tb05530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karpusas M, Hsu YM, Wang J, et al. Crystal structure of an extracellular fragment of human CD40 ligand. Structure. 1995;3:1031–9. doi: 10.1016/s0969-2126(01)00239-8. [DOI] [PubMed] [Google Scholar]
- 9.Bajorath J. Detailed comparison of two molecular models of the human CD40 ligand with an X-ray structure and critical assessment of model-based mutagenesis and residue mapping studies. J Biol Chem. 1998;273:24603–9. doi: 10.1074/jbc.273.38.24603. [DOI] [PubMed] [Google Scholar]
- 10.Schönbeck U, Libby P. The CD40/CD154 receptor/ligand dyad. Cell Mol Life Sci. 2001;58:4–43. doi: 10.1007/PL00000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Kooten C, Banchereau J. CD40-CD40 ligand. J Leukoc Biol. 2000;67:2–17. doi: 10.1002/jlb.67.1.2. [DOI] [PubMed] [Google Scholar]
- 12.Durie FH, Foy TM, Masters SR, Laman JD, Noelle RJ. The role of CD40 in the regulation of humoral and cell-mediated immunity. Immunol Today. 1994;15:406–11. doi: 10.1016/0167-5699(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 13.Hayward AR, Levy J, Facchetti F, et al. Cholangiopathy and tumours of the pancreas, liver, and biliary tree in boys with X-linked immunodeficiency with hyper-IgM. J Immunol. 1997;158:977–83. [PubMed] [Google Scholar]
- 14.Levy J, Español-Boren T, Thomas C, et al. Clinical spectrum of X-linked hyper-IgM syndrome. J Pediatr. 1997;131:47–54. doi: 10.1016/s0022-3476(97)70123-9. [DOI] [PubMed] [Google Scholar]
- 15.Seyama K, Kobayashi R, Hasle H, et al. Parvovirus B19-induced anemia as the presenting manifestation of X-linked hyper-IgM syndrome. J Infect Dis. 1998;178:318–24. doi: 10.1086/515633. [DOI] [PubMed] [Google Scholar]
- 16.Minegishi Y, Lavoie A, Cunningham-Rundles C, et al. Mutations in activation-induced cytidine deaminase in patients with hyper-IgM syndrome. Clin Immunol. 2000;97:203–10. doi: 10.1006/clim.2000.4956. [DOI] [PubMed] [Google Scholar]
- 17.Revy P, Muto T, Levy Y, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–75. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 18.Ferrari S, Giliani S, Insalaco A, et al. Mutations of CD40 gene cause an autosomal recessive form of immunodeficiency with hyper IgM. Proc Natl Acad Sci USA. 2001;98:12614–9. doi: 10.1073/pnas.221456898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zonana J, Elder ME, Schneider LC, et al. A novel X-linked disorder of immune deficiency and hypohidrotic ectodermal dysplasia is allelic to incontinentia pigmenti and due to mutations in IKK-gamma (NEMO) Am J Hum Genet. 2000;67:1555–62. doi: 10.1086/316914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain A, Ma CA, Liu S, Brown M, Cohen J, Strober W. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat Immunol. 2001;2:223–8. doi: 10.1038/85277. [DOI] [PubMed] [Google Scholar]
- 21.O'Gorman MRG, Zaas D, Paniagua M, Corrochano V, Scholl PR, Pachman LM. Development of a rapid whole blood flow cytometry procedure for the diagnosis of X-linked hyper-IgM syndrome patients and carriers. Clin Immunol Immunopathol. 1997;85:172–81. doi: 10.1006/clin.1997.4422. [DOI] [PubMed] [Google Scholar]
- 22.Seyama K, Kira S, Ishidoh K, Souma S, Miyakawa T, Kominami E. Genomic structure and PCR-SSCP analysis of the human CD40 ligand gene: its application to prenatal screening for X-linked hyper-IgM syndrome. Hum Genet. 1996;97:180–5. doi: 10.1007/BF02265262. [DOI] [PubMed] [Google Scholar]
- 23.García Rodríguez MC, López Granados E, Ferreira Cerdán A, Fontán Casariego G. Analysis of Bruton's tyrosine kinase gene in Spain. Hum Mutat. 2001;18:84. doi: 10.1002/humu.1155. [DOI] [PubMed] [Google Scholar]
- 24.Splawski JB, Nishioka J, Nishioka Y, Lipsky P. CD40 ligand is expressed and functional on activated neonatal T cells. J Immunol. 1996;156:119–27. [PubMed] [Google Scholar]
- 25.Farrington M, Grosmaire LS, Nonoyama S, et al. CD40 ligand expression is defective in a subset of patients with common variable immunodeficiency. Proc Natl Acad Sci USA. 1994;91:1099–103. doi: 10.1073/pnas.91.3.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thon V, Wolf HM, Sasgary M, et al. Defective integration of activating signals derived from the T cell receptor (TCR) and costimulatory molecules in both CD4+ and CD8+ T lymphocytes of common variable immunodeficiency (CVID) patients. Clin Exp Immunol. 1997;110:174–81. doi: 10.1111/j.1365-2249.1997.tb08314.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oliva A, Scala E, Quinti I. IL-10 production and CD40L expression in patients with common variable immunodeficiency. Scand J Immunol. 1997;46:86–90. doi: 10.1046/j.1365-3083.1997.d01-95.x. [DOI] [PubMed] [Google Scholar]
- 28.Notarangelo L, Peitsch MC, et al. CD40Lbase: a database of CD40L gene mutations causing X-linked hyper IgM syndrome. Immunol Today. 1996;17:511–6. doi: 10.1016/0167-5699(96)30059-5. [DOI] [PubMed] [Google Scholar]
- 29.Seyama K, Nonoyama S, Gangsaas I, et al. Mutations of the CD40 ligand gene and its effect on CD40 ligand expression in patients with X-linked hyper-IgM syndrome. Blood. 1998;92:2421–34. [PubMed] [Google Scholar]
- 30.Shapiro MB, Senapathy P. RNA splice junction of different classes of eukaryotes: sequence statistics and functional implication in gene expression. Nucl Acids Res. 1987;15:7155–74. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garber E, Su L, Ehrenfels B, Karpusas M, Hsu YM. CD154 variants associated with hyper-IgM syndrome can form oligomers and trigger CD40-mediated signals. J Biol Chem. 1999;274:33545–50. doi: 10.1074/jbc.274.47.33545. [DOI] [PubMed] [Google Scholar]
- 32.Seyama K, Osborne WRA, Ochs HD. CD40 ligand mutants responsible for X-linked hyper-IgM symdrome associate with wild-type CD40 ligand. J Biol Chem. 1999;274:11310–20. doi: 10.1074/jbc.274.16.11310. [DOI] [PubMed] [Google Scholar]
- 33.Barnhart B, Ford GS, Bhushan A, Song C, Covey LR. A polymorphic CD40 ligand (CD154) molecule mediates CD40-dependent signalling but interferes with the ability of soluble CD40 to functionally block CD154: CD40 interactions. Immunology. 2000;99:54–61. doi: 10.1046/j.1365-2567.2000.00943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su L, Garber EA, Hsu YM. CD154 variant lacking tumour necrosis factor homologous domain inhibits cell surface expression of wild-type protein. J Biol Chem. 2001;276:1673–6. doi: 10.1074/jbc.C000674200. [DOI] [PubMed] [Google Scholar]
- 35.Bajorath J, Chalupny NJ, Marken JS, et al. Identification of residues on CD40 and its ligand which are critical for the receptor–ligand interaction. Biochemistry. 1995;34:1833–44. doi: 10.1021/bi00006a003. [DOI] [PubMed] [Google Scholar]
- 36.Wang Y, Geer LY, Chappey C, Kans JA, Bryant SH. Cn3D: sequence and structure views for Entrez. Trends Biochem Sci. 2000;25:300–2. doi: 10.1016/s0968-0004(00)01561-9. [DOI] [PubMed] [Google Scholar]
- 37.Singh J, Garber E, Van Vlijmen H, et al. The role of polar interactions in the molecular recognition of CD40L with its receptor CD40. Protein Sci. 1998;7:1124–35. doi: 10.1002/pro.5560070506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bajorath J, Syama K, Nonoyama S, Ochs HD, Aruffo A. Classsification of mutations in the human CD40 ligand, gp39, that are associated with X-linked hyper-IgM syndrome. Protein Sci. 1996;5:531–4. doi: 10.1002/pro.5560050316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Belmont JW. Genetic control of X inactivation and processes leading to X-inactivation skewing. Am J Hum Genet. 1996;58:1101–8. [PMC free article] [PubMed] [Google Scholar]
- 40.de Saint Basile G, Tabone MD, Durandy A, Phan F, Fischer A, Le Deist F. CD40 ligand expression deficiency in a female carrier of the X-linked hyper IgM syndrome as a result of X chromosome lyonization. Eur J Immunol. 1999;29:367–73. doi: 10.1002/(SICI)1521-4141(199901)29:01<367::AID-IMMU367>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 41.Duplantier JE, Seyama K, Day NK, et al. Immunologic reconstitution following bone marrow transplantation for X-linked Hyper IgM syndrome. Clin Immunol. 2001;98:313–8. doi: 10.1006/clim.2000.4994. [DOI] [PubMed] [Google Scholar]