Abstract
Hepatitis C virus (HCV) infection has been detected in a large proportion of patients with mixed cryoglobulinaemia (MC). Circulating ‘free’ non-enveloped HCV core protein has been demonstrated in HCV-infected patients, and this suggests its possible involvement in the formation of cryoprecipitable immune complexes (ICs). Thirty-two anti-HCV, HCV RNA-positive patients with type II MC were evaluated. Non-enveloped HCV core protein, HCV RNA sequences, total IgM, rheumatoid factor (RF) activity, IgG and IgG subclasses, C3 and C4 fractions, C1q protein and C1q binding activity were assessed in both cryoprecipitates and supernatants. Non-enveloped HCV core protein was demonstrated in 30 of 32 (93·7%) type II MC patients. After separation of cold-precipitable material, the protein was removed completely from supernatant in 12 patients (40%), whereas it was enriched in the cryoprecipitates of the remaining 18. In addition, HCV RNA and IgM molecules with RF activity were concentrated selectively in the cryoprecipitates. Differential precipitation was found for both total IgG and IgG subclasses, as they were less represented in the cryoglobulins and no selective enrichment was noted. Immunological characterization of HCV core protein-containing cryoprecipitating ICs after chromatographic fractionation showed that the IgM monoclonal component had RF activity, whereas anti-HCV core reactivity was confined to the IgG fraction. C1q enrichment in addition to high avidity of ICs for C1q binding in the cryoprecipitates suggest that complement activation may occur through the C1q protein pathway. The present data demonstrate that non-enveloped HCV core protein is a constitutive component of cryoprecipitable ICs in type II MC patients.
Keywords: cryoglobulinaemia, hepatitis C virus, HCV core protein, immune complexes
Introduction
Cryoglobulins are cold-precipitable immunoglobulins (Ig) associated frequently with the development of vascular, renal and neurological lesions [1]. While single (type I) cryoglobulins contain a monoclonal Ig and occur predominantly in patients with malignant proliferative disorders of the immune system, mixed cryoglobulins (MC) are accounted for by two or more Ig isotypes, with (type II) or without (type III) a monoclonal component [2]. MC have been detected in patients with chronic inflammatory, autoimmune, lymphoproliferative and infectious diseases [3], although they are often defined as ‘essential’ due to the apparent lack of concomitant diseases. During the last 10 years, however, a growing body of evidence has defined a striking relation between MC and hepatitis C virus (HCV) infection [4–6].
HCV is an enveloped, positive-strand RNA virus with a genome of approximately 9600 nucleotides, encoding a polyprotein precursor of about 3000 amino acids [7]. It is cleaved by viral and host proteases, resulting in a series of proteins which include the capsid, two envelope proteins (E1 and E2) and seven (perhaps more) non-structural proteins [8]. The putative HCV virion consists of the viral envelope and an inner core. Virions often circulate in the form of immune complexes (ICs) bound to Ig [9,10].
A remarkable feature of HCV infection is its high propensity to become chronic and to develop cirrhosis in almost one-quarter of infected individuals [11]. The incidence of HCV infection in MC ranges from 40% to 100% in the reported case series [12] and in different geographical areas [13], though improved diagnostic techniques have shown increasing prevalence rates in the same area in the course of time [14]. In Italy, more than 80% of MC patients are infected with HCV [15]. MC, on the other hand, occurs in 20–50% of prospective series of chronically HCV-infected patients [16,17].
The primary role of HCV in the mechanism of cryoprecipitation is suggested by its selective concentration in cryoglobulins [5]. RNase-resistant HCV enrichment of cryoprecipitates indicates that intact viral particles are involved directly in their formation [18]. Density gradient fractionation analysis has shown that ICs isolated from either cryoprecipitates or supernatants have heterogeneous composition and different biological properties [19].
HCV nucleocapsid protein devoid of enveloped proteins has been detected in the bloodstream of HCV-infected patients [20–22] and may represent a good indicator of viral load [23]. Non-enveloped HCV core protein has been reported to be secreted from transfected hepatoma cell lines in cultures [24] and in HCV core protein transgenic mice [25]. Observation of non-enveloped nucleocapsids in the serum of HCV-positive patients with normal ALT levels and without signs of progressive liver disease suggests that they are overproduced during virogenesis [22]. The core protein has been found in the serum of most HCV chronic carriers with active liver disease and almost half of those with inactive disease [26]. In addition, serum levels of HCV core protein change following antiviral therapy and become undetectable in responsive patients [27].
Thus, it seems likely that HCV core protein participates in the formation of ICs [28] and suppresses T-cell response by interacting with the globular domain of C1q complement receptor (gC1qR) [29]. This interaction may play a key role in determining complement activation, a crucial interdependent regulator of the size and solubility of immune aggregates [30].
We illustrate here the detection of ‘free’ non-enveloped HCV core protein in type II MC patients, and discuss its role in cryoprecipitation and its clinical significance.
MATERIALS AND METHODS
Patients
Starting from January 1999, 32 patients with the purpura-weakness arthralgia syndrome were referred to the Department of Biomedical Sciences and Clinical Oncology of the University of Bari Medical School. All patients were positive for antibodies to HCV and HCV RNA, and negative for HBsAg and anti-HIV antibodies. Epidemiological data including age at time of liver biopsy, duration of liver disease, and exposure to risk factors are reported in Table 1. After giving their informed consent, all patients provided a liver biopsy specimen taken with the Menghini needle. No patient had received corticosteroids, interferon or other systemic treatments. The study was approved by the University Ethics Committee.
Table 1.
Epidemiological, histological, virogical, laboratory and clinical characteristics of the 32 cryoglobulinaemic patients
| Epidemiology | |
| Sex: M/F (ratio) | 7/25 (0·28) |
| Mean age (years) | 57 ± 11 |
| Presumed duration of disease (years) | 7 ± 6 |
| Blood/blood product transfusion (%) | 18 (56·2) |
| Liver histology | |
| Chronic active hepatitis (%) | 30 (93·7) |
| Inflammation index | 5·4 ± 2·6 |
| Fibrosis index | 2·9 ± 1·6 |
| Cirrhosis (%) | 2 (6·3) |
| Virology | |
| Serum HCV RNA (IU/ml) | 419 800 ± 293,600 |
| HCV genotype: | |
| 1b (%) | 14 (44) |
| 2a/2c (%) | 18 (56) |
| Anti-HCV (%) | 32 (100) |
| HBsAg | 0 |
| Anti-HIV | 0 |
| Laboratory | |
| Cryocrit (%) | 13 ± 22 |
| Rheumatoid factor activity (n.v. ≤ 40 IU/ml) | 520 ± 1103 |
| Monoclonal component IgMk (%) | 32 (100) |
| Immunoglobulins (mg/dL) | |
| IgM (n.v. 40–230) | 422 ± 291 |
| IgG (n.v. 700–1600) | 1320 ± 531 |
| Complement fractions (mg/dL) | |
| C3 (n.v. 93–188) | 116 ± 32 |
| C4 (n.v. 15–48) | 10 ± 7 |
| ALT levels (n.v. ≤ 30 IU/l) | 68 ± 31 |
| Clinical signs (%) | |
| Arthralgia | 31 (96·8) |
| Cutaneous vasculitis | 25 (78·1) |
| Peripheral neuropathy | 19 (59·4) |
| Raynaud's phenomenon | 6 (18·7) |
| Renal failure | 5 (15·6) |
| Nephrotic proteinuria | 2 (6·2) |
HCV core protein detection
A prototype ELISA kit for HCV core antigen detection (Ortho trak-C assay) was kindly provided by Ortho Clinical Diagnostics (Raritan, NJ, USA). ‘Free’ non-enveloped HCV core protein determination was carried out. A pretreatment antigen–antibody dissociation step was performed with detergent-containing reagents [0·3% TritonX 100; 1,5% 3-((3-cholamidopropyl)-dimethyl ammonium)-1-propanesulphonate, and 15% sodium dodecyl sulphate]. One hundred µl samples were mixed with 50 µl of dissociating buffer and incubated at 56°C for 30 min. Samples were then allowed to cool for 10 min. Duplicate wells coated with monoclonal antibodies directed to different regions of the HCV core antigen were incubated with pretreated samples at room temperature (r.t.) for 60 min under shaking at 900 r.p.m. Antigen–antibody complexes formed on the microwell surface were detected by an anticore monoclonal antibody (Fab fragments) conjugated to horseradish peroxidase (POD) in the second incubation (30 min at r.t., without shaking). Colour was thus developed with o-phenylenediamine (OPD) and hydrogen peroxide, and the product was read at 492 nm. Eight quantitative control calibrations at 0, 1, 1·5, 15, 50, 100, 200 and 800 pg/ml were included in the same kit to generate a calibration curve. The calibrators did not undergo the pretreatment step.
In each run the assay was validated if the following criteria were met: blank well with OD < 0·030; standard level 0 with OD < 0·050; standard level 100 with OD > 0·900. Each reactive sample was accepted if confirmed in a separate test. Specificity of the reaction included the following controls: a panel of 20 well-characterized normal subjects; five HBsAg chronic carriers with histologically proven chronic hepatitis; four patients with rheumatoid arthritis and high levels of rheumatoid factor activity; two with autoimmune chronic active hepatitis. Furthermore, HCV-related recombinant proteins (E1, E2, NS3, NS4, NS5; kindly provided by Dr S. Moroney, Ortho Clinical Diagnostics) at a concentration of 1 ng/ml were included. In all cases OD was found below that of calibrator at 1 pg/ml (0·044 ± 8; mean ± s.d.).
Chromatographic studies
Gel filtration was carried out with a Pharmacia (Uppsala, Sweden) automated fast-protein liquid chromatography system on a Superose 6 column (30 × 1·0 cm, i.d.). Two hundred µl of redissolved cryoprecipitates were injected and eluted with 150 mm NaCl, 50 mm phosphate buffer (pH 7·2) or with 0·2 m acetate buffer (pH 4·5) at 37°C. Total protein was monitored at 280 nm. IgG, IgM and RF activity were monitored by a Behring nephelometer (Dade Behring, Marburg, Germany).
Isolation of specific immune complexes from HCV core protein coupled to CNBr-Activated Sepharose 4B
Cyanogen bromide (CNBr)-activated Sepharose 4B (Pharmacia) swollen in 1 mm HCl was washed on a sintered glass filter and rewashed with coupling buffer (0·1 m sodium bicarbonate, pH 8·5, containing 0·5 m sodium chloride). Recombinant HCV core protein (aa 1–120, kindly provided by Dr S. Moroney, Ortho Clinical Diagnostics) was dissolved in coupling buffer at a final concentration of 5 mg/ml and immediately added to CNBr-activated Sepharose 4B. The suspension was incubated overnight at 4°C during gentle end-over-end rotation. Residual active groups of CNBr-activated Sepharose 4B were blocked with 0·2 m glycine, pH 8·0, for 2 h at r.t. Excess uncoupled proteins were washed off with coupling buffer followed by acetate buffer (0·1 m, pH 4·0, containing 0·5 m sodium chloride). The blocking agent was then washed away by extensive use of the coupling buffer. CNBr-activated Sepharose 4B gel was packed into a chromatographic column and equilibrated with coupling buffer. One hundred µl of redissolved cryoprecipitate was mixed with an equal volume of phosphate buffered saline (PBS) and applied on the top of the column. After extensive washing, immunoadsorbents were eluted with glycine-HCl buffer (0·1 m, pH 2·5). The final eluates were pooled, concentrated and buffer replaced with PBS.
Slot-dot blotting
Aliquots of dialysed against PBS were dotted onto PVDF membrane strips (Millipore Product Division, Bedford, MA, USA) at a concentration of 100 ng protein each. The PVDF strips were then blocked with 5% skimmed milk at 21°C for 2 h and incubated subsequently with AP-conjugated goat antihuman IgG, IgM antibodies (Dako, Copenhagen, Denmark) and a mouse monoclonal antibody against 17·109 XId (anti-17·109 XId MoAb). This antiserum, kindly provided by Dr D. Carson (San Diego, La Jolla, CA, USA) is directed against 17·109 antigen, an l-chain-associated XId expressed on human RFs of type I. The 17·109 epitope is present on human IgM, and not on IgG or IgA lacking RF activity. It is expressed weakly on isolated kappa-chain tryptic peptides [31].
ELISA for anti-HCV core reactivity
ELISA for anti-HCV core antibodies was performed on purified chromatographic fractions. Recombinant HCV core protein was adsorbed passively to microtitre plates at a concentration of 1 µg/ml in 0·05 m sodium carbonate buffer pH 9·6. Samples were added at dilution of 1 : 10 in PBS with 0·05% Tween 20 and 1% bovine serum albumin. Bound IgG or IgM was indicated by incubation with mouse antihuman IgG or IgM Fab fragments conjugated with POD (Dako). The plates were then developed by addition of the substrate OPD and by hydrogen peroxide.
Only reactions exceeding by more than 3 s.d. the mean OD of anti-HCV negative sera (n = 40) at 492 nm were regarded as reactive. The 3 s.d. assured a high specificity that was corroborated further by an avidity test. Increasing amounts of core protein (1 ng/ml−1 mg/ml) were added to the wells before samples. The mixtures were incubated for 60 min at 37°C and developed as described above using the conjugate and the substrate. All samples tested for inhibition were used at the same Ig concentration (0·5 mg/ml). This approach allowed us to define specific inhibition of the reaction with core protein, whereas no significant changes were obtained by using either HCV-related (E1, E2, NS3, NS4, NS5) or unrelated (HBs or HBc) proteins.
Measurement of immunological parameters
Concentrations of IgM, IgG, C3 and C4 fractions, RF activity (Olympus Diagnostics GmbH, Hamburg, Germany), IgG1, IgG2, IgG3 and IgG4 subclasses (NAS IgG1-4, Dade Behring), C1q and C1q binding activity (N Latex CIC, Dade Behring) were measured by nephelometry.
Virological parameters
HCV RNA in serum samples, supernatants and cryoprecipitates was determined using a reverse transcription–polymerase chain reaction (RT-PCR) test (Amplicor HCV, Roche Diagnostic Systems Inc., Branchburg, NY, USA) and quantified with the Quantiplex HCV RNA kit (Chiron Corp., Emeryville, CA, USA) based on a quantitative branched DNA signal amplification assay [32].
HCV genotypes were determined with the line probe assay (Inno-LIPA HCV, Innogenetics N.V., Zwijnaarde, Belgium). This identifies HCV types and subtypes. Measures to prevent contamination were used at all times [33]. HCV antibodies were assayed with a second generation ELISA and confirmed by recombinant immunoblotting assay (Ortho Clinical Diagnostics).
Liver histology
Biopsy specimens were evaluated under code by the pathologist who established the histological diagnosis. Scheuer's criteria [34] were used to define both the grade of inflammation and the stage of fibrosis. Liver disease was considered mild when the histological index score was equal to or lower than 4, and severe when it was higher.
Cryoprecipitate preparation
Blood samples were kept ar 37°C for 2 h in a glass tube. After centrifugation for 20 min at 4000 r.p.m. (LKB Bromma 2161 Midispin) at r.t., sera were stored at 4°C for 7 days. The cryoprecipitate was separated by centrifugation for 30 min at 4000 r.p.m and 4°C. A Wintrobe tube was used to quantify the cryocrit as the percentage of the total volume. After separation, cryoglobulins were washed five times with cold PBS. After the last wash, the cryoprecipitate was resuspended in 0·2 ml of PBS containing Trasylol (100 U/ml) and sodium azide (0·2% w/v) and redissolved at 37°C. Cryoprecipitates were kept at −70°C until use. Cryoprecipitates diluted in 0·5 m NaCl were fractionated by high-resolution gel electrophoresis to type cryoglobulins. Individual monoclonal bands were identified by immunofixation after electrophoretic separation, using a cellulose acetate strip impregnated with antibodies specific for heavy and light chains (Dako).
Statistical analyses
Differences between data were expressed as mean ± s.d. and evaluated by Mann–Whitney U-test and Fisher's exact test, with P < 0·05 as the significance cut-off.
RESULTS
All patients were evaluated prior to therapy. Their epidemiological, histological, virological and clinical data are set out in Table 1. Circulating non-enveloped HCV core protein was found in 30 of 32 (93·7%) unfractionated sera of the present series. According to the calibration curve, its mean level was 255·5 ± 176·8 pg/ml. Regression analysis failed to reveal a direct relationship between circulating HCV core protein levels and cryocrit values (r = 0·1).
Analysis of distribution of HCV core protein and HCV RNA sequences in the cryoprecipitates and corresponding supernatants is shown in Table 2. HCV core protein was detected in all cryoprecipitates, but in only 18 (60%) supernatants. Mean levels of non-enveloped HCV core protein in the cryoprecipitates were significantly higher than in the supernatants (318·5 ± 194 versus 86·4 ± 96·7 pg/ml, respectively; P = 0·003). A similar behaviour was demonstrated for HCV RNA genomic sequences. Mean levels in the cryoprecipitates (1789 313 ± 2341 818 IU/ml) differed significantly from those in the supernatants (284 809 ± 426 987 IU/ml) (P = 0·02). However, no obvious relation was found between levels of non-enveloped HCV core protein and those of HCV RNA sequences in the cryoprecipitates (r = 0·27).
Table 2.
Levels of non-enveloped HCV core protein and HCV RNA sequences in supernatants and cryoprecipitates
| Non-enveloped ‘free’ HCV core protein | HCV RNA sequences | |||||
|---|---|---|---|---|---|---|
| Positive (%) | Negative (%) | Mean ± s.d. (pg/ml) | Positive (%) | Negative (%) | Mean ± s.d. (IU/ml) | |
| Supernatants (30) | 18 (60) | 12 (40) | 86·4 ± 96·7 P = 0·003 | 28 (93·3) | 2 (6·7) | 284 809 ± 426 987 P = 0·02 |
| Cryoprecipitates (30) | 30 (100) | 0 | 318·5 ± 194 | 30 (100) | 0 | 1789 313 ± 2 341 818 |
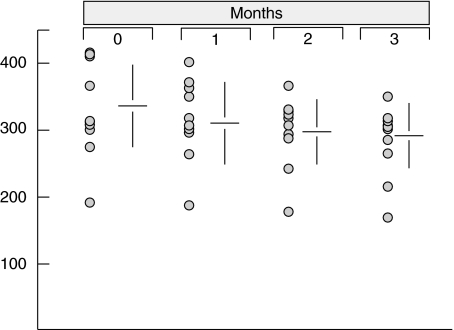
As determined with the reference curve, the detection threshold of the ELISA system was 1 pg/ml. Each specimen was tested in duplicate and its reactivity was confirmed in a separate assay. Reproducibility was also defined by a control study performed to verify the possible core protein shedding through virion degradation in the cryoprecipitates during a long storage period at 4°C. As reported in Fig. 1, stable results were obtained in 10 purified cryoglobulins stored for at least 3 months.
Fig. 1.
Time-related effect on the detection of non-enveloped HCV core protein in the cryoprecipitates maintained at 4°C.
Assessment of the test specificity was also carried out. HCV core protein and HCV RNA sequences were determined in cryoprecipitates and supernatants of HCV-negative cryoglobulinaemic patients. In none of them (three patients with ‘essential’ type II MC, two with HIV-positive type III MC, two with HBV-positive type III MC, five with Waldenström's macroglobulinaemia and five with IgG multiple myeloma) were non-enveloped HCV core protein and HCV RNA sequences detected.
Immunological parameters in the cryoprecipitates and corresponding supernatants are reported in Table 3. Despite similar IgM mean levels in the two phases of serum samples (195·4 ± 132·8 mg/dl in cryoprecipitates and 194·3 ± 217·2 mg/dl in supernatants), a significantly higher concentration of RF activity in the cryoproteins (508·1 ± 341·2 IU/ml) compared with supernatants (309 ± 288·9 IU/ml) was noted (P = 0·01). In sharp contrast, total IgG and IgG subclasses were enriched in the supernatants as compared with cryoprecipitates. A different distribution was indeed found for total IgG (1219·8 ± 473·7 versus 85 ± 138·2 mg/dl, P < 0·005) and for IgG1 (954·6 ± 370 versus 91·8 ± 112·5 mg/dl, P < 0·001), IgG2 (280·3 ± 70 versus 21·5 ± 18·9 mg/dl, P < 0·001), IgG3 (42 ± 30·7 versus 7·1 ± 5·3 mg/dl, P = 0·002), and IgG4 (36·8 ± 47 versus 23·4 ± 15·5 mg/dl, P = 0·04), indicating that a limited number of IgG molecules are involved in cryoprecipitation. The discrepancy we found between total IgG and IgG1 subclass in the cryoprecipitates may suggest that IgG have possibly been underestimated. Indeed, due to the nature of the samples, nephelometry may define overlapping values for these parameters in the cold-precipitated proteins. A slight, although not significant, enrichment of IgG4 was noticed in the cryoprecipitates.
Table 3.
Immunological parameters in cryoprecipitates and supernatants
| Cryoprecipitates | Supernatants | P | |
|---|---|---|---|
| IgM (mg/dl) | 194·3 ± 217·2 | 195·4 ± 132·8 | n.s. |
| RF activity (IU/ml) | 508·1 ± 341·2 | 309 ± 388·9 | = 0·01 |
| IgG (mg/dl) | 85 ± 138·2 | 1219 ± 473·7 | <0·005 |
| Subclasses (mg/dl) | |||
| IgG1 | 91·8 ± 112·5 | 954·6 ± 370 | <0·001 |
| IgG2 | 21·5 ± 18·9 | 280·3 ± 70 | <0·001 |
| IgG3 | 7·1 ± 5·3 | 42 ± 30·7 | = 0·002 |
| IgG4 | 23·4 ± 15·5 | 36·8 ± 47 | = 0·04 |
| C3 (mg/dl) | 2 ± 3·3 | 127·3 ± 44·9 | <0·001 |
| C4 (mg/dl) | 0·06 ± 0·25 | 10·6 ± 11·2 | <0·001 |
| C1q (mg/dl) | 49 ± 37·4 | 5·5 ± 6·8 | <0·001 |
| C1q binding activity(Agg IgG µg/ml) | 6·5 ± 9·1 | 1·2 ± 1·5 | = 0·03 |
Complement-related factors were also considered. C3 and C4 fractions were significantly lower in the cryoprecipitates than in the supernatants (2 ± 3·3 versus 127·3 ± 44·9 mg/dl, P < 0·001 and 0·06 ± 0·25 versus 10·6 ± 11·2 mg/dl, P < 0·001). In contrast, C1q and C1q binding activity, which reflects the ability of ICs to interact with C1q protein, were significantly higher in the cryoprecipitates (49 ± 37·4 versus 5·5 ± 6·8 µg/ml, P < 0·001 and 6·5 ± 9·1 versus 1·2 ± 1·5 µg/ml, P = 0·03, respectively).
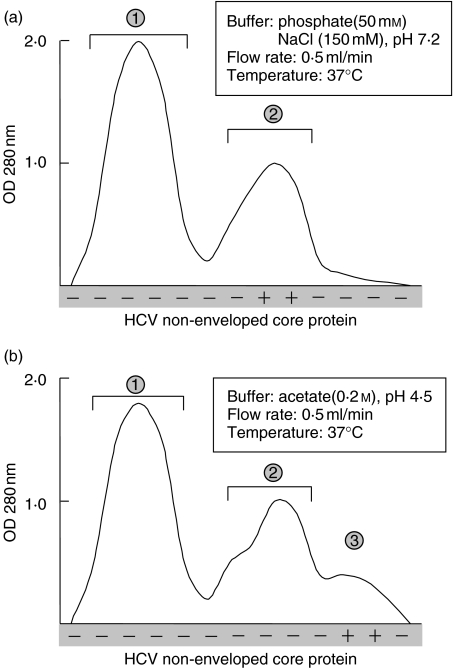
To characterize HCV core protein-containing ICs, three samples reactive to this protein in both supernatant and cryoprecipitate were purified and fractionated on a Superose 6 column at 37°C. The elution profile of HCV nucleocapsid protein was defined and compared to IgG, IgM and RF activity. Non-enveloped core protein consisted of a single peak which was eluted soon after peak 1. The elution profile of the same sample under dissociating conditions, shown in Fig. 2b, indicates that non-enveloped core protein was eluted soon after the IgG-containing fractions.
Fig. 2.
Elution profile of redissolved cryoprecipitate on Superose-6 gel-permeation under non-dissociating (a) and dissociating (b) conditions. (1) IgM peak; (2), IgG peak; (3) HCV core protein peak. Each peak was collected and studied for RF activity and anti-HCV core reactivity.
Fractions corresponding to each peak were pooled, concentrated by ultrafiltration and changed to PBS under extensive dialysis. The peaks containing IgM and IgG, respectively, were tested for anti-HCV core reactivity as well as for RF activity. Results showed that peak 1 had RF activity and no anticore reactivity, whereas peak 2 displayed anticore reactivity and no RF activity. The specificity of anti-HCV core protein reactivity of IgG molecules was assessed by ELISA. Addition of increasing amounts of HCV core protein resulted in the suppression of their binding to the HCV core-sensitized plates, whereas no changes were noted when irrelevant proteins were used. The same feature was observed when they were used plates sensitized with anticore antibodies. Pre-incubation of IgG (peak 2) with purified core protein (peak 3) induced specific inhibition of the reaction.
Intact IgM-IgG complexes were recovered when passing cryoprecipitates through CNBr-activated Sepharose 4B coupled with HCV core protein. Blotting analyses of the eluates confirmed the presence of IgG molecules and IgM with RF activity.
Taken together, these data suggest that the structure of these cryoprecipitable ICs comprises HCV nucleocapsid protein specifically bound to IgG, which in turn is linked to IgM through its Fc portion.
To assess this conclusion, further experiments were carried out. Serum samples from three HCV-positive non-cryoglobulinaemic patients with consistent titres of circulating HCV core protein were mixed at 1 : 1 ratio with redissolved HCV-negative cryoglobulins. After cold storage, cryoprecipitates were purified and titres of HCV core protein were defined and compared with supernatants. Results reported in Table 4 demonstrate consistent enrichment of HCV core protein in the cryoprecipitates, suggesting strongly that its tendency to aggregate and precipitate probably occurs in the presence of IgM with RF activity. This assumption was supported convincingly by core protein enrichment of spun down pellets when the same sera were mixed with serum from a patient with rheumatoid arthritis and high titres of RF activity. No significant differences were found between pellets and the upper phases when they were mixed with a normal serum lacking RF activity.
Table 4.
.Recovering of HCV core protein after cryoprecipitation from HCV-positive sera mixed with HCV-negative samples
| HCV non-enveloped core protein (pg/ml) | |||
|---|---|---|---|
| (1) | (2) | (3) | |
| 1 HCV-negative cryoglobulins | |||
| Supernatant | 86 | 101 | 53 |
| Cryoprecipitate | 262 | 244 | 198 |
| 2 RF-positive serum | |||
| Upper phase | 126 | 110 | 93 |
| Pellet | 194 | 221 | 188 |
| 3 Normal serum | |||
| Upper phase | 219 | 185 | 156 |
| Pellet | 140 | 156 | 151 |
A further experiment was carried out mixing cryoglobulin-purified IgM (peak 1) with cryoglobulin-derived HCV nucleocapsid protein (peak 3). After overnight incubation at 37°C, increasing amounts of purified IgG (peak 2) were added and the mixture stored in the cold for 7 days. After centrifugation, pellets and the upper phases were tested for HCV core protein. Results depicted in Fig. 3 showed that cold precipitation of HCV core protein was correlated directly with the IgG concentration in the mixture. At concentration of 1 µg/ml of IgG, more than 80% of core protein was recovered in the pellet. Because purified IgG had specific anticore reactivity, it can be inferred that cryoprecipitation is function of its selective binding to the antigen in the presence of IgM molecules with RF activity. This was visible in the dynamics of the curve, in that no HCV core protein was found in the pellet in the absence or very low concentration of IgG. In addition, no significant changes occurred when an irrelevant IgG was added to the system.
Fig. 3.
Recovery of HCV core protein after cold-precipitation in the presence of increasing amounts of IgG with specific anticore reactivity ( ) or irrelevant IgG (○). HCV core protein was premixed with purified IgM with RF activity.
) or irrelevant IgG (○). HCV core protein was premixed with purified IgM with RF activity.
DISCUSSION
Cryoglobulins are found frequently in HCV infection [15] and constitute a striking feature in the variegated clinical spectrum of HCV chronic carriers [35] A typical picture of cryoglobulinaemic vasculitis can be recognized in approximately 10% of MC patients, who are at higher risk of malignant lymphoproliferative disorders, especially in the Mediterranean basin [36]. This suggests that the chronic lymphoproliferative process driven by HCV may progress towards a frank lymphoid neoplasia. However, MC should not be considered an in situ or occult B-cell lymphoma, in that recent evidence indicates that B-cell clonal expansion does not display the molecular features of a true neoplastic process [37].
The cryoglobulinaemic syndrome is probably the consequence of a pathogenetic noxa that acts upon the host's immune system, resulting in an altered regulation of peripheral immune response [38]. Cryoglobulins are the product of virus–host interactions, on which interdependent regulators confer biological activities and potential pathogenicity. Our experiments were set out to clarify the composition of cold-precipitating ICs and determine the involvement of HCV core protein as the relevant ligand.
Non-enveloped HCV core protein was detected in the bloodstream of 93·7% of our 32 type II MC patients and was found to precipitate by cold-separation of serum samples in most of them. The selective increase of HCV core protein and HCV RNA probably reflects the amount of HCV particles and strongly indicates that both constitute major antigen components of cold-insoluble ICs.
Regression analysis was not consistent with a direct relation between their concentrations in the cryoprecipitates. This, presumably, points to the presence of further viral components and the heterogeneous composition of ICs with cryoprecipitating properties. It was therefore not surprising that there was no direct relation between the amount of cryoprecipitated core protein and cryocrit levels.
As defined by immunofixation, Ig composition of cold-insoluble ICs included polyclonal IgG and monoclonal IgM. After the formation of cryoprecipitates and their removal by centrifugation, essentially half of the total IgM remained in the soluble fraction, suggesting that there were two or more IgM populations with different affinity for IgG molecules. Differences in the capacity to cryoprecipitate were also found for IgM RF molecules, in that almost one-third remained in solution. In contrast, a very small proportion of IgG molecules were involved in cryoprecipitation. Almost 10% of total IgG were identified in the cryoglobulins, suggesting specific interactions between the Ig components. All IgG subclasses were represented.
Chromatographic experiments allowed to confirm previous reports [10], in that cryoprecipitable ICs were composed of IgG and IgM. In immunoadsorbent experiments, both IgG and IgM were bound to HCV core protein coupled to CNBr-activated Sepharose 4B. Because IgM had RF activity and IgG showed specific reactivity against HCV core protein, it was concluded that cold-insoluble ICs are formed primarily by IgM RF linked to IgG, which is in turn bound to HCV core protein. Thus, our data indicate that HCV core protein is cold-precipitated in the context of ICs. Cold-dependent insolubility of HCV core protein seems the result of IgM RF activity, which acts as incomplete cryoglobulin, in that it precipitates at low temperature in the presence of IgG with specific anticore reactivity. The dynamics of cold-dependent insolubility demonstrates that the addition of an irrelevent IgG to an IgM RF/HCV core protein mixture was unable to precipitate the protein. This implies that a potentially functional RF repertoire may be selected positively by IgG anticore antibodies. Thus, it can be argued that cryoprecipitated IgM RFs represent distinct high-affinity molecules directed against preferential self-antigens [39]. Following their binding and when exposed to cold, RF molecules are subjected to a conformational change that is probably responsible for their precipitation [40].
Normal mean levels of C3 and C4 in the soluble phase corresponded to very low amounts (if any) in the cryoprecipitate, suggesting the existence of two virtually distinct microenvironments in which complement is differently activated. Complement is known to be a major interdependent regulator of IC size and composition. Complement binding to nascent ICs may decrease their size and maintain them in solution [30]. Compared with supernatant, significant differences of C1q and C1q binding activity were shown in unsolubilized ICs. Efficient engagement of C1q protein by cryoglobulins may be an important pathogenetic mechanism involved in the cryoglobulin-related pathway. In this context, it has been demonstrated recently that HCV core protein interacts directly with the globular domain of C1q receptor (gC1qR) (29). HCV core–gC1qR interaction has been assumed to play a critical role in modulating the T-cell immune response [41,42]. HCV core-induced inhibition of the cell responsiveness may represent a pathogenetic mechanism unable to suppress B-cell clone(s) which produce RF autoantibodies, generated by chronic antigen challenge in HCV-related type II MC [36]. The wide expression of gC1qR on the surface of both circulating blood cells [43] and endothelial cells [44] may favour their specific binding to HCV core protein-containing ICs. HCV core deposition has indeed been reported in the skin [45] and kidney [18] of MC patients.
In conclusion, our study indicates that non-enveloped HCV core protein is a constitutive antigen component of cryoglobulins in HCV-infected patients. Whether this protein is required for generating cryoglobulin-mediated tissue injury in vivo is being investigated currently.
Acknowledgments
We are indebted to Dr Doriana Lamanna and Mr Vittorio Padolecchia for their skilfull technical assistance; to Dr Sean Moroney (Ortho Clinical Diagnostics, Raritan, NJ, USA) for providing us with recombinant HCV proteins, and to Dr Dennis A. Carson (San Diego, La Jolla, CA, USA) for the generous gift of antiserum to 17·109 XId. This study was supported in part by ‘Associazione Italiana per la Ricerca sul Cancro’ (AIRC), by Consiglio Nazionale delle Ricerche (CNR), Project ‘Terapia preclinica molecolare in oncologia’, and by a grant from the Italian Ministry of University and Scientific and Technologic Research, National Project ‘Chronic liver damage induced by hepatitis C virus’.
REFERENCES
- 1.Gorevic PD, Kassab HJ, Levo Y, et al. Mixed cryoglobulinemia. clinical aspects and long-term follow-up of 40 patients. Am J Med. 1980;60:287–308. doi: 10.1016/0002-9343(80)90390-3. [DOI] [PubMed] [Google Scholar]
- 2.Miescher PA, Huang YP, Izui S. Type II cryoglobulinaemia. Semin Hematol. 1995;32:80–5. [PubMed] [Google Scholar]
- 3.Gorevic PD, Frangione B. Mixed cryoglobulinemia cross-reactive idiotypes. Implications for the relationship of MC to rheumatic and lymphoproliferative diseases. Semin Hematol. 1991;28:79–94. [PubMed] [Google Scholar]
- 4.Ferri C, Greco F, Longombardo G, et al. Antibodies to hepatitic C virus in patients with mixed cryoglobulinemia. Arthritis Rheum. 1991;34:1606–10. doi: 10.1002/art.1780341221. [DOI] [PubMed] [Google Scholar]
- 5.Agnello V, Chung RT, Kaplan LM. A role for hepatitis C virus infection in type II cryoglobulinemia. N Engl J Med. 1992;327:1490–5. doi: 10.1056/NEJM199211193272104. [DOI] [PubMed] [Google Scholar]
- 6.Dammacco F, Sansonno D. Antibodies to hepatitis C virus in essential mixed cryoglobulinaemia. Clin Exp Immunol. 1992;87:352–6. doi: 10.1111/j.1365-2249.1992.tb03001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–62. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 8.Major ME, Feinstone SM. The molecular organization of hepatitis C. Hepatology. 1997;25:1527–38. doi: 10.1002/hep.510250637. [DOI] [PubMed] [Google Scholar]
- 9.Hijikata M, Shimizu YK, Kato H, et al. Equilibrium centrifugation studies of hepatitis C virus: evidence for circulating immune complexes. J Virol. 1993;67:1953–8. doi: 10.1128/jvi.67.4.1953-1958.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sansonno D, Iacobelli AR, Cornacchiulo V, et al. Immunochemical and biomolecular studies of circulating immune complexes isolated from patients with acute and chronic hepatitis C virus infection. Eur J Clin Invest. 1996;26:465–75. doi: 10.1046/j.1365-2362.1996.162317.x. [DOI] [PubMed] [Google Scholar]
- 11.Liang TJ, Rehermann B, Seeff LD, Hoofnagle JH. Pathogenesis, natural history, treatment and prevention of hepatitis C. Ann Intern Med. 2000;132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 12.Cacoub P, Fabiani FL, Musset L, et al. Mixed cryoglobulinaemia and hepatitis C virus. Am J Med. 1994;96:124–32. doi: 10.1016/0002-9343(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 13.Pawlotsky JM, Roudot-Thoraval F, Simmonds P, et al. Extrahepatic immunologic manifestations in chronic hepatitis C and hepatitis C virus serotypes. Ann Intern Med. 1995;122:169–73. doi: 10.7326/0003-4819-122-3-199502010-00002. [DOI] [PubMed] [Google Scholar]
- 14.Peña LR, Nand S, De Maria N, Van Thiel DH. Hepatitis C virus infection and lymphoroliferative disorders. Dig Dis Sci. 2000;45:1854–60. doi: 10.1023/a:1005557506221. [DOI] [PubMed] [Google Scholar]
- 15.Dammacco F, Sansonno D, Piccoli C, Tucci FA, Racanelli V. The cryoglobulins: an overview. Eur J Clin Invest. 2001;31:628–38. doi: 10.1046/j.1365-2362.2001.00824.x. [DOI] [PubMed] [Google Scholar]
- 16.Dammacco F, Sansonno D, Cornacchiulo V, et al. Hepatitis C virus infection and mixed cryoglobulinaemia: a striking association. Int J Clin Lab Res. 1993;23:45–9. doi: 10.1007/BF02592281. [DOI] [PubMed] [Google Scholar]
- 17.Lunel F, Musset L, Cacoub P, et al. Cryoglobulinaemia in chronic liver disease: role of hepatitis C and liver damage. Gastroenterology. 1994;106:1291–30. doi: 10.1016/0016-5085(94)90022-1. [DOI] [PubMed] [Google Scholar]
- 18.Sansonno D, Gesualdo L, Monno C, Schena FP, Dammacco F. Hepatitis C virus-related proteins in kidney tissue from hepatitic C virus-infected patients with cryoglobulinemic membranoproliferative glomerulonephritis. Hepatology. 1997;25:1237–44. doi: 10.1002/hep.510250529. [DOI] [PubMed] [Google Scholar]
- 19.Kanto T, Hayashi N, Takehara T, et al. Density analysis of hepatitis C virus particle population in the circulation of infected hosts: implications for virus neutralization or persistence. J Hepatol. 1995;22:440–8. doi: 10.1016/0168-8278(95)80107-3. [DOI] [PubMed] [Google Scholar]
- 20.Tanaka T, Yasui K, Ohta Y, Hasegawa A, Tanaka S, Kahawa H. Simple fluorescent enzyme immunoassay for detection and quantification of hepatitis C viremia. J Hepatol. 1995;26:742–5. doi: 10.1016/0168-8278(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 21.Aoyagi K, Ohue C, Iida K, et al. Development of a simple and highly sensitive enzyme immunoassay for hepatitis C virus core antigen. J Clin Microbiol. 1999;37:1802–8. doi: 10.1128/jcm.37.6.1802-1808.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maillard P, Krawczynski K, Nitkiewicz J, et al. Non-enveloped nucleocapsids of hepatitis C virus in the serum of infected patients. J Virol. 2001;75:8240–50. doi: 10.1128/JVI.75.17.8240-8250.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bouvier-Alias M, Patel K, Dahari H, et al. Clinical utility of total HCV core antigen quantification: a new indirect marker of HCV replication. Hepatology. 2002;36:211–8. doi: 10.1053/jhep.2002.34130. [DOI] [PubMed] [Google Scholar]
- 24.Sabile A, Perlemuter G, Bono F, et al. Hepatitis C virus core protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology. 1999;30:1064–76. doi: 10.1002/hep.510300429. [DOI] [PubMed] [Google Scholar]
- 25.Large MK, Kittlesen DY, Hann YS. Suppression of host immune response by the core protein of hepatitis C virus: possible implications for hepatitis C virus persistance. J Immunol. 1999;162:931–8. [PubMed] [Google Scholar]
- 26.Komatsu F, Takasaki K. Determination of hepatitis C virus (HCV) core protein using a novel approach for quantitative evaluation of HCV viremia in anti-HCV positive patients. Liver. 1999;19:375–80. doi: 10.1111/j.1478-3231.1999.tb00065.x. [DOI] [PubMed] [Google Scholar]
- 27.Kurtz JB, Boxall F, Qusir N, Shirley J, Coleman D, Chandler C. The diagnostic significance of an assay for ‘total’ hepatitis C core antigen. J Virol Meth. 2001;96:127–32. doi: 10.1016/s0166-0934(01)00327-5. [DOI] [PubMed] [Google Scholar]
- 28.McLauchlan J. Properties of the hepatitis C virus core protein: a structural protein that modulates cellular processes. J Viral Hepatol. 2000;7:2–14. doi: 10.1046/j.1365-2893.2000.00201.x. [DOI] [PubMed] [Google Scholar]
- 29.Kittlesen DJ, Chianese-Bullock KA, Yao ZQ, Braciale TJ, Hahn T. Interaction between complement receptor gC1qR and hepatitis C virus core protein inhibits T-lymphocyte proliferation. J Clin Invest. 2000;106:1239–49. doi: 10.1172/JCI10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindahl G, Sjobring U, Johnsson E. Human complement regulators: a major target for pathogenic microorganisms. Curr Opin Immunol. 2000;12:44–51. doi: 10.1016/s0952-7915(99)00049-7. [DOI] [PubMed] [Google Scholar]
- 31.Carson DA, Fong SA. A common idiotope on human rheumatoid factors identified by a hybridoma antibody. Mol Immunol. 1983;20:1081–7. doi: 10.1016/0161-5890(83)90116-5. [DOI] [PubMed] [Google Scholar]
- 32.Urdea MS, Horn T, Fultz TJ. Branched DNA amplification multimers for the sensitive direct detection of human hepatitis C viruses. Nucl Acids Res Symp Sem. 1991;24:197–200. [PubMed] [Google Scholar]
- 33.Kwok S, Higuchi R. Avoiding false positive with PCR. Nature. 1989;339:237–8. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 34.Scheuer PJ. Classification of chronic viral hepatitis: a need for reassesment. J Hepatol. 1991;13:372–4. doi: 10.1016/0168-8278(91)90084-o. [DOI] [PubMed] [Google Scholar]
- 35.Gumber SC, Chopra S. Hepatitis C. Multifaceted disease. Ann Intern Med. 1995;123:615–20. doi: 10.7326/0003-4819-123-8-199510150-00008. [DOI] [PubMed] [Google Scholar]
- 36.Dammacco F, Sansonno D, Piccoli C, Racanelli V, D’Amore FP, Lauletta G. The lymphoid system in hepatitis C virus infection. autoimmunity, mixed cryoglobulinaemia, and overt B-cell malignancy. Semin Liver Dis. 2000;20:143–57. doi: 10.1055/s-2000-9613. [DOI] [PubMed] [Google Scholar]
- 37.Racanelli V, Sansonno D, Piccoli C, D’Amore FP, Tucci FA, Dammacco F. Molecular characterization of B cell clonal expansions in the liver of chronically hepatitis C virus-infected patients. J Immunol. 2001;167:21–9. doi: 10.4049/jimmunol.167.1.21. [DOI] [PubMed] [Google Scholar]
- 38.Tighe H, Warnatz K, Brinson D, et al. Peripheral deletion of rheumatoid factor B cells after abortive activation by IgG. Proc Natl Acad Sci USA. 1997;94:646–51. doi: 10.1073/pnas.94.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang DF, Olee T, Masuho Y, Matsumoto Y, Carson DA, Chen PP. Sequence analyses of three immunoglobulin G anti-virus antibodies reveal their utilization of autoantibody-related immunoglobulin Vh genes, but not V lambda genes. J Clin Invest. 1992;90:2197–208. doi: 10.1172/JCI116105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stone MJ, Fedak JE. Studies on monoclonal antibodies. II. Immune complex (IgM-IgG) cryoglobulinaemia: the mechanism of cryoprecipitation. J Immunol. 1974;113:1377–85. [PubMed] [Google Scholar]
- 41.Yao ZQ, Nguyen DT, Hiotellis AI, Hahn YS. Hepatitis C core protein inhibits human T lymphocyte responses by a complement-dependent regulatory pathway. J Immunol. 2001;167:5264–72. doi: 10.4049/jimmunol.167.9.5264. [DOI] [PubMed] [Google Scholar]
- 42.Yao ZQ, Ray S, Eisen-Vandervelde A, Waggoner S, Hahn YS. Hepatitis C virus: immunosuppression by complement regulatory pathway. Viral Immunol. 2001;14:277–95. doi: 10.1089/08828240152716547. [DOI] [PubMed] [Google Scholar]
- 43.Eggleton P, Ghebrehiwet B, Sastry KN, et al. Identification of a gC1q-binding protein on the surface of human neutrophils. J Clin Invest. 1995;95:1569–78. doi: 10.1172/JCI117830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lim BL, Reid KBM, Ghebrehiwet B, Peerschke EIB, Leigh LAE, Preissner KT. The binding for globular heads of complement C1q, gC1qR. Functional expression and characterization as a novel vitronectin binding factor. J Biol Chem. 1996;271:26739–44. doi: 10.1074/jbc.271.43.26739. [DOI] [PubMed] [Google Scholar]
- 45.Sansonno D, Cornacchiulo V, Iacobelli AR, Di Stefano R, Lospalluti M, Dammacco F. Localization of hepatitis C virus antigens in liver and skin tissues of chronic hepatitis C virus-infected patients with mixed cryoglobulinaemia. Hepatology. 1995;21:305–12. [PubMed] [Google Scholar]