Abstract
Autoantibodies to smooth muscle (SMA) and nuclear components (ANA) arise in the natural course of chronic infection with hepatitis C virus. In view of the growing evidence for ‘molecular mimicry’ as a mechanism of autoimmunity we investigated whether cross-reactive immune reactions between host smooth muscle/nuclear components and HCV antigens may contribute to the formation of SMA and ANA in chronic HCV infection. Computer-assisted protein database search methods were used to identify three smooth muscle (smoothelin698–717, myosin1035–1054, vimentin69–88) and three nuclear (matrin722–741, histone H2A11–30, replication protein A133-152) host antigens with the highest local sequence similarity to the HCV polyprotein and 20-mer peptides corresponding to these regions were constructed. Sera from 51 children with chronic HCV infection [median age: 8 (2–16); 27 boys], 26 SMA positive and five ANA positive, were tested for reactivity to the synthesized HCV peptides and their human homologues by enzyme linked immunosorbent assay (ELISA). Sera from patients with HBV infection and chronic liver disease of different aetiologies were used as controls. ‘Double reactivity’ to HCV peptides and smooth muscle/nuclear homologues was associated strongly with HCV infection (P < 0·001 for both). Humoral cross-reactivity was established as the basis for double recognition by competition ELISA. Double-reactivity to smooth muscle and HCV peptide antigens correlated with SMA positivity by indirect immunofluouresence (P = 0·05). Of 15 patients double-reactive to myosin1035–1054 and its HCV homologue, 13 recognized whole myosin by immunoblot. These results suggest that ANA and SMA in chronic HCV infection may arise, at least in part, as a consequence of cross-reactive immune responses to HCV and host smooth muscle/nuclear antigens.
Keywords: antibodies, antigens/peptides/, epitopes, autoimmunity, infectious immunity-virus
INTRODUCTION
Infection with hepatitis C virus (HCV) becomes persistent in 80% of patients and leads to chronic liver disease (CLD) in a substantial proportion [1]. A large population-based study demonstrated convincingly a correlation between the presence of non-organ-specific autoantibodies (NOSA), mainly antismooth muscle (SMA) and antinuclear (ANA) antibodies, and chronic liver disease in HCV-positive individuals [2]. We and others have demonstrated the development of SMA and ANA to be part of the natural course of chronic infection with HCV independent of treatment with interferon (IFN)-α [3–6]. Despite the repeated demonstration of ANA and SMA as a prominent feature of chronic HCV infection, the mechanisms responsible for their genesis remain poorly understood.
‘Molecular mimicry’ between viral and self-antigens leading to immunological cross-reactivity and the emergence of autoimmunity is well documented, both in experimental systems and human disease [7–10]. Two powerful models of pathogen-driven autoimmunity provide further strong support for molecular mimicry as an important mechanism in the abrogation of self-tolerance. First, immunological cross-reactivity between outer surface protein A of Borrelia burgdorferi and human leucocyte function-associated antigen-1 has been demonstrated convincingly to be central in the pathogenesis of treatment-resistant Lyme arthritis [11]. In a second study of a murine model of herpes stromal keratitis, corneal infection with herpes virus type-1 (HSV-1) leads to cross-reactive cellular autoimmunity between the UL6 protein of HSV-1 and corneal antigens, resulting in the destruction of corneal tissue [12]. Moreover, we have provided evidence for humoral cross-reactivity between hepatitis B virus DNA polymerase and human smooth muscle and nuclear components as a mechanism for the emergence of ANA and SMA in patients with chronic hepatitis B virus (HBV) infection [13].
We hypothesized that molecular mimicry between HCV and human smooth muscle and nuclear antigens may contribute to the genesis of SMA and ANA in chronic HCV infection. By scanning protein databases for regional sequence similarities between the HCV polyprotein and putative antigenic targets of ANA and SMA, homologous sequences were identified and peptides corresponding to these regions were constructed and tested as targets of a cross-reactive immune response.
MATERIALS AND METHODS
Patients
Fifty-one patients with chronic liver disease due to HCV infection were investigated (median age: 8 years, range 2–16). All patients were HCV RNA (Amplicor, Hoffmann la Roche, Basel, Switzerland) and anti-HCV antibody (United Biomedical Inc., Hauppage, New York, USA and Sanofi Pasteur, Marnes-la-Coquette, France) positive. Twenty-nine patients were treated with IFN-α and 22 were untreated.
The autoantibody profile of these patients has been reported elsewhere [3]. Autoantibodies to nuclear (ANA), smooth muscle (SMA), liver kidney microsomal type 1 (LKM1), mitochondrial, liver cytosolic antigen type 1 and gastric parietal cell (GPC) were tested at a screening dilution of 1/10 in phosphate buffer saline (PBS) using frozen rat liver, kidney and stomach as substrate, as described previously [14]. At the time of investigation, 27 patients were ANA and/or SMA positive: one was ANA positive (titre: 1/10), 22 were SMA positive (titre range: 1/10–1/40; median: 1/10) and four were ANA/SMA double-positive (titre range: 1/10–1/40; median: 1/10).
Sera from 92 patients HCV negative patients with other chronic liver disorders were used as pathological controls (median age: 10·5 years, range 2–25). Of these, 24 had chronic hepatitis B virus (HBV) infection. All were HBV DNA (dot-blot assay; Abbott, Chicago, IL, USA) and HBeAg positive (microparticle enzyme immunoassay, Abbott). Two patients were ANA positive (both at a titre of 1/40), eight were SMA positive (titre range: 1/10–1/40; median: 1/10) and two were ANA/SMA double-positive (titre of 1/10). Thirty-six had autoimmune liver disease: 24 autoimmune hepatitis (AIH), diagnosed according to international criteria [15] (12 ANA and/or SMA positive and 12 LKM1 positive) and 12 ANA/SMA positive sclerosing cholangitis [autoimmune sclerosing cholangitis (ASC)] with characteristic cholangiographic changes [16]. Autoantibody titres more than 1/10 were observed in 33 patients. Twelve patients (six AIH, six ASC) were ANA (titre range: 1/40–1/10 240; median: 1/640) and SMA positive (titre range: 1/20–1/2560; median: 1/160), six (three AIH and three ASC) were ANA positive (titre range: 1/80–1/5120; median: 1/480), four (two AIH, two ASC) were SMA positive (1/10 in one, 1/40 in two and 1/640 in one) and 11 were LKM1 positive (titre range: 1/20–1/10 240; median: 640). Ten children with Wilson's disease [17], 11 with Alagille's syndrome and 11 with alpha 1 antitrypsin deficiency (AATD) (PIZZ phenotype by isoelectric focusing) were also tested. Six patients with Wilson's disease were autoantibody positive, four for ANA (1/10 in one, 1/40 in two and 1/640 in one) and two for SMA (both at 1/10).
Sera from 24 healthy children [median age: 9 (2–14); 16 boys] age-matched with the children with chronic HCV infection were tested as controls.
The study was undertaken following approval by the local ethics committee.
Protein database search and analysis
HCV polyprotein sequences representing the most common genotypes in our patient cohort were extracted from the SWISSPROT and PIR protein databases and comprised (listed by accession number): PIR: A39166 (subtype 1a), PIR: A39253 (subtype 1b), SWISSPROT: P26660 (subtype 2a), SWISSPROT: P26661 (subtype 2b) and PIR: A41546 (subtype 1a). Each polyprotein sequence was serially divided into 20 amino acid segments, each overlapping the preceding segment by five residues using a custom-made algorithm written in perl. The resulting 20 amino acid sequences were used to perform a blast local homology search of a subdatabase of all human smooth muscle and nuclear protein sequences registered in the PIR and SWISSPROT databases. Six HCV polyprotein/human nuclear and smooth muscle sequence pairs with the highest local sequence similarity were selected for synthesis (Fig. 1). The physicochemical properties of the selected peptides were assessed using the antigenicity indices of Hopp and Woods [18], Parker [19] and Welling [20], using the PBIL server at http://www.pbil.ibcp.fr.
Fig. 1.
Amino acid sequence homology between the hepatitis C virus polyprotein, sequences A39166 (genotype 1a), A41546 (genotype 1a), A39253 (genotype 1b), P26660 (genotype 2a), P26661 (genotype 2b), and (a) human nuclear proteins (matrin-3, histone H2A, replication protein A) and (b) smooth muscle proteins (smoothelin, smooth muscle myosin, vimentin). The sequences in bold represent homologous peptides constructed for testing by ELISA. Amino acids are shown in standard single-letter code; a colon (:) indicates identical residues; a full stop (.) indicates conservative substitutions; a dash (–) indicates identical residues to the HCV subtype that was synthesized.
Peptide synthesis
Twelve 20-mer amino acid peptides containing the relevant homologues between the HCV polyprotein and the antigenic targets of ANA and SMA (Fig. 1) and a 20-mer randomly generated control peptide, HEDYVNQSLRPTPLEISVRA, were synthesized using fmoc chemistry (431 A Synergy, Perkin Elmer, Warrington, UK) and cleaved as described previously [21]. Peptide purity was then checked by high performance liquid chromatography (Perkin Elmer).
Elisa
Reactivity to the control and synthetic peptides was determined using a previously established enzyme linked immunosorbent assay (ELISA). Briefly, peptides diluted in phosphate buffer saline (PBS) to a concentration of 50 µg/ml were coated onto 96-well ELISA microtitre plates (Flow Laboratories, Herts, UK) overnight at 4°C. Plates were washed three times in 1% Tween-20 (Sigma Chemical Co, Poole, Dorset, UK), incubated with 5% bovine serum albumin for 1 h and followed by the addition of 100 µl of patient and control sera, diluted 1/100 in PBS-Tween, for 2 h at 37°C. After washing, 100 µl of horseradish peroxidase conjugated rabbit antihuman IgG (DAKO Ltd, Bucks, UK) were added, diluted 1/2000 in PBS-Tween and incubated for 1 h at 37°C. The reaction was developed using 100 µl of substrate [0·4 mg/ml o-phenylediamine in citrate phosphate buffer (pH 5·0) containing 4 µl of 3% hydrogen peroxide], terminated with 50 µl of 3 m H2SO4 and optical density read in a Molecular Devices (Hayward Heath, West Sussex, UK) microplate reader at 490 nm. Patient and control sera were tested in triplicate. The highest value of mean OD490[test peptide]/OD490[control peptide] + 3 standard deviations for the 24 healthy children was determined as 1·4, and reaction for a given peptide was considered positive when OD490[test peptide]/OD490[control peptide] was more than 1·5.
To ensure consistency between assays four sera, including two with negative and two with positive reactivity, were used as reference controls in each assay. Mean coefficient of variation values ranged from 1·1% to 8·5% (intra-assay) and 5% to 12% (interassay).
Inhibition studies
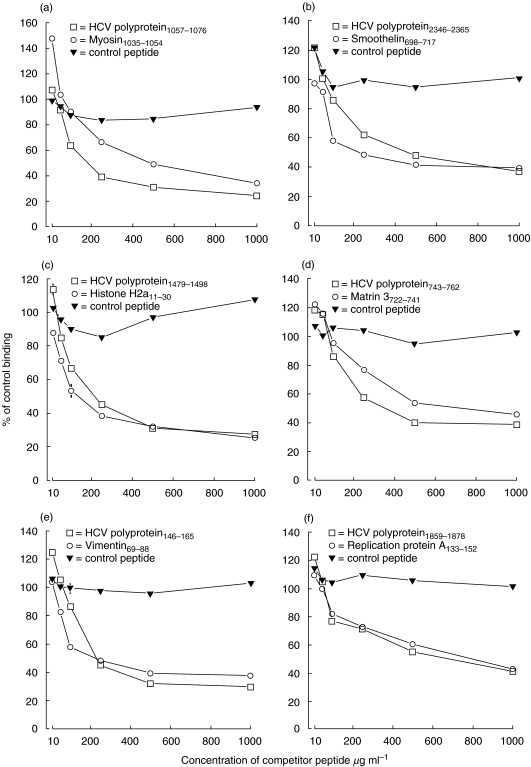
Competition ELISAs were performed using HCV polyprotein peptides as competitor in the liquid phase (Fig. 2). HCV polyprotein peptide was coated onto 96-well ELISA microtitre plates (Flow Laboratories) at a concentration of 50 µg/ml and incubated overnight at 4°C. Non-specific reactive sites were blocked by incubating with 5% BSA in PBS at 37°C for 1 h. Solutions of HCV polyprotein peptides, the corresponding self-homologue and the control peptide were prepared at concentrations of 10, 50, 100, 250, 500 and 1000 µg/ml in PBS. Test serum was diluted in these solutions to a final dilution of 1/100 and following incubation at 37°C for 2 h, 100 µl aliquots of the peptide/antibody mixture were transferred to the wells of the precoated 96-well ELISA microtitre plates in triplicate. This was performed for all HCV polyprotein/self-peptide pairs (Fig. 1). Antibody detection was then carried out under identical conditions to those described above.
Fig. 2.
(a–f) Inhibition studies. Inhibition of antibody binding to HCV polyprotein peptide by preincubation of serum with the HCV polyprotein peptide (□), corresponding self-homologue (○) or control peptide (▾). Antibody binding is represented as a percentage (± s.e.m.) of binding to the HCV polyprotein peptide in the absence of a competitor peptide. Where error bars are not shown, errors are smaller than the data symbols.
Immunoblot
Antibodies to human myosin were detected by immunoblot using sera of 15 HCV-positive patients who had double reactivity to HCV1057–1076 and corresponding self-homologue, myosin1035–1054 by ELISA. Myosin was loaded onto 7·5% polyacrylamide gels (Bio-Rad Laboratories, Hemel Hempstead, UK) (1 µg/well). Following electrophoresis proteins were blotted onto nitrocellulose filters in a semidry electrophoretic transfer cell (Bio-Rad Laboratories) and non-specific binding was blocked by 5% skimmed milk in TNT buffer (10 mmol/l Tris buffer pH 8·0 containing 0·15 mmol/l NaCl and 0·05% Tween 20) for 1 h. Filter strips were incubated with the patients’ sera for 2 h at a dilution of 1/200 and antigens targeted by the patients’ antibodies were visualized using peroxidase-conjugated rabbit antihuman IgG (Dako Ltd, Bucks, UK). To test antigen and disease specificity of the immunoblot assay and its correlation with reactivity by ELISA, serum from 11 patients with chronic HCV infection, nine patients with chronic HBV infection, four patients with type II AIH positive for LKM-1 antibodies, three patients with AATD, two patients with type I AIH, three patients with ASC and three healthy individuals, all negative for reactivity to myosin1035–1054 by ELISA, were investigated for reactivity to native myosin by immunoblot (Fig. 3).
Fig. 3.
Immunoblot analysis of serum reactivity against smooth muscle myosin. Myosin (1 µg/well) was subjected to SDS-PAGE, and sera were applied at 1/200 dilution. Lanes 1–4, HCV-positive patients double-reactive to HCV1057–1076 and myosin1035–1054 peptides, a band is present at ∼200 Kda corresponding to myosin; lane 5, HCV-positive patient reactive to HCV1057–1076 but not to myosin1035–1054; lane 6, HBV-positive patient unreactive to HCV and myosin peptides; lane 7, healthy subject unreactive to HCV and myosin peptides; lanes 5–7 are negative.
Statistical analysis
Comparison between categorical values was performed using the χ2 test or Fisher's exact probability test for small numbers as appropriate. A probability (P) value less than 0·05 was considered significant.
RESULTS
Protein database search
The protein database search revealed six human nuclear and smooth muscle proteins that had high amino acid sequence similarity to the HCV polyprotein. Three human nuclear proteins, namely, histone H2A.Z (SWISSPROT: P17317), matrin 3 (SWISSPROT: P43243) and replication protein A (SWISSPROT: P15927), and three human smooth muscle proteins, namely (accession number in parentheses), smooth muscle myosin (SWISSPROT: P35749), vimentin (SWISSPROT: P08670) and smoothelin (SWISSPROT: P53814), were identified with sequence similarities ranging from 70% to 90% with their respective HCV polyprotein homologues, spanning 10–12 amino acids (Fig. 1). All selected HCV and human peptide sequences displayed favourable antigenicity indices and correlated well across the three algorithms used (data not shown).
ELISA: reactivity to HCV polyprotein peptides
Reactivity to one of the six HCV polyprotein peptides (Table 1) was significantly more common in chronic HCV-positive patients [51 (100%)] compared with patients with other chronic liver disorders [23 (25%)] (nine with chronic HBV infection, five with LKM1 AIH, four with ANA/SMA AIH, three with ASC, one with Wilson's disease and one with AATD) or in healthy controls [0%] (P < 0·001 for all).
Table 1a.
Frequency, n (%), of reactivity to the HCV polyprotein and identified nuclear homologues in patients with chronic HCV infection, other chronic liver disorders and in normal healthy children
| Chronic HCV (n = 51) | Chronic HBV (n = 24) | ANA/SMA AIH (n = 12) | LKM1 AIH (n = 12) | ANA/SMA ASC (n = 12) | Wilson’s disease (n = 10) | Alagille’s syndrome (n = 11) | AATD (n = 11) | Healthy controls (n = 24) | |
|---|---|---|---|---|---|---|---|---|---|
| HCV-pol1479–1498 | 41 (80) | 2 (8) | 1 (8) | 3 (25) | 0 | 0 | 0 | 0 | 0 |
| Histone11–30 | 25 (49) | 2 (8) | 3 (25) | 0 | 3 (25) | 0 | 0 | 0 | 0 |
| HCV-pol1479–1498 and histone11–30 | 19 (37) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HCV-pol743–762 | 20 (39) | 3 (12) | 1 (8) | 0 | 0 | 1 (10) | 0 | 0 | 0 |
| Matrin722–741 | 39 (76) | 13 (54) | 3 (25) | 1 (8) | 0 | 1 (10) | 0 | 1 (9) | 0 |
| HCV-pol743–762 and matrin722–741 | 17 (33) | 0 | 0 | 0 | 0 | 1 (10) | 0 | 0 | 0 |
| HCV-pol1859–1878 | 35 (69) | 4 (16) | 1 (8) | 4 (32) | 2 (17) | 0 | 0 | 1 (9) | 0 |
| Replication protein133–152 | 32 (63) | 3 (12) | 0 | 0 | 3 (25) | 0 | 0 | 0 | 0 |
| HCV-pol1859–1878 and replication protein133–152 | 20 (39) | 0 | 0 | 0 | 2 (17) | 0 | 0 | 0 | 0 |
n= number of patients; HCV = hepatitis C virus infection; HBV = hepatitis B virus infection; AIH = autoimmune hepatitis; ANA = anti nuclear antibody; SMA = smooth muscle antibody; LKM1 = liver kidney microsomal type 1 autoantibody; ASC = autoimmune sclerosing cholangitis; AATD = alpha 1 antitrypsin deficiency.
The majority (80%) of the patients with chronic HCV infection showed reactivity to the HCV polyprotein1479–1498 peptide, an amino acid sequence in the non-structural (NS) protein 3 region of the HCV. The frequency of binding to the other individual HCV polyprotein peptides – HCV polyprotein698–717 (amino acid region located in the core), HCV polyprotein743–762 (E2 envelope), HCV polyprotein1057–1076 (NS3 region), HCV polyprotein1859–1878 (NS4B), HCV polyprotein2346–2365 (NS5A) − was 35%, 39%, 43%, 69% and 71%, respectively. Thirty-six (71%) HCV positive patients were reactive to at least three of the six HCV polyprotein peptides. Infection with a particular HCV genotype was not associated with reactivity to a specific HCV polyprotein peptide.
Of the 92 pathological controls, 16 (five with chronic HBV infection, four with ANA/SMA AIH, three with LKM1 AIH, three with ASC and one with AATD) were reactive to one HCV polyprotein peptide, six (four with chronic HBV infection, one with LKM1 AIH, one with Wilson's disease) were reactive with two HCV polyprotein peptides and one (LKM1 AIH) was positive for three of the six HCV polyprotein peptides. Four LKM1 positive AIH patients were reactive to the HCV polyprotein1859–1878 peptide.
Reactivity to nuclear peptides
Reactivity to either the histone H2A11–30, matrin 3722–741 or replication protein133–152 peptides was significantly more common in chronic HCV positive patients [49 (96%)] compared with patients with other chronic liver disorders [29 (32%)] (15 with chronic HBV infection, six with ANA/SMA AIH, five with ASC, one with LKM1 AIH, one with Wilson's disease and one with AATD) or in healthy controls (P < 0·001 for all). Of the 49 patients with chronic HCV infection who were reactive to one of the three nuclear peptides, five (10%) were ANA positive at a titre ≥ 1/10 whilst 44 (90%) were ANA negative. Of the 29 reactive patients with other chronic liver disorders, 12 (four with ANA/SMA AIH, four with ASC, three with chronic HBV infection and one with Wilson's disease) were ANA positive and 17 were ANA negative.
Reactivity to smooth muscle peptides
Reactivity to myosin1035–1054, vimentin69–88 and smoothelin698–717 peptides was significantly more common in HCV-positive patients [43/51 (84%)] compared with patients with other chronic liver disorders [33/92 (36%)] or in healthy controls (0%). Of the 43 chronic HCV-infected patients who were reactive either to myosin1035–1054, vimentin69–88 and smoothelin698–717 peptides, 23 (53%) were SMA positive while 20 (47%) were SMA negative. Of the 33 patients with other chronic liver disorders (nine with ASC, seven with chronic HBV infection, seven with ANA/SMA AIH, four with Alagille's syndrome, three with Wilson's disease, two with LKM1 AIH and one with AATD) reactive to one of the smooth muscle peptides, 12 were SMA positive and 21 were SMA negative.
Five (42%) patients with ASC were reactive to the smoothelin698–717 peptide. Four (33%) patients with ANA/SMA positive AIH and seven (58%) with ASC were reactive to the myosin1035–1054 peptide. Two (8%) patients with chronic HBV infection reacted with the myosin1035–1054 peptide compared with 30 (59%) patients with chronic HCV infection.
Double reactivity to HCV polyprotein peptides and self-homologues
Double reactivity to the HCV polyprotein and to the histone H2A11–30, matrin 3722–741 and replication protein A133–152 peptides was noted in 19 (37%), 17 (33%) and 20 (39%) patients with chronic HCV infection, respectively, compared with three (3%) patients with other chronic liver disorders (two with ASC, one with Wilson's disease) and none of the healthy controls (P < 0·001 for all). Among the patients with chronic HCV infection, double reactivity to the human nuclear and smooth muscle peptides and their corresponding HCV homologues was observed similarly in autoantibody-positive and autoantibody-negative patients (Table 3). Double reactivity to the HCV polyprotein and to myosin1035–1054, vimentin69–88 and smoothelin698–717 peptides was observed in 19 (37%), eight (16%) and 27 (53%) patients, respectively, with chronic HCV infection compared with none of the patients with other chronic liver disorders or in healthy controls (P < 0·001 for all).
At a cut-off titre of 1/20 for SMA, double reactivity to one of the HCV polyprotein and smooth muscle peptide pairs was significantly more common in SMA positive than in SMA negative patients with chronic HCV infection [10/11 (91%) versus 24/40 (60%), P= 0·05].
Viral genotype and reactivity to HCV polyprotein peptides and self-homologues
Viral genotype was available for 46 of the 52 patients with HCV infection. Forty-two of the 46 (91%) patients were infected by genotypes 1 or 2. Although no broadly applicable correlations were observed between HCV genotype and peptide reactivity, the frequency of antibodies directed to smoothelin698–717 tended to be higher in patients infected by HCV genotype 1 than those infected by other genotypes (72%versus 50%, P= 0·18), while antibodies directed to replication protein133–152 were more frequent in patients infected by HCV genotype 1 than those infected by other genotypes (64%versus 30%, P= 0·056). As both smoothelin698–717 and replication protein133–152 share greater homology with genotype 1 HCV peptides than genotype 2 peptides, this would be consistent with antibody cross-reactivity between HCV and self-epitopes arising as a function of sequence similarity.
Interferon treatment and reactivity to HCV polyprotein peptides and self-homologues
The prevalence of autoantibodies to nuclei (ANA), smooth muscle (SMA), gastric parietal cell (GPC) and/or liver kidney microsomal type 1 (LKM-1) was not statistically different between the treated and untreated patients with chronic HCV infection, although they tended to be higher in treated versus untreated patients (90%versus 68%, P= 0·12). However, conventional autoantibody titres (ANA, SMA, GPC, LKM-1) were similar in treated and untreated patients. Among the cross-reactive antibodies directed to the 12 peptide homologues, the titres of antibodies against myosin1035–1054 and its viral homologue HCV-pol1057–1076 were significantly higher in untreated patients than in treated patients. The titre of antibodies against the other 10 peptides was similar in the two subgroups (Table 2). As viral load is lower in the treated population this would suggest, at least in the case of smooth-muscle myosin, that cross-reactivity to self-antigens is driven by HCV antigen dose.
Table 2.
Titres of antibodies to HCV-polyprotein peptides and self-homologues in treated and untreated patients with HCV infection
| Antibody titre (OD490 ± s.d.) | ||||
|---|---|---|---|---|
| Tissue origin of self-homologue | Peptide homologues | Treated | Untreated | P (treated versus untreated) |
| Nuclear | Histone11–30 | 2·11 ± 0·61 | 2·09 ± 0·49 | 0·97 |
| HCV-pol1479–1498 | 2·33 ± 0·63 | 2·72 ± 0·95 | 0·14 | |
| Matrin722–741 | 2·16 ± 0·63 | 2·54 ± 1·01 | 0·16 | |
| HCV-pol743–762 | 1·89 ± 0·43 | 1·98 ± 0·34 | 0·62 | |
| Replication protein133–152 | 2·22 ± 0·7 | 2·34 ± 0·87 | 0·69 | |
| HCV-pol1859–1878 | 2·13 ± 0·5 | 2·16 ± 0·36 | 0·87 | |
| Smooth muscle | Myosin1035–1054 | 1·81 ± 0·29 | 2·33 ± 0·48 | 0·001 |
| HCV-pol1057–1076 | 1·67 ± 0·21 | 2·11 ± 0·42 | 0·005 | |
| Vimentin69–88 | 1·70 ± 0·27 | 1·95 ± 0·39 | 0·19 | |
| HCV-pol146–165 | 2·05 ± 0·43 | 1·76 ± 0·22 | 0·11 | |
| Smoothelin698–717 | 1·84 ± 0·34 | 1·88 ± 0·35 | 0·8 | |
| HCV-pol2346–2365 | 1·70 ± 0·17 | 1·66 ± 0·19 | 0·52 | |
Bold = P < 0·05.
Inhibition studies
Antibody binding to the individual HCV polyprotein peptides was inhibited by 60–80% by preincubation with the HCV polyprotein peptide or self-homologue. No inhibition was observed with the control peptide (Fig. 2).
Immunoblot
Using myosin as target antigen, a ∼200 KD band was observed in 13 of 15 sera of patients with chronic HCV infection who were double-positive to the HBV-pol1057–1076 peptide and the human homologue, myosin1035–1054, by ELISA. None of the 32 sera from patients with chronic HCV/HBV infection, autoimmune hepatitis, and other chronic liver disease, negative for reactivity to myosin1035–1054 by ELISA, recognized native myosin on Western blot (Fig. 3).
DISCUSSION
We provide evidence for immunological cross-reactivity as a mechanism for the generation of SMA and ANA in chronic HCV infection. Our argument rests on six key criteria: (1) local sequence homology between HCV and human smooth-muscle and nuclear components, which provides a structural framework for cross-reaction; (2) the potential of homologous HCV peptides to serve as B-cell epitopes; (3) simultaneous recognition of HCV and self-peptides by HCV sera; (4) specificity of the antiself response to HCV infection; (5) the capacity of HCV peptides to competitively inhibit antibody recognition of homologous self-peptides; and (6) demonstration of reactivity to whole self-antigen as a further measure of the physiological specificity of the autoimmune response.
Computer-assisted protein database search revealed six human proteins with high local sequence similarities with the HCV polyprotein: three nuclear proteins, namely, histone H2A.Z [22], matrin 3 [23] and replication protein A [24] and three smooth muscle proteins, namely, smooth-muscle myosin [25], vimentin [26] and smoothelin [27]. Of the nuclear proteins, histone H2A.Z and matrin 3 serve as structural components within the nucleus, while replication protein A is involved in DNA replication. The smooth muscle protein myosin is involved in muscle contraction while smoothelin and vimentin are cytoskeletal proteins. Smoothelin is exclusively expressed in contractile smooth muscle cells and to date is the most specific marker for smooth muscle described.
Of the six HCV polyprotein peptides selected for investigation, HCV1479–1498 is recognized most commonly in our patient sample (80%). This region was investigated first by Yuki et al., who identified an almost identical peptide sequence, HCV1479–1497, to be an important antigenic region of the HCV NS3 antigen [28]. This finding was confirmed subsequently by the identification of a major humoral epitope, HCV1460–1532, reported to be recognized by 50% of HCV positive blood donors [27]. The HCV NS5a peptide, HCV2346–2365, is the second most commonly recognized epitope in our study (71%). It lies close to the carboxyl-end of a previously reported major humoral epitope of the NS5 antigen, HCV2284–2329, that was found to harbour the majority of antibody reactivity directed towards the NS5 antigen [29]. However, this study did not investigate the downstream region which contains HCV2346–2365, a highly hydrophilic sequence, that we demonstrate to be an extension of the major NS5 epitope. Although HCV146–165 is reported to be a subdominant humoral epitope of the core antigen, remarkably it contains, in its entirety, the dominant T-cell helper (Th1) epitope of the core antigen, HCV141–160 [30]. Thus, it is conceivable that a cross-reactive T-cell helper response to HCV146–165 could give rise, via T–B reciprocity [31], to antibodies that cross-react with vimentin69–88. HCV peptides HCV743–762 (E2), HCV1859–1878 (NS4b) and HCV1057–1076 (NS3), investigated in our study, lie within regions that, to our knowledge, have not been investigated at high resolution for the presence of linear epitopes and therefore represent preliminary evidence for the antigenic potential of these regions. The crystal structure of the NS3 protease locates HCV1057–1076 (homologous to myosin1035–1054) within the relatively solvent accessible A1 strand at the NH-terminal region of the protein close to His57 (part of the ‘catalytic triad’ of the enzyme), allowing the possibility of antibody binding to this region within the native protein [32].
Reactivity to the nuclear and smooth-muscle peptides homologous to HCV antigens is strongly associated with chronic HCV infection (P < 0·001 for all), suggesting that these autoantibodies are generated as a specific response to HCV infection. Moreover, double-reactivity to HCV and homologous self-peptides is found only in the the context of HCV infection. The reduction in cross-reactive antibody titres on IFN treatment observed with myosin (Table 2) suggests that, at least for this antigen, cross-reactivity is driven by HCV antigen load. Antibody recognition of self-peptide homologues is cross-reactive with the corresponding HCV peptides, as demonstrated by inhibition studies. Double-reactivity to a smooth-muscle peptide and its HCV homologue was significantly more common in those patients positive for SMA by immunofluorescence (P = 0·05). Finally, the capacity of sera cross-reactive to smooth-muscle myosin and its HCV homologue to recognize specifically whole smooth-muscle myosin is demonstrated by immunoblot. It will be important to confirm recognition of whole self-antigens directly by LKM1 antibodies specific for HCV viral homologues using competition assays.
Unexpectedly, a high proportion (54%) of patients with chronic infection caused by another hepatotropic virus, hepatitis B virus, reacted to the matrin 3722–741 self-peptide but not to its HCV homologue – HCV polyprotein743–762 (12%). This raises the possibility of cross-reactivity between matrin 3722–741 and a homologous region of HBV. A local homology search of the HBV polyprotein using matrin 3722–741 as template reveals amino acid sequence 303–308 (303LENGIK) of matrin 3 shares four identities and one conserved substitution with the HBV DNA polymerase residues 132–137 (132LDKGIK) but has only three identities with the HCV polyprotein residues 752–758 (752LENLVI). Thus, cross-reactivity between the homologous regions of HBV DNA polymerase and matrin 3 may explain the high reactivity to the matrin 3722–741 peptide in HBV-positive patients but not its HCV homologue. Similarly, cross-reactive immunity may also account for the observed reactivity of ANA/SMA positive type 1 AIH patients to the myosin1035–1054 peptide, as myosin (282ATQQEL) shares homologous sequences with actin (313AMQQKL), a major autoantigen in ANA/SMA positive type 1 AIH [33]. [Amino acid annotation in the text: amino acids in standard single-letter code; bold: identical residues; underlined: conservative substitutions.] Although ANA/SMA positive ASC patients also had a high reactivity to myosin1035–1054 (58%) and smoothelin698–717 (42%) peptides, but not to their HCV polyprotein homologues, a similar analysis cannot be performed for this subgroup as the target autoantigens of ANA/SMA positive ASC remain to be characterized. Regardless of the mechanism of production of antibodies to self-components in chronic HBV infection, AIH or ASC, in contrast to the chronic HCV patient group, the absence of double-reactivity to the respective HCV homologues underscores the role of structural similarity as the basis of reactivity to HCV and self-homologues in the context of HCV infection.
While autoimmunity to smooth muscle and nuclear components may arise through a cross-reactive mechanism induced by HCV infection, progression to overt autoimmune disease is not observed. The presence of strongly suppressive mechanisms preventing the development of autoimmune disease, despite humoral autoimmunity, is borne out in a transgenic murine model that constitutively expresses hepatitis B virus surface antigen (HBsAg) (on hepatocytes) under control of the mouse albumin promoter. These transgenic mice do not develop spontaneous autoimmunity although repetitive vaccination with HbsAg/CFA/HBV leads to a T-dependent antisAg ‘autoantibody’ response. T cell tolerance, however, remains intact and overt autoimmune disease does not ensue [34]. Our finding that cross-reactive humoral immunity between HCV and self contributes to the generation of autoimmunity to smooth-muscle and nuclear components together with the recent demonstration, by Lenzi et al. [2], that NOSA are strongly associated with active viral replication and heightened hepatic dysfunction in chronic HCV infection suggests that induction of autoimmunity may allow viral escape, as the anti-HCV response is diverted from the pathogen to host components, possibly contributing to the establishment of persistent infection.
Table 1b.
Frequency, n (%), of reactivity to the HCV polyprotein and identified smooth muscle homologues in patients with chronic HCV infection, other chronic liver disorders and in normal healthy children
| Chronic HCV (n = 51) | Chronic HBV (n = 24) | ANA/SMA AIH (n = 12) | LKM1 AIH (n = 12) | ANA/SMA ASC (n = 12) | Wilson’s disease (n = 10) | Alagille’s syndrome (n = 11) | AATD (n = 11) | Healthy controls (n = 24) | |
|---|---|---|---|---|---|---|---|---|---|
| HCV-pol1057–1076 | 22 (43) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Myosin1035–1054 | 30 (59) | 2 (8) | 4 (33) | 0 | 7 (58) | 2 (20) | 3 (27) | 0 | 0 |
| HCV-pol1057–1076 and myosin1035–1054 | 19 (37) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HCV-pol146–165 | 18 (35) | 4 (16) | 0 | 1 (8) | 1 (8) | 0 | 0 | 0 | 0 |
| Vimentin69–88 | 19 (37) | 2 (8) | 0 | 1 (8) | 2 (17) | 1 (10) | 0 | 1 (9) | 0 |
| HCV-pol146–165 and vimentin69–88 | 8 (16) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HCV-pol2346–2365 | 36 (71) | 0 | 1 (8) | 0 | 0 | 1 (10) | 0 | 0 | 0 |
| Smoothelin698–717 | 32 (63) | 3 (12) | 3 (25) | 1 (8) | 5 (42) | 0 | 1 (9) | 0 | 0 |
| HCV-pol2346–2365 and smoothelin698–717 | 27 (53) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
n = number of patients; HCV = hepatitis C virus infection; HBV = hepatitis B virus infection; AIH = autoimmune hepatitis; ANA = anti nuclear antibody; SMA = smooth muscle antibody; LKM1 = liver kidney microsomal type 1 autoantibody; ASC = autoimmune sclerosing cholangitis; AATD = alpha 1 antitrypsin deficiency.
Acknowledgments
At the time of this study, GVG was in support of a grant from Schering Plough Limited (Hertfordshire, UK). GMV is supported by the Children's Liver Disease Foundation (CLDF), Digbeth, Birmingham, UK. The healthy children were recruited through the CLDF, Gillingham and Kent branch, UK.
REFERENCES
- 1.Van der Poel CL, Cuypers HT, Reesink HW, et al. Confirmation of hepatitis C virus infection by new four-antigen recombinant immunoblot assay. Lancet. 1991;337:317–9. doi: 10.1016/0140-6736(91)90942-i. [DOI] [PubMed] [Google Scholar]
- 2.Lenzi M, Bellentani S, Saccoccio G, et al. Prevalence of non-organ-specific autoantibodies and chronic liver disease in the general population: a nested case–control study of the Dinysos cohort. Gut. 1999;45:435–41. doi: 10.1136/gut.45.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gregorio GV, Pensati P, Iorio R, Vegnente A, Mieli-Vergani G, Vergani D. Autoantibody prevalence in children with liver disease due to chronic hepatitis C virus (HCV) infection. Clin Exp Immunol. 1998;112:471–6. doi: 10.1046/j.1365-2249.1998.00574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abuaf N, Lunel F, Giral P, et al. Non-organ specific autoantibodies associated with chronic C virus hepatitis. J Hepatol. 1993;18:359–64. doi: 10.1016/s0168-8278(05)80281-8. [DOI] [PubMed] [Google Scholar]
- 5.Clifford BD, Donahue D, Smith L, et al. High prevalence of serological markers of autoimmunity in patients with chronic hepatitis C. Hepatology. 1995;21:613–9. [PubMed] [Google Scholar]
- 6.Bortolotti F, Vajro P, Balli F, et al. Non-organ specific autoantibodies in children with chronic hepatitis C. J Hepatol. 1996;25:614–20. doi: 10.1016/s0168-8278(96)80228-5. [DOI] [PubMed] [Google Scholar]
- 7.Kammer AR, van der Burg SH, Grabscheid B, et al. Molecular mimicry of human cytochrome P450 by hepatitis C virus at the level of cytotoxic T cell recognition. J Exp Med. 1999;190:169–76. doi: 10.1084/jem.190.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atkinson MA, Bowman MA, Campbell L, Darrow BL, Kaufman DL, Maclaren NK. Cellular immunity to a determinant common to glutamate decarboxylase and coxsackie virus in insulin-dependent diabetes. J Clin Invest. 1994;94:2125–9. doi: 10.1172/JCI117567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Misko IS, Cross SM, Khanna R, Elliott SL, Schmidt C, Pye SJ, Silins SL. Cross-reactive recognition of viral, self, and bacterial peptide ligands by human class I-restricted cytotoxic T lymphocyte clonotypes: implications for molecular mimicry in autoimmune disease. Proc Natl Acad Sci USA. 1999;96:2279–84. doi: 10.1073/pnas.96.5.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oldstone MB. Molecular mimicry and immune-mediated diseases. Faseb J. 1998;12:1255–65. doi: 10.1096/fasebj.12.13.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gross DM, Forsthuber T, Tary-Lehmann M, et al. Identification of LFA-1 as a candidate autoantigen in treatment-resistant Lyme arthritis. Science. 1998;281:703–6. doi: 10.1126/science.281.5377.703. [DOI] [PubMed] [Google Scholar]
- 12.Zhao ZS, Granucci F, Yeh L, Schaffer PA, Cantor H. Molecular mimicry by herpes simplex virus type-1: autoimmune disease after viral infection. Science. 1998;279:1344–7. doi: 10.1126/science.279.5355.1344. [DOI] [PubMed] [Google Scholar]
- 13.Gregorio GV, Choudhuri K, Ma Y, Vegnente A, Mieli-Vergani G, Vergani D. Mimicry between the hepatitis B virus DNA polymerase and the antigenic targets of nuclear and smooth muscle antibodies in chronic hepatitis B virus infection. J Immunol. 1999;162:1802–10. [PubMed] [Google Scholar]
- 14.Homberg JC, Stelly N, Andreis I, Abuaf N, Saadoun F, Andre J. A new antimitochondria antibody (anti-M6) in iproniazid-induced hepatitis. Clin Exp Immunol. 1982;47:93–102. [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarez F, Berg PA, Bianchi FB, et al. International autoimmune hepatitis group report: review of criteria for diagnosis of autoimmune hepatitis. J Hepatol. 1999;31:929–38. doi: 10.1016/s0168-8278(99)80297-9. [DOI] [PubMed] [Google Scholar]
- 16.Gregorio GV, Portmann B, Karani J, et al. Autoimmune hepatitis/sclerosing cholangitis overlap syndrome in childhood: a 16-year prospective study. Hepatology. 2001;33:544–53. doi: 10.1053/jhep.2001.22131. [DOI] [PubMed] [Google Scholar]
- 17.Martins da Costa C, Baldwin D, Portmann B, Lolin Y, Mowat AP, Mieli-Vergani G. Value of urinary copper excretion after penicillamine challenge in the diagnosis of Wilson's disease. Hepatology. 1992;15:609–15. doi: 10.1002/hep.1840150410. [DOI] [PubMed] [Google Scholar]
- 18.Hopp TP, Woods KR. A computer program for predicting protein antigenic determinants. Mol Immunol. 1983;20:483–9. doi: 10.1016/0161-5890(83)90029-9. [DOI] [PubMed] [Google Scholar]
- 19.Parker JM, Guo D, Hodges RS. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data. correlation of predicted surface residues with antigenicity and X-ray-derived accessible sites. Biochemistry. 1986;25:5425–32. doi: 10.1021/bi00367a013. [DOI] [PubMed] [Google Scholar]
- 20.Welling GW, Weijer WJ, van der Zee R, Welling-Wester S. Prediction of sequential antigenic regions in proteins. FEBS Lett. 1985;188:215–8. doi: 10.1016/0014-5793(85)80374-4. [DOI] [PubMed] [Google Scholar]
- 21.Choudhuri K, Gregorio GV, Mieli-Vergani G, Vergani D. Immunological cross-reactivity to multiple autoantigens in patients with liver kidney microsomal type 1 autoimmune hepatitis. Hepatology. 1998;28:1177–81. doi: 10.1002/hep.510280502. [DOI] [PubMed] [Google Scholar]
- 22.Hatch CL, Bonner WM. The human histone H2A.Z gene. Sequence and regulation. J Biol Chem. 1990;265:15211–8. [PubMed] [Google Scholar]
- 23.Belgrader P, Dey R, Berezney R. Molecular cloning of matrin 3. A 125-kilodalton protein of the nuclear matrix contains an extensive acidic domain. J Biol Chem. 1991;266:9893–9. [PubMed] [Google Scholar]
- 24.Erdile LF, Wold MS, Kelly TJ. The primary structure of the 32-kDa subunit of human replication protein A. J Biol Chem. 1990;265:3177–82. [PubMed] [Google Scholar]
- 25.Matsuoka R, Yoshida MC, Furutani Y, et al. Human smooth muscle myosin heavy chain gene mapped to chromosomal region 16q12. Am J Med Genet. 1993;46:61–7. doi: 10.1002/ajmg.1320460110. [DOI] [PubMed] [Google Scholar]
- 26.Ferrari S, Battini R, Kaczmarek L, et al. Coding sequence and growth regulation of the human vimentin gene. Mol Cell Biol. 1986;6:3614–20. doi: 10.1128/mcb.6.11.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Claeys H, Volckaerts A, Mertens W, Liang Z, Fiten P, Opdenakker G. Localization and reactivity of an immunodominant domain in the NS3 region of hepatitis C virus. J Med Virol. 1995;45:273–81. doi: 10.1002/jmv.1890450306. [DOI] [PubMed] [Google Scholar]
- 28.Yuki N, Hayashi N, Kasahara A, et al. Antibodies to a putative hepatitis C virus polyprotein in Jananese patients with chronic liver disease. J Med Virol. 1992;38:86–91. doi: 10.1002/jmv.1890380203. [DOI] [PubMed] [Google Scholar]
- 29.Zhang ZX, Chen M, Sonnerborg A, Sallberg M. Antigenic structure of the complete nonstructural (NS) 2 and 5 proteins of hepatitis C virus (HCV): anti-HCV NS2 and NS5 antibody reactivities in relation to HCV serotype, presence of HCV RNA, and acute HCV infection. Clin Diagn Lab Immunol. 1994;1:290–4. doi: 10.1128/cdli.1.3.290-294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Iwata K, Wakita T, Okumura A, et al. Interferon gamma production peripheral blood lymphocytes to hepatitis C virus core protein in chronic hepatitis C infection. Hepatology. 1995;22:1057–64. doi: 10.1016/0270-9139(95)90609-6. [DOI] [PubMed] [Google Scholar]
- 31.Shirai M, Arichi T, Chen M, et al. T cell recognition of hypervariable region-1 from hepatits C virus envelope protein with multiple class II MHC molecules in mice and human: preferential help for induction of antibodies to the hypervariable region. J Immunol. 1999;162:568–76. [PubMed] [Google Scholar]
- 32.Barbato G, Cicero DO, Cordier F, et al. Inhibitor binding induces active site stabilization of the HCV NS3 protein serine protease domain. EMBO J. 2000;19:1195–206. doi: 10.1093/emboj/19.6.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czaja AJ, Cassani F, Cataleta M, Valentini P, Bianchi FB. Frequency and significance of antibodies to actin in type 1 autoimmune hepatitis. Hepatology. 1996;24:1068–73. doi: 10.1002/hep.510240515. [DOI] [PubMed] [Google Scholar]
- 34.Wirth S, Guidotti LG, Ando K, Schlicht HJ, Chisari FV. Breaking tolerance leads to autoantibody production but not autoimmune liver disease in hepatitis B virus envelope transgenic mice. J Immunol. 1995;154:2504–15. [PubMed] [Google Scholar]