Abstract
Loss of deoxyribonuclease I (Dnase1) function is associated with systemic lupus erythematosus (SLE) in humans and mice; however, no coding mutations in Dnase1 are found in polygenic murine models. Instead, both MRL-lpr strains and NZB/W F1 hybrids are homozygous for T89I missense in the macrophage-DNASE, desoxyribonuclease I-like 3 (Dnase1l3). By in vitro expression studies, this substitution decreases this enzyme's nuclease activity against free DNA by only approximately twofold; however, the mutation has a greater effect on the capacity of media conditioned with Dnase1l3 to confer a barrier to liposomal gene transfection to HeLa cells. The 89I substitution decreases the Dnase1l3 barrier function in vitro by eightfold (P < 0·01). In splenocytes and BM-derived macrophages of SLE mice, while cellular Dnase1l3 levels are induced relative to C57BL/6 (control) mice, levels of FD-nuclease activity are similar. Finally, media conditioned by MRL and NZB/W F1 macrophages, relative to control, contains a weak interferon-gamma (IFN-γ) inducible Dnase1l3-associated barrier to transfection. This barrier function is hypothesized to reflect the inability of SLE mice to degrade membrane-enveloped DNA-associated antigens, such as apoptotic bodies, which are predicted to stimulate the characteristic autoimmunity of SLE. Our results for these two generally independent models strongly suggest that Dnase1l3 deficiency increases the susceptibility of these mice to polygenic SLE.
Keywords: DNase, SLE, lupus, MRL, NZB/W F1, genetics
INTRODUCTION
For over 40 years, deficiency in a deoxyribonuclease, mainly Dnase I or Dnase1, has been proposed to predispose to the autoimmune disease, SLE [1]. In support of this hypothesis, decreased serum Dnase activity was found in human SLE [2,3] and (NZB X NZW) F1 (NZB/W) mice [4]. Recently, Dnase–/– mice were found to develop SLE manifestations, such as immune complex deposition, glomerulonephritis and elevated antinuclear antibody titres [5]. Finally, a heterozygous null mutation in Dnase1 was found in two probably related SLE patients with high anti-dsDNA titres [6]. On the other hand, in NZB/W F1 mice and humans, Dnase1 enzyme trials suggested only marginal benefit [7,8]. More importantly, Dnase1 mutations have not been found commonly in human SLE [2], and coding mutations are absent from the syntenic murine locus in SLE models studied here.
We hypothesize that deficiency of the homologous but distinct Dnase1l3 (Dnase1-like 3) enzyme is common in SLE [9]. This macrophage-secreted enzyme shares the serum compartment with Dnase1 and is probably the faint 28 kD DNASE remaining in serum of Dnase1–/– mice [5]. Unlike Dnase1, Dnase1l3 can confer a significant barrier to transfection of liposome-associated DNA, a novel enzyme function termed barrier to liposomal (BT) activity [10]. While BT activity is nuclease-dependent, it requires more than just the ability to degrade naked or ‘free’ DNA (FD-nuclease activity), because Dnase1l3 truncated for 21 C-terminal amino acids has abundant FD-nuclease activity but no BT activity. This report finds that the missense Dnase1l3 variant present in two SLE models is associated with only a mild defect in FD nuclease activity, but with a greater defect in vitro and macrophage secreted BT activity.
METHODS
Murine stocks
Seven to 8-week-old mice were obtained for our studies (Jackson Laboratories, Bar Harbor, ME, USA). Animals were housed locally for 10 days prior to tissue harvest. One mouse each from NZB, NZW, MRL-lpr, DBA/2, BXSB-Yaa and BL/6 was used for RNA isolation from liver (for reverse transcription-polymerase chain reaction (RT-PCR) of Dnase1l3 cDNA) and kidney (for RT-PCR of Dnase1 cDNA). DNA from other strains, including BXD recombinant inbreds, was obtained from Jackson Laboratories. For the studies of splenocyte and macrophage protein and FD-nuclease assays, we used three mice each of C57BL/6 (BL/6, female), MRL-lpr (female) and BXSB-Yaa (male) strains, as well NZB/W F1 hybrids. Finally, for BT assays, MRL and BL/6 strains, as well as NZB/W F1 hybrids, were used.
Reagents
IFN-γ was obtained from Genzyme (Cambridge, MA, USA); chemicals and restriction enzymes from Fisher Scientific (Hampton, NH, USA); recombinant mouse GM-CSF and IL-4, from R&D Systems (Minneapolis, MN, USA); and Taq polymerase from Promega (Madison, WI, USA). Deionized water was purified further with a Milli-Q system (Millipore, Bedford, MA, USA).
Sequencing and genotyping
Dnase1l3 coding sequence was amplified from liver cDNAs. The cloned cDNAs were subjected to double-stranded sequencing of both strands. Amplification primers flanking the entire coding sequence of Dnase1l3 and Dnase1 cDNA have been described previously [10]. The Dnase1 cDNAs were reverse-transcribed from kidney total RNAs, and all were identical. BL/6 Dnase1l3 sequence was identical to NM 007870 (wild-type).
The genotype at base pair (bp) 438 of Dnase1l3 was confirmed for all strains by amplifying the surrounding 48 nucleotide genomic fragment surrounding this base pair with 5′-AATGG AAATTCAAGAAGAAGCAC-3′ and 5′-CGAGAACTAATC ACATAGTTCAAT-3′. The latter primer is mutated (A441T) to create an amplimer susceptible to SspI (AATATT) cleaved in 89I (mutant) strains, but not in wild-type strains. After SspI digest, products were analysed by 6% acrylamide gels. BXD recombinant inbreds, derived from parental strains discordant at this mutation were genotyped to determine the chromosomal location of Dnase1l3 (http://www.informatics.jax.org).
Immunoblotting experiments
Our polyclonal antisera against a murine Dnase1l3 peptide has been described previously [10]. Boiled protein extracts (100 µg) were separated electrophoretically by 12% SDS-PAGE and electro-transferred to nitrocellulose (Sigma) in tissue studies, or Immobilon-P (Millipore) in cell culture studies. Protein concentration was determined either by BCA Protein Assay (Pierce) or equalized by immunoblots of β-actin expression. Membranes were blocked for 2 h in PBS-0·1% Tween-20 containing 5% non-fat dry milk and 4% goat serum, then incubated overnight at 4°C with either anti-Dnas1l3 antisera at 1 : 4000 or the anti-green fluorescent protein (GFP) at 1 : 7000. ECL detection (Amersham-Pharmacia, Piscataway, NJ, USA) was performed and results recorded by radiography. Images and photographs were scanned as 600 dpi TIFF images, and analysed by measuring pixel area and intensity of traced bands, using Scion Image 4·02 software (http://www.scioncorp.com/).
Enzyme activity assays
Zymograms and DNASE radial diffusion assays (RDAs) were used to measure FD nuclease activity by previously described methods [10]. All eukaryotic transfections were performed with FuGENE 6 polycationic liposomal reagent (Roche, Indianapolis, IN, USA).
FD nuclease assays
To compare the in vitro activity of wild-type and 89I, Bl/6 Dnase1l3 cDNA (AA 11–305) was amplified with primers 5′-CCACGCCTGAGCTCCCTGCTGCTCT-3′ and 5′-GAGCGTG GTACCTAGGAGCGATTG-3′ (reverse), and cloned in-frame downstream of a prokaryotic GST cassette (pGEX4T) (Amersham-Pharmacia). Site-directed mutagenesis was used to synthesize the T89I allele using the QuikChange kit (Stratagene, La Jolla, CA, USA). Double-strand sequencing was used to confirm inserts. Transformed BL-21 cells at an OD 600 of 0·7 were induced by 0·05 mm IPTG for 3 h at 25 degrees. GST-fusions were purified from Escherichia coli lysates using glutathione-sepharose beads (Amersham-Pharmacia). FD nuclease activity was measured by zymogram, while protein levels were compared by immunoblot.
The cloning of Dnase1l3 into the eukaryotic expression pcDNA3·1 vector (Invitrogen) was described previously [10]. Mutant counterparts of this clone were created by site-directed point mutagenesis. Lipofection of wild-type and 89I pcDNA3·1-Dnase1l3 vectors (3 µg purified using Promega Wizard Plus DNA purification system) was performed into HeLa. Nuclease activity was determined after 36 h by RDAs of conditioned supernatants [2% fetal calf serum (FCS)]. The relative result from zymograms of cell lysates and media gave similar results to those for prokaryotic studies. Dnase1l3 protein levels were compared using immunoblots of cell lysates. Immunoblots with antiβ-actin demonstrated equal loading of total protein per lane.
Assays of murine cells
For tissue assays, three mice were sacrificed for each strain or model (Table 1 and Fig. 3,4). Serum samples were obtained by repeat tail bleeds (n = 3 per mouse). Splenocytes were isolated by filtering the minced organ through cell strainers (Becton Dickinson, Franklin Lakes, NJ, USA), then treating with red blood cell (RBC) lysis buffer (Sigma, St. Louis, MO, USA), and culturing for 24–36 h at 2 million cells/well (6-well Falcon) in RPMI with 10% FCS. Half the splenocyte population was used for immunoblot isolation; RDAs were measured after 24–36 h in culture. Bone marrow (BM) cultures were isolated similarly by flushing femurs and tibia, then plated using 2 ml RPMI containing 10% FBS, 2 mm HEPES, 50 µmβ-mercaptoethanol, 20 ng/ml granulocyte macrophage-colony stimulating factor (GM-CSF) and 1 ng/ml interleukin (IL)-4. The cells were incubated with media changes and removal of non-adherent cells every 3 days. After 7 days of culture, a homogeneous population of adherent cells was obtained. Immunoblots for each sample were performed in duplicate.
Table 1.
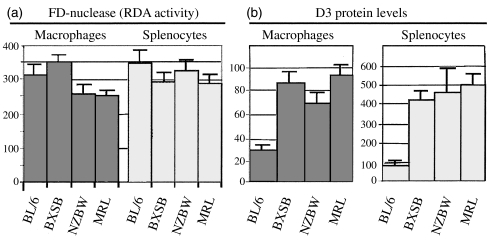
Dnase1l3 levels and FD nuclease activity; data from three lupus models (BXSB, NZB/WF1 and MRL-lpr) and one control strain (C57BL)
| Macrophages | Splenocytes | |||
|---|---|---|---|---|
| Mice | Dnase1l3 Levels | FD nuclease | Dnase1l3 Levels | FD nuclease |
| C57BL/6 | 30 ± 3‡ | 314 ± 33§ | 83 ± 22‡ | 346 ± 34 |
| BXSB | 86 ± 8 | 351 ± 25 | 422 ± 53 | 290 ± 25 |
| NZB/W | 70 ± 7 | 256 ± 32 | 463 ± 128 | 326 ± 23 |
| MRL | 92 ± 9 | 251 ± 20 | 503 ± 64 | 286 ± 23 |
All values are expressed in pixels; mean ± standard deviation. Statistical ignificance:
P < 0·01 for both macrophage and splenocyte Dnase1l3 levels of BL/6 compared to all lupus mice using Wilcoxon rank sum test.
P < 0·01 for C57BL macrophage RDA activity versus NZ and MRL-lpr (but not BXSB) mice. In addition, P < 0·01 was found for the comparison between BXSB and both NZ and MRL. Both calculations used Wilcoxon rank sum test.
Fig. 3.
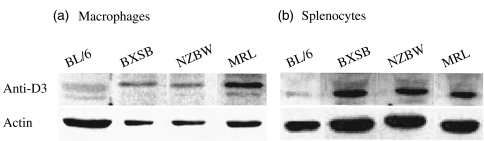
Representative anti-Dnase1l3 Western blots of splenocytes and macrophage cell lysates. Anti-Dnase1l3 (upper) and anti-β-actin (lower) westerns of cell lysates from (a) BM-derived macrophages and (b) splenocytes. The Western blots show induced levels of Dnase1l3 in all SLE models relative to BL/6. Expression results were normalized relative to actin expression and (n = 3) represented in Fig. 4 and Table 1.
Fig. 4.
Macrophage protein levels of Dnase1l3 and secreted FD-nuclease activity from BXSB, NZBW, MRL and BL6. This figure represents graphically the mean pixel volumes from Table 1. (a) FD-nuclease activity (RDA) secreted by macrophages was lower in NZBW and MRL-lpr mice relative to BL/6 (n = 15, both P < 0·01). (b) Immunoblot levels of Dnase1l3 protein in cell lysates from both populations were significantly elevated (n = 11 blots, P < 0·01). Thus, relative to their induced levels of Dnase1l3, macrophages from 89I SLE mice were deficient in secreted DNASE activity.
Barrier to liposomal transfection assays
BT assays have been described previously [10]. To prepare Dnase-conditioned media, 2 µg of either pcDNA3·1-Dnase1l3 plasmid or its mutagenized (89I) counterpart were lipofected into 1–2 × 106 HeLa cells. At 24 h, the media was replaced with DMEM/2% FCS, and conditioned for 36–48 h. A second set of naive HeLa cells are next incubated in Dnase-conditioned media, and then these cells lipofected with 2 µg of N1-eGFP plasmid (Clontech, Palo Alto, CA, USA). FD-nuclease activity in conditioned supernatant is measured by RDAs or zymograms using purified salmon sperm DNA (Sigma). GFP transfection efficiency was estimated by immunoblot densitometry of GFP expression or assayed by flow cytometry of 20 000 transfected cells using a Becton-Dickinson FACS Vantage flow cytometer.
BM-derived macrophages were isolated as described previously, plated on plastic 6-well Falcon tissue culture dishes in 2 ml RPMI containing 10% FBS, 2 mm HEPES and 50 µm β-mercaptoethanol. After 24 h, the media was replaced with 2 ml RPMI containing 10% FBS ± IFN-γ and incubated for an additional 48 h. BT activity in macrophage-conditioned media was assayed as described above. NZB/W F1 and MRL mice were used to examine BT assays. Note, the previous macrophage and splenocyte experiments used MRL-lpr mice.
Statistical analysis
The results (pixel volumes) for each experiment were paired and analysed by Wilcoxon signed rank tests.
RESULTS
Sequence analysis of Dnase1l3 and Dnase1
Dnase1 coding sequences were invariant in the six strains studied. For Dnase1l3, the only difference detected in coding sequences was the transition ACA438ATA[438 C > T] present in Dnase1l3 from MRL-lpr (Fig. 1), DBA/2, NZB, and NZW strains. The alteration encodes the non-conservative substitution of threonine at amino acid 89 by isoleucine (T89I) (Fig. 1). This change was confirmed by SspI restriction analysis of the amplified 48 bp genomic fragment using primers described in Methods. Using this genotyping assay, the mutation was also confirmed in the four strains above, as well as C3H and A strains. The SM, AKR, LG, 129, BALB/c, BXSB-Yaa and C57BL/10 strains were wild-type. Genotypes of BXD (C57BL × DBA/2) recombinant inbreds were fully concordant with markers DNASE14Byu1 and DNASE14Mit99, consistent with location on mouse chromosome 14, as expected from syntenic human 3p21 locus (data not shown).
Fig. 1.
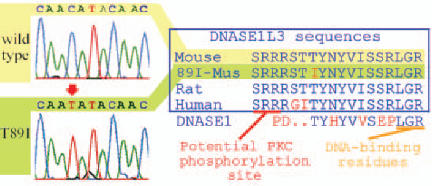
Sequence and alignment of mutant and normal DNASEs. Left register depicts wild-type (a, yellow) and 89I (b, green) sequence traces from BL/6 and MRL-lpr, respectively. The ACA to ATA transition at bp 438 encodes the non-conservative substitution isoleucine for threonine as shown in the (c). (c) Alignment shows alteration superimposed occurs relative to other Dnase1l3 and annotated bovine DNASE 1. Full alignments of human DNASEs can be found in (9).
Comparison of FD nuclease activity of wild-type (89T) and 89I D
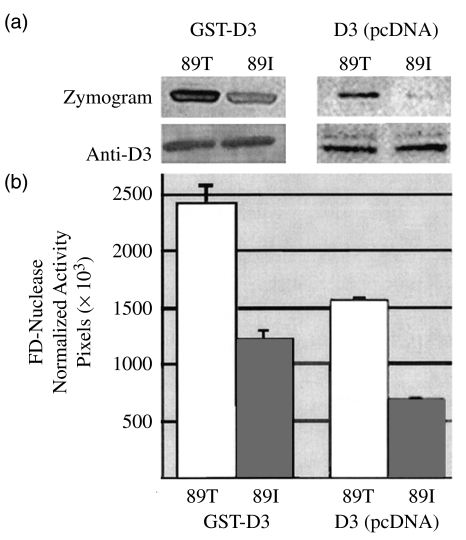
The glutathione-agarose-purified GST-fusion for the 89I (mutant) Dnase1l3 showed 54% of the zymographic FD nuclease activity shown by the 89T (wild-type) enzyme (Fig. 2a, n = 8, P < 0·01). Similar, but not significant results (57%) were obtained after normalized expression in HeLa cells (n = 6 observations, 28517 ± 2007 pixels versus 16322 ± 937 pixels; P > 0·05). The nuclease activity on zymograms was normalized relative to the nearly equivalent immunoblotted Dnase1l3 levels in lysates. A twofold difference was generally also observed when comparing RDAs of media conditioned by transfected cells and by Dnase1l3-GFP fusions (GFP fused in frame at C-terminus) and immunoblotted with anti-GFP (data not shown). Thus, in vitro studies in eukaryotic and prokaryotic systems confirm the 89I substitution decreases FD nuclease activity by about 50%.
Fig. 2.
FD-nuclease activities of 89T and 89I Dnase1l3 s. (a) Corresponding zymograms and anti-Dnase1l3-peptide Western blots of Dnase1l3-GST fusions expressed in a prokaryotic system (E. coli, left) and Dnase1l3 expressed in eukaryotic cells (HeLa, right). Anti-Dnase1l3 immunoblots detect, respectively, the expected column-purified approximately 56 kD GST-fusions (left) or approximately 28 kD enzymes in lysates of cells transfected with pcDNA3·1-Dnase1l3. Zymograms (top) were performed on parallel and equal loads of the immunoblotted samples. While protein levels are nearly equal, the 89I enzyme has 53% the nuclease activity of the wild-type enzyme (n = 8, mean values 2419 ± 154 versus 1211 ± 84 pixels; Wilcoxon signed rank sum, P < 0·01). Zymogram activity was normalized relative to Dnase1l3-expression by western. (b) Graph of results.
FD nuclease and cellular DNASE1L3 levels in murine SLE models and BL/6
Two bands of 28 and 34 kD are immunodetected with anti-Dnase1l3 peptide antisera in cell lysates from both splenocytes and bone-marrow-derived macrophages; the bands represent the enzyme before and after signal peptide cleavage. In conditioned media zymograms, only a single 28 kD band is observed. To determine the expression levels of Dnase1l3 in mice, we immunoblotted cellular lysates from splenocytes and BM macrophages from BXSB-Yaa, BL/6, MRL-lpr and NZB/W F1 mice. RDAs were used to measure FD nuclease activities of media conditioned by these cells.
The most striking observation was induction of Dnase1l3 in cellular protein lysates from splenocytes (four- to fivefold) and macrophages (two- to threefold) in all three lupus models relative to BL/6 (Fig. 3). Similar levels of induction can be obtained in control BL/6 macrophages, but only after treatment with PMA or IFN-γ (data not shown). The latter stimulus also leads to a parallel increase in secreted FD nuclease activity and 28 kD Dnase activity (data not shown). In media conditioned by SLE splenocytes and macrophages, there was a relative deficiency of FD-nuclease activity; hence, while the Dnase1l3 enzyme levels were increased, the expected parallel increases in FD nuclease activity was not observed (Figs 3,4 and Table 1). In related observations, among the strains/models studied, no significant differences were found in urine FD nuclease activity by RDAs, which is attributed to Dnase1. In addition, we could not reconfirm a significant deficiency of serum FD nuclease activity in SLE strains relative to BL/6 mice [4] (data not shown).
Comparison of BT activity of wild-type and mutant
The discovery of Dnase1l3-specific BT activity led to us to re-evaluate the difference between wild-type and mutant enzyme found in media conditioned by Dnase1l3-lipofected HeLa (Fig. 5). By BT assays, the 89I enzyme had an approximately eightfold defect in barrier to transfection activity (n = 8, P-value <0·01); in other words, the mutant enzyme was much less efficient in blocking the entry of lipofected DNA. Activity was normalized relative to cellular levels of Dnase1l3 protein in the FD nuclease activity in the conditioned media was similar in 89I and 89T, probably because of a nearly twofold higher level of enzyme in 89I transfected cell lysates. Undiluted conditioned media in these experiments, which achieves similar levels of FD nuclease activity by RDA to those seen in IFN-γ induced macrophage-conditioned media, conferred a near complete barrier to transfection activity with both 89I and 89T, but this was only true with 1 : 10 dilutions of media conditioned with the wild-type enzyme.
Fig. 5.
BT assays of in vitro-expressed mutant (89I) and wild-type Dnas1l3. Dnase1l3 protein levels as assessed in lysates of HeLa cells transfected with nuclease-expressing plasmids. (a) Representative lysates used to condition media show approximately twofold higher levels of the mutant protein by anti-Dnase1l3 immunoblot. Actin levels were nearly equivalent (data not shown). (b) Representative RDAs of conditioned media show near equivalent FD-nuclease activity. The twofold higher level of 89I expression appears to equalize RDA activity. (c) However, when the respective conditioned media are added to naive HeLa and assayed for the presence of BT activity, media with wild-type 89T enzyme provides a nearly eightfold greater barrier to liposomal transfection of GFP. (d) Graphic demonstration of the results of anti-GF Western blots performed for BT assays. As depicted, media conditioned with wild-type enzyme blocks transfection significantly more than media conditioned with 89I Dnase1l3.
Comparison of BT activity present in macrophage-conditioned media from NZB/W F, MRL, and BL/6
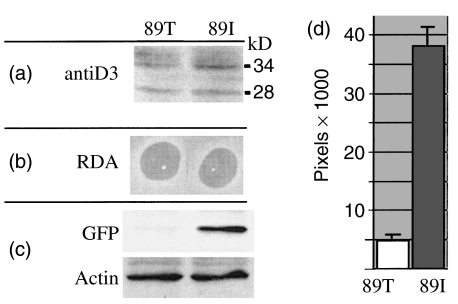
To determine if the relative deficiency of FD nuclease activity seen in media conditioned by SLE macrophages translated into a decreased barrier to transfection activity, we evaluated the ability of media conditioned by BM-derived macrophages from MRL-lpr, NZB/W F1 mice and wild-type C57BL to confer barrier to transfection activity to HeLa cells (Fig. 6). Media was conditioned for 48 h by adherent cells after IFN-γ stimulation. The numbers of adherent cells obtained from both NZB/W F1 and BL/6 mice (n = 2 mice, each marrow isolate split into two wells) were equivalent by visual inspection, and also suggested by actin immunoblotting. Media conditioned by macrophages from MRL and NZB/W F1 mice has a clear deficiency of barrier to transfection activity. These results support our hypothesis that deficient BT and DNASE1L3 activity are related and predispose to SLE in mice.
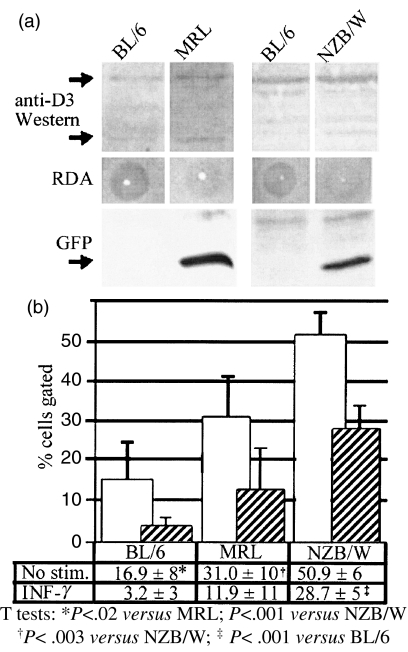
Fig. 6.
BT assays of macrophage-conditioned supernatants of BL/6, MRL-Mp, NZB/W. (a) Despite slightly higher levels of Dnase1l3 protein (upper 34 kD and lower 28 kD band) in cell lysates from SLE macrophages (n = 2 mice), the FD nuclease activity in macrophage-conditioned media was lower than that of BL/6. Lower register shows an anti-GFP Western blot of lipofected HeLa cells exposed to macrophage-conditioned media. Media conditioned by cells from SLE mice with 89I mutation had less capacity to block GFP gene transfection; hence, SLE macrophages had a deficiency of BT activity. (b) Percentage of gated cells that were GFP positive by flow cytometry of transfected HeLa cells are depicted by graphs with ± standard deviation (n = 4 trials). Media conditioned by unstimulated macrophages from both MRL-Mp and NZB/W mice fail to erect a barrier relative to BL/6. NZB/W mice had less BT activity than MRL mice. Even after IFN-γ induction, these differences were maintained; however, BT activity remained significantly diminished only in NZB/W mice.
DISCUSSION
Initially, our findings did not identify a clear Dnase1l3 deficiency in SLE mice, as the defect in nuclease activity against free-DNA was mild and the induced levels of enzyme were expected to compensate for the enzymatic defect. In addition, the 89I alteration was present in non-lupus-prone strains, and prior screens had not linked this locus to SLE in MRL and NZ models. The discovery of a nuclease-dependent, but distinct, enzymatic barrier function of Dnase1l3 prompted a re-evaluation. The in vitro defect in BT activity of the 89I enzyme is not likely to be compensated by the two- to fivefold induction of protein seen in SLE macrophages and splenocytes. In addition, in two independent SLE prone 89I mice, the MRL and NZB/W F1, a dramatic defect of Dnase1l3-associated macrophage-secreted BT activity was observed relative to the non-lupus-prone, wild-type C57BL/6 mice. The presence of this mutation and defect in two independently derived models strongly suggest Dnase1l3 deficiency increases their susceptibility to SLE.
We propose that the activity measured by the BT assay reflects a biological function that protects against SLE. The postulated Dnase1l3 function differs from the janitorial activities assigned to the lysosomal Dnase1, which appears to target naked or free-DNA [11,12]. BT-like functions are predicted to target membrane-associated DNA, for degradation, and prevent this material from inciting autoimmunity against itself and associated components. We propose Dnase1l3 functions as a macrophage-conferred ‘cellular goalie’ defending cells from potentially mutagenic [13] or antigen-associated cell-exogenous nucleosomal DNA. We propose that Dnase1l3 levels are induced in SLE mice to cope for increased load of undegraded material.
We speculate that the main in vivo target of Dnase1l3 is apoptotic debris, but could include other membrane-associated DNA particles, such as viruses. The phosphatidylserine lipids exposed on apoptotic body surfaces resembles liposomal transfection reagents, and this may flag this material for Dnase1l3 and macrophage phagocytosis [14]. Dnase1l3 dysfunction may also compound defects seen in SLE for macrophage phagocytosis [15,16] and clearance of apoptotic bodies [17].
Why the T89I substitution decreases BT more than FD nuclease activity is unknown. When the final 22 residues of Dnase1l3 are fused to Dnase1, the chimeric enzyme shows substantial BT activity, suggesting that the sequence of the upstream DNASE did not substantially affect BT activity [10]; however, as the T89 residue is conserved between both nucleases (Fig. 1), these experiments do not address specifically the importance of this residue. The mutation could possibly affect a potential protein kinase C (PKC) phosphorylation site (SRR) present in both Dnases (Fig. 1).
Previous studies of Dnases in SLE mice focused on defects of serum FD nuclease activity [4]. We generally could not confirm significant trends in the differences and, in addition, sera from MRL, NZB/W and BL/6 all showed similar capacities to interfere with liposomal transfection (data not shown). This suggests that serum DNASE-associated activity may not be useful to predict SLE susceptibility, perhaps because it reflects the combined action of multiple enzymes. The putative SLE-protective effects of Dnase1l3, however, may be derived via plasma, and hence humoral augmentation of DNASE1L3 activity may protect against SLE.
It is unclear why neither Dnase1l3 nor Dnase1 have been identified as susceptibility loci by genetic screens in mice [18]. The presumed absence of 89I allele from the NZ-derived lupus-prone congenic NZM2410 and its presence in non-lupus-prone strains, such as C3H and DBA/2, suggest this locus is neither necessary nor sufficient by itself to cause SLE. Disease promotion by the 89I allele probably requires other factors, perhaps antigen overproduction or defects in IFN-γ signalling. In the human disease, DNASE1L3 defects may play a greater role, since significant association to the 3p21 DNASE1L3 locus have been reported in African Americans with SLE [19]. The recent observations that peripheral leukocytes from SLE patients commonly express high levels of IFN-γ inducible genes, yet have a paradoxical repression of DNASE1L3 support our prediction that deficiency of this gene product is common and associated with SLE progression [20].
Based on the findings, a number of predictions can be made. The induced Dnase1l3 protein levels and the associated relative deficiency of macrophage-conditioned FD nuclease activity in the 89T BXSB mice, suggests a secondary of this nuclease is present. The 89I allele is predicted to excacerbate disease in lupus-prone congenics derived from NZ × C57BL/6 crosses [21], and may have synergistic lupus-promoting effects with other antigen clearance defects such as murine DNASE1 [5], C1q [17] or SAP null [22] mice. Finally, we predict human patients with SLE will also have primary or secondary DNASE1L3 abnormalities.
Acknowledgments
AW and MCS were supported by grants from the Lupus Research Institute Novel Research Grant Program (http://www.lupusresearchinstitute.org) and Central Research Committee at Southern Illinois School of Medicine. We acknowledge the help received from Dr Rita Trammell in the Department of Surgery at Southern Illinois School of Medicine and Anna Travelstead from Flow Cytometry facility.
REFERENCES
- 1.Klemperer PG, Lee S. Nucleic acid depolymerization in systemic lupus erythematosus. J Mt Sinai Hosp. 1949;16:61. [PubMed] [Google Scholar]
- 2.Tew MB, Johnson RW, Reveille JD, et al. A molecular analysis of the low serum deoxyribonuclease activity in lupus patients. Arthritis Rheum. 2001;44:2446–7. doi: 10.1002/1529-0131(200110)44:10<2446::aid-art409>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 3.Chitrabamrung S, Rubin RL, Tan EM. Serum deoxyribonuclease I and clinical activity in systemic lupus erythematosus. Rheumatol Int. 1981;1:55–60. doi: 10.1007/BF00541153. [DOI] [PubMed] [Google Scholar]
- 4.Macanovic M, Lachmann PJ. Measurement of deoxyribonuclease I (DNase) in the serum and urine of systemic lupus erythematosus (SLE)-prone NZB/NZW mice by new radial enzyme diffusion assay. Clin Exp Immunol. 1997;108:220–6. doi: 10.1046/j.1365-2249.1997.3571249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Napirei M, Karsunky H, Zevnik B, et al. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–81. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- 6.Yasutomo K, Horiuchi T, Kagami S, et al. Mutation of DNASE1 in people with systemic lupus erythematosus. Nat Genet. 2001;28:313–4. doi: 10.1038/91070. [DOI] [PubMed] [Google Scholar]
- 7.Davis JC, Jr, Manzi S, Yarboro C, et al. Recombinant human Dnase I (rhDNase) in patients with lupus nephritis. Lupus. 1999;8:68–76. doi: 10.1191/096120399678847380. [DOI] [PubMed] [Google Scholar]
- 8.Verthelyi D, Dybdal N, Elias KA, et al. DNAse treatment does not improve the survival of lupus prone (NZB x NZW) F1 mice. Lupus. 1998;7:223–30. doi: 10.1191/096120398678920037. [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez AM, Rodin D, Nomura H, et al. Identification, localization, and expression of two novel human genes similar to deoxyribonuclease I. Genomics. 1997;42:507–13. doi: 10.1006/geno.1997.4748. [DOI] [PubMed] [Google Scholar]
- 10.Wilber A, Lu M, Schneider MC. Deoxyribonuclease I-like III Is an inducible macrophage barrier to liposomal transfection. Mol Ther. 2002;6:35–42. doi: 10.1006/mthe.2002.0625. [DOI] [PubMed] [Google Scholar]
- 11.Nishikawa A, Nanda A, Gregory W, et al. Identification of amino acids that modulate mannose phosphorylation of mouse DNase I, a secretory glycoprotein. J Biol Chem. 1999;274:19309–15. doi: 10.1074/jbc.274.27.19309. [DOI] [PubMed] [Google Scholar]
- 12.Walport MJ Lupus. DNase and defective disposal of cellular debris. Nat Genet. 2000;25:135–6. doi: 10.1038/75963. [DOI] [PubMed] [Google Scholar]
- 13.Holmgren L, Szeles A, Rajnavolgyi E, et al. Horizontal transfer of DNA by the uptake of apoptotic bodies. Blood. 1999;93:3956–63. [PubMed] [Google Scholar]
- 14.Fadok VA, Xue D, Henson P. If phosphatidylserine is the death knell, a new phosphatidylserine-specific receptor is the bellringer. Cell Death Differ. 2001;8:582–7. doi: 10.1038/sj.cdd.4400856. [DOI] [PubMed] [Google Scholar]
- 15.Laderach D, Bach JF, Koutouzov S. Nucleosomes inhibit phagocytosis of apoptotic thymocytes by peritoneal macrophages from MRL+/+ lupus-prone mice. J Leukoc Biol. 1998;64:774–80. doi: 10.1002/jlb.64.6.774. [DOI] [PubMed] [Google Scholar]
- 16.Licht R, Jacobs CW, Tax WJ, et al. No constitutive defect in phagocytosis of apoptotic cells by resident peritoneal macrophages from pre-morbid lupus mice. Lupus. 2001;10:102–7. doi: 10.1191/096120301672276558. [DOI] [PubMed] [Google Scholar]
- 17.Nash JT, Taylor PR, Botto M, et al. Immune complex processing in C1q-deficient mice. Clin Exp Immunol. 2001;123:196–202. doi: 10.1046/j.1365-2249.2001.01459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kono DT, Theofilopoulos AN. Genetic susceptibility to spontaneous lupus in mice. In: Theofilopoulos AN, editor. Genes and Genetics of Autoimmunity. Switzerland: Karger; 1999. p. 72. [DOI] [PubMed] [Google Scholar]
- 19.Moser KL, Neas BR, Salmon JE, et al. Genome scan of human systemic lupus erythematosus: evidence for linkage on chromosome 1q in African-American pedigrees. Proc Natl Acad Sci USA. 1998;95:14869–74. doi: 10.1073/pnas.95.25.14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baechler EC, Batliwalla FM, Karypis G, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–5. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morel L, Croker BP, Blenman KR, et al. Genetic reconstitution of systemic lupus erythematosus immunopathology with polycongenic murine strains. Proc Natl Acad Sci USA. 2000;97:6670–5. doi: 10.1073/pnas.97.12.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bickerstaff MC, Botto M, Hutchinson WL, et al. Serum amyloid P component controls chromatin degradation and prevents antinuclear autoimmunity. Nat Med. 1999;5:694–7. doi: 10.1038/9544. [DOI] [PubMed] [Google Scholar]