Abstract
Toll-like receptors (TLR) are signal molecules essential for the cellular response to bacterial cell wall components. Different functional effective polymorphisms for the TLR 4 gene (Asp299Gly; Thr399Ile) and for the TLR 2 gene (Arg677Trp, Arg753Gln) have recently been described that are associated with impaired lipopolysaccharide signal transduction. A total of 122 patients with chronic periodontal disease and 122 healthy unrelated controls were genotyped for the Asp299Gly and Thr399Ile polymorphism of the TLR 4 gene and the Arg677Trp and Arg753Gln mutation of the TLR 2 gene. The mutations were identified with polymerase chain reaction followed by restriction fragment length polymorphism (RFLP) analysis. The prevalence of the Asp299Gly and the Thr399Ile mutant allele was 4·1% (10/244) and 4·5% (11/244) among periodontitis patients. For the healthy controls the prevalence was 3·3% (8/244) for the Asp299Gly (P = 0·810) and 3·7% (9/244) for the Thr399Ile mutant allele (P = 0·819). The Arg753Gln mutant allele was found in 2·9% (7/244) of the periodontitis subjects as compared to 4·1% (10/244) in the control group (P = 0·622). The Arg677Trp mutant allele was not found in any of the study subjects. Unlike in ulcerative colitis there was not observed an association between chronic periodontitis and the various mutations of the TLR 2 and 4 gene.
Keywords: periodontitis, TLR2, TLR4, receptor, pathogen-associated molecular patterns (PAMPs), lipopolysaccharide, proinflammatory
INTRODUCTION
The host immune system detects invading pathogens primarily through an array of pattern-recognition receptors. These receptors recognize conserved pathogen associated molecular patterns (PAMPs) that are typically shared by large groups of microorganisms, i.e. bacterial lipopolysaccharide [1]. The Toll-like receptors are members of an evolutionary conserved interleukin-1 super-family of transmembrane receptors that recognize pathogen associated molecular patterns (for review see [2]). The cytoplasmatic domain of the Toll-like receptors show high similarity to the intracellular portion of the interleukin-1 (IL-1) receptor family and is termed Toll/IL-1R (TIR) domain [3]. The extracellular domain of the Toll-like receptors bears leucine-rich repeats (LRRs).
For infections with gram-negative bacteria, lipopolysaccharide is the main source of inflammation and Toll-like receptor-4 is crucial in mediating its effects including the activation of the inflammatory molecular cascade via the NFκB pathway [4]. The recognition and transduction of the lipopolysaccharide signal through the Toll-like receptor-4 requires the presence of several additional molecules, particularly the lipopolysaccharide-binding protein (LBP) and the CD14 receptor and the MD-2 protein [2]. The TLR-4 protein is expressed on macrophages, cardiomyocytes, airway epithelia, endothelial cells and in many other tissues [5].
Toll-like receptor 2 was shown to be involved primarily in the recognition of peptidoglycans and lipoteichonic acid of gram-positive bacteria [6,7]. Moreover, TLR 2 is specifically involved in the recognition of the periodontopathogenic bacteria Porphyromonas gingivalis. It was previously suggested that TLR 2 activity upon stimulation by P. gingivalis is related to the lipopolysaccharide [8]. However, according to more recent data the Toll-like receptor 2 recognizes unknown cell wall components of P. gingivalis rather than the lipopolysaccharide itself [9].
Recently, two common cosegregating missense mutations, Asp299Gly and Thr399Ile, affecting the extracellular domain of the TLR4 protein have been characterized [7]. Both mutations lead to an attenuated efficacy of lipopolysaccharide signalling and a reduced capacity to elicit inflammation. Consistently, a significantly reduced risk for atherosclerosis and an increased risk for gram-negative infections were found for individuals carrying these mutations [10,11]. Also for the TLR2 gene two single nucleotid polymorphisms (Arg677Trp and Arg753Gln) have been identified that abrogate the ability of TLR2 to mediate a response to bacterial cell wall components [12].
Albeit the gram-negative bacterial infection was regarded as the primary pathophysiological factor in periodontitis it is commonly accepted that periodontitis has also a genetic background [13]. Specifically members of the monocyte/macrophage system along with the NFκB inflammatory pathway are crucial for the establishment of destructive periodontal disease [14]. Hence, functional relevant polymorphisms of genes that are involved in the stimulation and regulation of lipopolysaccharide mediated inflammatory processes are excellent candidates for the elucidation of the genetic background of the periodontal pathogenesis [15].
We hypothesized that the two polymorphisms of the Toll-like receptor-4 gene (Asp299Gly and Thr399Ile) and Toll-like receptor-2 gene (Arg677Trp and Arg753Gln) are implicated in the pathogenesis of chronic periodontitis. The current study compared the allele frequency and genotype distribution for these mutations among periodontitis patients and healthy control subjects.
MATERIALS AND METHODS
Patient population
Patients with severe medical disorders including diabetes mellitus, immunological disorders, increased risk for bacterial endocarditis and pregnant females were excluded from the study. The study protocol was approved by the ethical committee of the medical faculty of the Ludwig-Maximilians University (No. 290/01). Written informed consent was provided by all participants prior to their enrolment.
Periodontitis group
One hundred and twenty-two patients were recruited from the Department of Periodontology, Ludwig-Maximilians University (Munich, Germany). The male to female ratio was 54:46%. The median ± SD age within the periodontitis group was 52·9 ± 12·3 years (age range 25–74 years). In the periodontitis group all patients presented the diagnosis of generalized chronic periodontitis. The diagnosis of periodontitis was made on basis of a standard evaluation procedure:
determination of probing pocket depth measured with a Michigan type ‘O’probe at 6 locations on each tooth (mesio-buccal, mid-buccal, disto-buccal, mesio-lingual, mid-lingual, disto-lingual);
determination of furcation involvement by means of a Naber type probe;
bleeding on probing registered as present or absent;
bone loss estimated by orthopantomographs.
The probing pocket depth was defined as the distance from the base of the periodontal pocket to the free gingival margin. The furcation involvement was assessed by horizontal probing from the furcation entrance to the base of the defect. The furcation involvement was graded according to the classification of Nyman et al. [16].
To be classified as having generalized periodontitis a patient had to present with the following characteristics:
a total of at least 15 teeth in situ;
≥ 8 teeth with a probing pocket depth of ≥5 mm at least at one location and/or a furcation involvement ≥ class II;
evidence of bone loss manifested as the distance between the alveolar crest and the cemento-enamel junction of ≥3 mm around the affected teeth.
Periodontitis patients were assigned to one of three groups of disease severity on basis of the following criteria:
mild group (maximum probing depth: 6 mm; attachment loss >30%: ≤5 teeth; no attachment loss >50%);
moderate group (maximum probing depth: 8 mm; attachment loss >30%: ≤8 teeth; attachment loss >50: ≤5 teeth);
severe group (maximum probing depth: >8 mm; attachment loss >30%: >8 teeth; attachment loss >50%: >5 teeth) (Table 1).
Table 1.
Criteria used for the classification of periodontitis patients according to the severity of disease (mild, moderate, severe)
| Classification of periodontal disease | |||
|---|---|---|---|
| Periodontitis (moderate) | Periodontitis (severe) | Periodontitis (mild) | |
| Maximum pocket depth | 6 mm | 8 mm | >8 mm |
| Attachment loss > 30% | ≤5 teeth | ≤8 teeth | ≤5 teeth |
| Attachment loss > 50% | – | ≤5 teeth | ≤8 teeth |
| Number of patients | 35 | 52 | 35 |
Control group
A total of 122 unrelated, ethnically matched, healthy Caucasian individuals were included in the control group. Control subjects had to meet the following criteria for inclusion: (1) ≥22 teeth in situ (2) not more than 1 site with probing pocket depth ≥3 mm (3) no furcation involvement at any tooth. The median age within the control group was 40 years (SD ± 13·3) and the age ranged from 18 to 73 years.
Blood samples and DNA isolation
Peripheral venous blood samples of 9 ml were drawn from each individual by standard venipuncture. Each blood sample was collected in sterile tubes containing 15% K3EDTA solution. DNA was isolated using partly the QIAamp® DNA Blood Midi Kit (Qiagen, Hilden, Germany), partly the salting out procedure [17].
Genotyping of the Toll-like receptor-2 and -4 gene
Determination of the TLR 2 and 4 gene mutations was accomplished with polymerase chain reaction (PCR) and restriction fragment length polymorphism. The total volume of the PCR was 25 µl (TLR2: 20 µl) containing 100 ng of genomic DNA, 1 × PCR-buffer (Qiagen, Hilden, Germany), 0·2 mm of each dNTP (Sigma, Taufkirchen, Germany), 0·75 units (TLR2: 0·5 units) of HotStarTaq™ DNA polymerase (Qiagen) and 7·5 pmol (TLR2: 5 pmol) of each primer (TIB MOLBIOL, Berlin, Germany). The final concentration of MgCl2 was 4 mm for Asp299Gly, 1·5 mm for Thr399Ile, and 3 mm for Arg753Gln and Arg677Trp. The PCR was performed in a thermocycler UNO-Thermoblock with an initial denaturation step (95°C for 15 min), 35 cycles (94°C for 30 s, 62°C (Asp299Gly), 60°C (Thr399Ile), or 65° (Arg677Trp and Arg753Gln) for 30 s, 72°C for 30 s), and a final extension step (72°C for 10 min). The restriction assay contained 1× restriction buffer, 20 units (TLR2 Arg677Trp: 12·5 units) of the respective restriction enzyme (New England Biolabs, Beverly, MD, USA) and 15 µl (TLR2: 20 µl) of the PCR product. It was incubated overnight at 37°C and analysed by electrophoresis on a 2·5% agarose gel. The primer sequences are presented in Table 2. Full length PCR products were digested into the restriction fragments as listed in Table 3. Additionally, one sample for each of the three possible genotypes had formerly been confirmed by sequencing and severed as standards in the restriction analysis. Sequencing was also performed for all individuals in which cosegregation of the mutant alleles did not occur. For sequencing PCR products overlapping both mutational sites of the TLR 2 and TLR 4 gene were amplified using the primers as presented in Table 2. The PCR was performed in a total volume of 100 µl and the final concentration of MgCl2 was 1·5 mm, the other components and concentrations were as described above. The PCR comprised an initial denaturation step of 15 min at 95°C, 35 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 65°C and extension for 60 s at 72°C, and a final extension step for 10 min at 72°C. Before sequencing the PCR products were purified using QIAquick PCR Purification Kit (Qiagen).
Table 2.
Sequences of primers as used for PCR
| Gene | Polymorphism | Primers | Annealing temp (°C) | MgCl2 |
|---|---|---|---|---|
| TLR4 | Asp299Gly | F: 5′- AGCATACTTAGACTACTACCTCCATG-3′ | ||
| R: 5′- GAGAGATTTGAGTTTCAATGTGGG-3′ | 62°C | 4·0 mm | ||
| TLR4 | Thr399Ile | F: 5′- GGTTGCTGTTCTCAAAGTGATTTTGGGAGAA-3′ | ||
| R: 5′- GGAAATCCAGATGTTCTAGTTGTTCTAAGCC-3′ | 60°C | 1·5 mm | ||
| TLR4 | Sequencing | F: 5′-ACAAATCTGCTCTAGAGGGCCTG-3′ | ||
| R: 5′-GCCATTTTCAAGACTTCGAGACTGG-3′ | 65°C | 1·5 mm | ||
| TLR2 | Arg677Trp | F: 5′-CCCCTTCAAGTTGTGGCTTCATAAG-3′ | ||
| R: 5′-AGTCCAGTTCATACTTGCACCAC-3′ | 65°C | 3·0 mm | ||
| TLR2 | Arg753Gln | F: 5′-CATTCCCCAGCGCTTCTGCAAGCTCC-3′ | ||
| R: 5′-GGAACCTAGGACTTTATCGCAGCTC-3′ | 65°C | 3·0 mm | ||
| TLR2 | Sequencing | F: 5′-CCCAGGAAAGCTCCCAGCAG-3′ | ||
| R: 5′-GGAACCTAGGACTTTATCGCAGCTC-3′ | 60°C | 1·5 mm |
Table 3.
Restriction enzymes and length of the restriction fragments
| Gene | Polymorphism | Restriction enzyme | Restriction temp °C | Length of the Restriction fragments: |
|---|---|---|---|---|
| TLR4 | Asp299Gly | Nco I | 37°C | Wild type (allele A):″ 188 bp |
| Asp299Gly (allele G):″168 bp + 20 bp | ||||
| TLR4 | Thr399Ile | Hinf I | 37°C | Wild type (allele C):″ 124 bp |
| Thr399Ile (allele T):″ 98 bp + 26 bp | ||||
| TLR2 | Arg677Trp | Mwo I | 60°C | Wild type (allele C):″″130 bp + 22 bp |
| Arg677Trp (allele T):″ 152 bp | ||||
| TLR2 | Arg753Gln | Msp I | 37°C | Wild type (allele G):″ 104 bp + 25 bp |
| Arg753Trp (allele A):″ 129 bp |
Statistical analysis
The Fisher's exact test was used to compare the distribution of the different mutations between the periodontitis affected individuals and the representative control group. The statistical procedure was performed at a level of significance of 5% (P < 0·05).
RESULTS
Genotyping of the Toll-like receptor-2 and -4 gene
Overall, within the group of patients with periodontal disease the allele frequency of the Asp299Gly mutation was 4·1% (10/244) (Fig. 1). that for the Thr399Ile mutation was 4·5% (11/244) (Fig. 2). In comparison, that in the control group was 3·3% (8/244) for the Asp299Gly mutant allele (P = 0·810) and 3·7% (9/244) for the Thr399Ile mutant allele (P = 0·819). Regarding the TLR2 gene the frequency of the Arg753Gln mutant allele was 2·9% (7/244) for periodontitis patients and 4·1% (10/244) for healthy controls (P = 0·622) (Fig. 3). The Arg677Trp mutant allele was not found in any of the periodontitis patients or healthy controls (Table 4; Fig. 4).
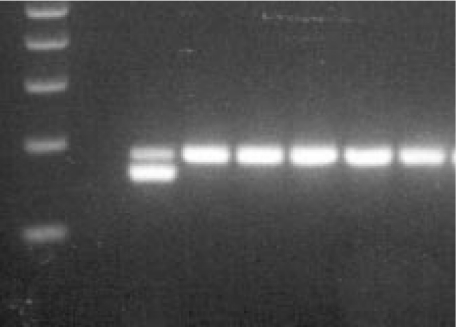
Fig. 1.
Agarose gel electrophoresis of the TLR4 Asp299Gly polymorphism: lane 1, 100 bp DNA ladder; lane 2, no DNA control; lane 3, restriction fragments in case of an individual heterozygous for the Asp299Gly mutation; lanes 4–8, restriction fragments in case of 5 wildtype individuals.
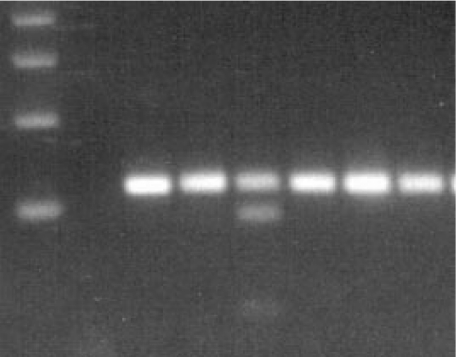
Fig. 2.
Agarose gel electrophoresis of the TLR4 Thr399Ile polymorphism: lane 1, 100 bp DNA ladder; lane 2, no DNA control; lane 5: restriction fragments in case of an individual heterozygous for the Thr399Ile mutation; lanes 3,4,6–8, restriction fragments in case of 5 wildtype individuals.
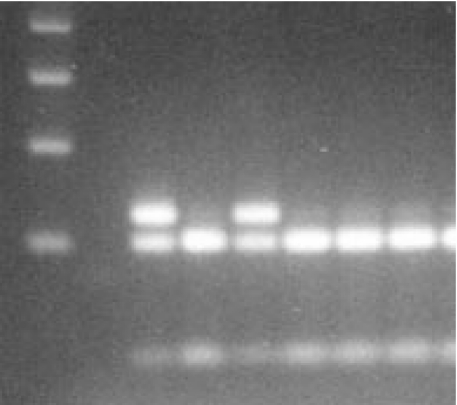
Fig. 3.
Agarose gel electrophoresis of the TLR2 Arg/53Gln polymorphism: lane 1, 100 bp DNA ladder; lane 2, no DNA control; lanes 3 & 5, restriction fragments in case of two individuals heterozygous for the Arg753Gln mutation; lanes 4,6–8, restriction fragments in case of 4 wildtype individuals.
Table 4.
Prevalence of the Toll-like receptor-4 mutant alleles (Asp299Gly, Thr399Ile) in individuals with periodontitis and the healthy control subjects
| Frequency of alleles 129 bp | ||||
|---|---|---|---|---|
| TLR4 | TLR2 | |||
| Asp299Gly | Wildtype | Arg677Trp | Wildtype | |
| Periodontitis (total) | 10 (4·1%) | 234 (95·9%) | 0 (0·0%) | 244 (100·0%) |
| Periodontitis (mild) | 3 (4·3%) | 67 (95·7%) | 0 (0·0%) | 70 (100·0%) |
| Periodontitis (moderate) | 6 (5·8%) | 98 (94·2%) | 0 (0·0%) | 104 (100·0%) |
| Periodontitis (severe) | 1 (1·4%) | 69 (98·6%) | 0 (0·0%) | 70 (100·0%) |
| Control | 8 (3·3%) | 236 (96·7%) | 0 (0·0%) | 244 (100·0%) |
| Thr399Ile | Wildtype | Arg753Gln | Wildtype | |
| Periodontitis (total) | 11 (4·5%) | 233 (95·5%) | 7 (2·9%) | 237 (97·1%) |
| Periodontitis (mild) | 4 (5·7%) | 66 (94·3%) | 3 (4·3%) | 67 (96·7%) |
| Periodontitis (moderate) | 6 (5·8%) | 98 (94·2%) | 0 (0·0%) | 104 (100·0%) |
| Periodontitis (severe) | 1 (1·4%) | 69 (98·6%) | 4 (5·7%) | 66 (94·3%) |
| Control | 9 (3·7%) | 235 (96·3%) | 10 (4·1%) | 234 (95·9%) |
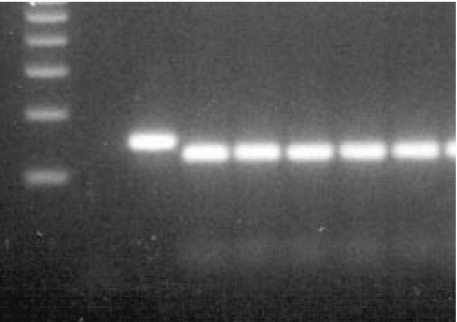
Fig. 4.
Agarose gel electrophoresis of the TLR2 Arg677Trp polymorphism: lane 1, 100 bp DNA ladder; lane 2, no DNA control; lane 3, PCR product without (before) restriction; lanes 4–8, restriction fragments in case of 5 wildtype individuals.
Hetrozygosity for the Asp299Gly mutation was found in 8·1% and 6·6% of the periodontitis and control subjects, respectively (P = 0·807). A total of 8·9% of the individuals with periodontal disease and 7·4% of the control subjects were heterozygous for the Thr399Ile mutation (P = 0·816). Regarding the Arg753Gln mutation of the TLR 2 gene heterozygosity was found in 5·7% of periodontitis patients and in 8·2% of the healthy controls (P = 0·615). None of the study subjects in the periodontitis or control group showed homozygosity for the Toll-like receptor-2 and -4 mutant alleles. Cosegregation between the mutant alleles of the Toll-like receptor-4 gene was found in 86% among periodontitis patients and in 89% of healthy controls.
Statistical analysis
The statistical analysis of the allele frequencies among both study populations are shown in Table 5.
Table 5.
Statistical analysis of data as obtained with Pearson's χ2-test and Fisher's exact test (P < 0·05)
| Statistical analysis TLR4 | TLR2 | |||
|---|---|---|---|---|
| Arg299Gly Allele frequency (versus control) | Thr399Ile Allele frequency (versus control) | Arg677Trp Allele frequency (versus control) | Arg753Gln Allele frequency (versus control) | |
| Periodontitis (total) | P = 0·810 | P = 0·819 | P = 1·000 | P = 0·622 |
| Periodontitis (mild) | P = 0·972 | P = 0·682 | P = 1·000 | P = 0·786 |
| Periodontitis (moderate) | P = 0·433 | P = 0·558 | P = 1·000 | P = 0·081 |
| Periodontitis (severe) | P = 0·681 | P = 0·573 | P = 1·000 | P = 0·803 |
DISCUSSION
According to current pathogenetic models periodontal disease is initiated and maintained in the first line by gram-negative bacterial infection of the gingival sulcus [18]. The periodontal infection together with the presence of specific pathogen associated molecular patterns (PAMPs), i.e. bacterial lipopolysaccharide, stimulates an inflammatory cascade that finally results in periodontal tissue destruction [15,19].
For the recognition and transduction of the lipopolysaccharide signal the transmembrane Toll-like receptor-4 plays a pivotal role [20,21]. Upon binding of lipopolysaccharide the Toll-like receptor-4 activates the NFκB-system ultimately leading to the synthesis and release of an array of proinflammatory cytokines, chemokines and costimulatory molecules that are potential stimulators of the periodontitis associated tissue destructive effects [15,22]. Consistently, the expression of the Toll-like receptor-4 in gingival tissue was shown to be specifically associated with severe periodontal disease [23].
Two recently identified mutations of the Toll-like receptor-4 gene (Asp299Gly and Thr399Ile) were reported to attenuate human responsiveness to lipopolysaccharide both, in-vitro and in-vivo [7,24]. Recent findings suggested that the carriage of these mutations is related to the risk for atherosclerosis and gram-negative infections [10]. A further study of our group provided evidence that genetic variation of the Toll-like receptor-4 contributes to the development of ulcerative colitis (unpublished data). Although the biological significance of these findings are yet not fully established it appears reasonable to assume that functional effective mutations in the Toll-like receptor-4 gene might also influence the individual susceptibility for periodontal disease.
In this investigation we found heterozygosity for the Asp299Gly and the Thr399Ile TLR4 allele roughly in 8% of the periodontitis patients and in 7% of the healthy controls. Except for one individual in each study group cosegregation was found in all heterozygous individuals. The data of previous studies revealed carriage rates for the Asp299Gly mutation ranging from 7% to 12% among healthy Caucasian subjects indicating that the results presented herein are in accordance with the majority of literature data [10,24,25].
Independently on the severity of disease the prevalence of the Toll-like receptor-4 mutations among periodontitis patients and healthy controls was not significantly different in this study. It can thus be assumed that neither the Asp299Gly nor the Thr399Ile mutation influence the individual susceptibility for periodontal disease. Several reasons might be responsible for the lack of association between the Toll-like receptor-4 polymorphism and periodontal disease.
First, the responsiveness of monocytes from donors that are hetrozygous for the Toll-like receptor-4 mutant alleles showed no deficit as compared to monocytes from wildtype individuals upon stimulation with lipopolysaccharide in-vitro [24]. Conversely, human monocytes homozygous for the Toll-like receptor-4 mutations are hyporesponsive against lipopolysaccharide [25]. It can thus be assumed that the susceptibility for peridontitis might be increased only for homozgous individuals. However, for none of the study subjects in the periodontitis group homozygosity for the Toll-like receptor-4 mutant alleles was found.
Second, TLR2 has been suggested as the most critical re-ceptor and signal transducer for PAMPs of one of the most common periodontopathic bacteria, Porphyromonas gingivalis [18]. Unlike the endodotoxine from most gram-negative bacteria P. gingivalis lipopolysaccharide was previously shown to bind exclusively to TLR 2 [8]. However, more recent data demonstrated that not the P. gingivalis lipopolysaccharide itself binds TLR 2 [9]. According to this study the TLR2 signalling is mediated primarily by yet unknown cell wall components of P. gingivalis. Hence, assuming that the infection with P. gingivalis has strong impact on the development of periodontal disease the attenuated signalling through the Toll-like receptor-4 in individuals carrying one or both mutations might apparently gain only minor overall relevance on the pathogenesis of periodontal disease. Consequently, mutations of the Toll-like receptor-2 gene that are associated with changes in signal transduction upon stimulation by pathogen molecular patterns might be more relevant than the mutations of the TLR-4 receptor. However, herein the prevalence of the TLR2 Arg677Trp and Arg753Gln mutation was also not significantly different between periodontitis patients and healthy controls
In conclusion, the functional effective mutations of the Toll-like receptor-2 and -4 gene that attenuate the response to PAMPs was equally prevalent among periodontitis patients with any disease severity and healthy control subjects. Functionally effective mutations of the Toll-like receptor-2 and -4 gene have apparently no influence on the individual susceptibility for periodontitis.
REFERENCES
- 1.Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–8. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 2.Takeda K, Akira S. Toll receptors and pathogen resistance. Cell Microbiol. 2003;5:143–53. doi: 10.1046/j.1462-5822.2003.00264.x. [DOI] [PubMed] [Google Scholar]
- 3.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:785–7. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 4.Medzhitov R, Preston-Hurlburt P, Janeway CA. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–7. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 5.Zarember KA, Godowski PJ. Tissue expression of human Toll-like receptors and differential regulation of Toll-like receptor mRNAs in leukocytes in response to microbes, their products, and cytokines. J Immunol. 2002;168:554–61. doi: 10.4049/jimmunol.168.2.554. [DOI] [PubMed] [Google Scholar]
- 6.Underhill DM, Ozinsky A, Smith KD, Aderem A. Toll-like receptor 2 mediates mycobacteria-induced proinflammatory signaling in macrophages. Proc Natl Acad Sci. 1999;96:14459–63. doi: 10.1073/pnas.96.25.14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arbour NC, Lorenz E, Schutte BC, et al. TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat Genet. 2000;25:187–91. doi: 10.1038/76048. [DOI] [PubMed] [Google Scholar]
- 8.Yoshimura A, Kaneko T, Kato Y, Golenbock DT, Hara Y. Lipopolysaccharides from periodontopathic bacteria Porphyromonas gingivalis and Capnocytophaga ochracea are antagonsits for human Toll-like receptor 4. Infect Immun. 2002;70:218–25. doi: 10.1128/IAI.70.1.218-225.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogawa T, Asai Y, Hashimoto M, Takeuchi O, Kurita T, Yoshikai Y, Miyake K, Akira S. Cell activation by porphyromonas gingivalis lipid A molecule through Toll-like receptor 4- and myeloid differentiation factor 88-dependent signalling pathway. Int Immunol. 2002;14:1325–32. doi: 10.1093/intimm/dxf097. [DOI] [PubMed] [Google Scholar]
- 10.Kiechl S, Lorenz E, Reindl M, Wiedermann CJ, Oberhollenzer F, Bonora E, Willeit J, Schwartz DA. Toll-like receptor 4 polymorphisms and artherogenesis. N Engl J Med. 2002;347:185–92. doi: 10.1056/NEJMoa012673. [DOI] [PubMed] [Google Scholar]
- 11.Agnese DM, Calvano JE, Hahm SJ, Coyle SM, Corbett SA, Calvano SE, Lowry SF. Human toll-like receptor 4 mutations but not CD14 polymoprhisms are associated with an increased risk of gram-negative infections. J Infect Dis. 2002;186:1522–5. doi: 10.1086/344893. [DOI] [PubMed] [Google Scholar]
- 12.Bochud PY, Hawn TR, Aderem A. Cutting edge: a toll-like receptor 2 polymorphism that is associated with lepromatous leprosy is unable to mediate mycobacterial signaling. J Immunol. 2003;170:3451–4. doi: 10.4049/jimmunol.170.7.3451. [DOI] [PubMed] [Google Scholar]
- 13.Page RC, Offenbacher S, Schroeder HE, Seymour GJ, Kornman KS. Advances in the pathogenesis of periodontitis: summary of developments, clinical implications and future directions. Periodontol 2000. 1997;14:216–48. doi: 10.1111/j.1600-0757.1997.tb00199.x. [DOI] [PubMed] [Google Scholar]
- 14.Page RC. Milestones in periodontal research and the remaining critical issues. J Periodont Res. 1999;34:331–9. doi: 10.1111/j.1600-0765.1999.tb02262.x. [DOI] [PubMed] [Google Scholar]
- 15.Graves DT, Jiang Y, Genco C. Periodontal disease: bacterial virulence factors, host response and impact on systemic health. Curr Opin Infect Dis. 2000;13:227–32. doi: 10.1097/00001432-200006000-00005. [DOI] [PubMed] [Google Scholar]
- 16.Nyman S, Lindhe J. Examination of patients with periodontal disease. In: Lindhe J, Karring T, Lang NP, editors. Clinical Periodontology and Implant Dentistry. Copenhagen: Munksgaard; 1997. pp. 383–95. [Google Scholar]
- 17.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucl Acids Res. 1998;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Socransky S, Haffajee A, Cugini M, Smith C, Kent RJ. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 19.Baker P, Dixon M, Evans R, Dufour L, Johnson E, Roopenian D. CD4 (+) T cells and the proinflammatory cytokines gamma interferon and interleukin-6 contribute to alveolar bone loss in mice. Infect Immunol. 1999;67:2804–9. doi: 10.1128/iai.67.6.2804-2809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qureshi ST, Lariviere L, Leveque G, Clermont S, Moore KJ, Gros P, Malo D. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr 4) J Exp Med. 1999;189:615–25. doi: 10.1084/jem.189.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poltorak A, He X, Smirnova I, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr 4gene. Science. 1998;282:2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 22.Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proeteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–80. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- 23.Mori Y, Yoshimura A, Ukai T, Lien E, Espevik T, Hara Y. Immunohistochemical localization of Toll-like receptors 2 and 4 in gingival tissue from patients with periodontitis. Oral Microbiol Immunol. 2003;18:54–8. doi: 10.1034/j.1399-302x.2003.180109.x. [DOI] [PubMed] [Google Scholar]
- 24.Erridge C, Stewart J, Poxton IR. Monocytes heterozygous for the Asp299Gly and Thr399Ile mutations in the Toll-like receptor 4 gene show no deficit inlipopolysaccharide signalling. J Exp Med. 2003;197:1787–91. doi: 10.1084/jem.20022078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmitt C, Humeny A, Becker CM, Brune K, Pahl A. Polymorphism of TLR4: rapid genotyping and reduced response to lipopolysaccharide of TLR4 mutant alleles. Clin Chem. 2002;48:1661–7. [PubMed] [Google Scholar]