Abstract
CD4+ T lymphocytes play an important role in the pathogenesis of systemic lupus erythematosus (SLE). To characterize the clonal expansion of CD4+ T cells in murine lupus models, we analysed the T cell clonality in various organs of young and nephritic MRL/lpr and NZB/W F1 mice using reverse transcription–polymerase chain reaction (RT-PCR) and subsequent single-strand conformation polymorphism (SSCP) analysis. We demonstrated that some identical T cell clonotypes expanded and accumulated in different organs (the bilateral kidneys, brain, lung and intestine) in nephritic diseased mice, and that a number of these identical clonotypes were CD4+ T cells. In contrast, young mice exhibited little accumulation of common clones in different organs. The T cell receptor (TCR) Vβ usage of these identical clonotypes was limited to Vβ2, 6, 8·1, 10, 16 and 18 in MRL/lpr mice and to Vβ6 and 7 in NZB/W F1 mice. Furthermore, some conserved amino acid motifs such as I, D or E and G were observed in CDR3 loops of TCRβ chains from these identical CD4+ clonotypes. The existence of systemically expanding CD4+ T cell clones in the central nervous system (CNS) suggests the involvement of the systemic autoimmunity in CNS lesions of lupus. FACS-sorted CD4+CD69+ cells from the kidney displayed expanded clonotypes identical to those obtained from the whole kidney and other organs from the same individual. These findings suggest that activated and clonally expanded CD4+ T cells accumulate in different tissues of nephritic lupus mice, and these clonotypes might recognize restricted T cell epitopes on autoantigens involved in specific immune responses of SLE, thus playing a pathogenic role in these lupus mice.
Keywords: animal models/studies-mice/rats, lupus/systemic lupus erythematosus, T cell receptor (TCR)
INTRODUCTION
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by the production of pathogenic autoantibodies and tissue deposition of immune complexes, which results in multiple organ damage. MRL/lpr and NZB/W F1 mice, that develop spontaneously a lupus syndrome closely resembling human SLE, are considered to be excellent models for investigating the pathogenesis of the human disease. It has been suggested that T cell-dependent antigen-specific immune responses play a key role in the pathogenesis of these two strains. Published data have documented the involvement of T cells, especially CD4+ T cells, in lupus in the mouse. This involvement includes: (1) the infiltration of affected tissues with predominantly CD4+ T cells [1,2]; (2) the development of lymphadenopathy but not autoimmune renal disease or autoantibodies in CD4+ T cell-deficient or MHC class II-deficient lupus mice [3,4]; and (3) improvement in the disease manifestations following treatment with anti-CD4 MoAb [5–7]. These results suggest that CD4+ T cells play an important role in the development of lupus.
Most of the studies have implicated CD4+ T cells as primarily responsible for pathogenic anti-DNA autoantibody production [3,8,9]. Although the participation and importance of CD4+ T cells in lupus mice has been emphasized, the pathogenic CD4+ T cell clonotypes that expanded in different sites of these murine models have not yet been identified and characterized. Also, the nature of antigen-specific T cells in SLE still remains poorly defined. Detection and characterization of the pathogenic T cell clonotypes that expanded in lupus mice may help to illustrate the contribution of CD4+ T cell clones to the antigen-specific immune responses in SLE and provide insight into the nature of the T cell Ags involved.
In order to address these questions and to investigate further the pathogenic role of CD4+ T cells clonally expanded in SLE, we compared the T cell clonality in different organs derived from young and nephritic NZB/W F1 as well as MRL/lpr mice using the established reverse transcription–polymerase chain reaction (RT-PCR)–single-strand conformation polymorphism (SSCP) study in combination with CDR3 sequence analysis of the T cell receptor (TCR) Vβ family. Our results showed that there were some identical T cell clonotypes expanded and accumulated in different organs in nephritic mice. Most of those identical clonotypes were CD4+ T cells. In contrast, few identical T cell clonotypes were found in young mice. In addition, some conserved amino acid motifs were observed in CDR3 loops of TCR Vβ from these identical CD4+ clonotypes.
MATERIALS AND METHODS
Mice
NZB/W F1 mice were purchased from Japan SLC (Shizuoka, Japan) and MRL/lpr mice were purchased from Charles River (Kanagawa, Japan). All the mice used were female. Female MRL/lpr mice develop severe nephritis at 12–24 weeks, and female NZB/W F1 mice develop severe nephritis at 24–48 weeks. Therefore, we selected 20–26-week-old MRL/lpr mice and 36–47-week-old NZB/W F1 mice with severe glomerulonephritis (continuous proteinuria > 300 mg/dl) accompanied by symptoms related to the central nervous system as nephritic mice. Six-week-old MRL/lpr mice and 12-week-old NZB/WF1 mice were used as young mice.
RNA isolation and cDNA synthesis
The spleen, peripheral lymph nodes (pLNs; axillary, inguinal and mesenteric), kidneys, brain, lung, small intestine and heart were collected from each mouse after perfusion of the mice with PBS of 20 ml. Total RNA was isolated from the above tissues using ISOGEN (Nippon Gene, Tokyo, Japan) according to the manufacturer's protocol. Eight to 10 µg of total RNA was converted to cDNA.
TCR RT-PCR/single-strand conformation polymorphism (SSCP) analysis
RT-PCR–SSCP studies of the TCR Vβ chain were performed as described previously [10,11]. Briefly, one-fiftieth of each cDNA reaction was mixed with each primer set (a common Cβ primer and a Vβ specific primer) at a final concentration of 1·2 pmol/l. PCRs were performed with dNTPs and Taq DNA polymerase (Takara Bio Inc, Shiga, Japan) for 35 cycles (95°C for 1·5 min, 54°C for 2 min and 72°C for 3 min) in a GeneAmp® PCR System 9700 (Applied Biosystems, Foster City, CA, USA). After electrophoresis in non-denaturing 4% polyacrylamide gels, the amplified product was transferred onto a nylon membrane (Perkin Elmer Co. Foster City, CA, USA) and then hybridized with a biotinylated Cβ oligonucleotide probe. Finally, the membrane was visualized by subsequent incubation with streptoavidin, biotinylated alkaline phosphatase and a chemiluminescent substrate system (Phototope™ Star Detection kit; New England Biolabs, Beverley, MA, USA).
Identification of TCR CDR3 for sequencing analysis
Representative subcloned DNA with different CDR3 sequences and RT-PCR products for each organ were subjected to SSCP electrophoresis at the same time to determine which of the T cell clonotypes with unique TCR Vβ CDR3 sequences migrate to the position on the gel where the discrete band of interest was shown by the former PCR product. Identified DNAs were then sequenced by an ABI 310 automated sequencer (Applied Biosystems, Foster City, CA, USA).
Preparation of single cell suspension
The spleen, kidneys, lung or peripheral lymph nodes (pLNs; axillary, inguinal and mesenteric) derived from the mice were minced and incubated in 5 ml of an enzyme preparation containing RPMI-1640 (Gibco Laboratories, Grand Island, NY, USA), 10% fetal calf serum, 1% penicillin, 0·1% collagenase and 20 µg/ml DNase (Sigma Chemical Co., St Louis, MO, USA) at 37°C for 30 min to obtain single cell suspensions.
Magnetic cell sorting
Single cell suspensions from pLNs were separated into CD4 depletion, CD8 depletion or CD4 and CD8 depletion populations using MACS® magnetic separation columns LD (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's instructions. In brief, cells were incubated with biotin-conjugated rat antimouse CD4 (PharMingen; clone RM4-5), biotin-conjugated rat antimouse CD8α (PharMingen; (clone 53–6·7)), or both of these antibodies for 10 min on ice, followed by Streptavidin MicroBeads (Miltenyi Biotec) for 15 min at 4°C. After washing, the mixtures of labelled and non-labelled cells were passed over an LD column inserted into a MidiMACS magnet (Miltenyi Biotec). The CD4-, CD8- or CD4- and CD8-depleted fraction was collected in the flow-through. The purity of the MACS-separated depletion population confirmed by flow cytometry was >94%.
Flow cytometric analysis and antibodies
For single cells obtained from the lung or kidneys of young and nephritic NZB/W F1 mice, the following MoAbs used for immunofluorescent staining were purchased from BD PharMingen (San Diego, CA, USA): FITC anti-mouse CD4 (GK1·5), PE antimouse CD69 (H1·2F3) and PE antimouse CD62L (MEL-14). After washing in 1 × phosphate buffered saline (PBS), cells were fixed with 1% paraformaldehyde in PBS and then stored at 4°C in the dark before flow cytometric analysis. Cytometry was performed on an EPICS XL Flow Cytometer (Coulter Electronics, Hialeah, FL, USA) using system ii™ software version 2·1 (Becton Dickinson). The gate was set for lymphocyte based on forward and side light-scatter characteristics, and 10 000 events were acquired per sample.
FACS purification of cell populations
Single cells prepared from the right kidney of nephritic NZB/W F1 mice were stained with FITC anti-CD4 MoAb, PE anti-CD40L MoAb and biotin anti-CD69 followed by allophycocyanin-conjugated streptavidin. Then CD4+CD69+ cells were sorted on a FACSVantage HG flow cytometer (Becton Dickision) after being gated on a CD4-positive population. The purity of the sorted population exceeded 97%.
RESULTS
Analyses of T cell clonality in various organs from young and nephritic murine models of lupus
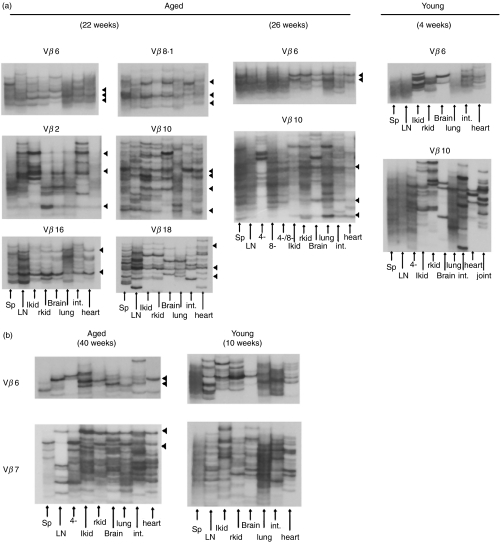
To compare the expanded T cell clonality in various organs from nephritic mice with lupus nephritis and young mice without obvious proteinuria, we performed the established RT-PCR–SSCP analysis which can discriminate T cell clones with different T cell receptor (TCR) Vβ motifs. The spleen, peripheral lymph nodes, bilateral kidneys, brain, lung, small intestine and heart were isolated from young and nephritic MRL/lpr or NZB/W F1 mice and were subjected to RT-PCR–SSCP analysis as described in Materials and Methods. Representative results are shown in Fig. 1. Although young lupus-prone mice without any manifestation of nephritis showed a few distinct expanded T cell clones in each individual organ, these clones were essentially restricted to the individual organ. In contrast, dominant T cell clonal expansions and accumulations with some limited TCR Vβ usage were observed in the kidneys and other organs of nephritic diseased mice, and intriguingly some identical clonotypes were demonstrated in the bilateral kidneys, brain, lung and intestine. The dominant T cell clonal expansions were essentially conserved in repeated electrophoresis of the new PCR products sythesized from the same cDNAs. The limited TCR Vβ usage was Vβ2, Vβ6, Vβ8·1, Vβ10, Vβ16, Vβ18 in MRL/lpr mice (Fig. 1a) and Vβ6, Vβ7 in NZB/W F1 mouse (Fig. 1b).
Fig. 1.
Analysis of T cell clonality in various organs from young and nephritic lupus mice by RT-PCR–SSCP. Different organs such as the spleen, LNs, bilateral kidneys, brain, lung, small intestine and heart isolated using the magnetic cell sorting method from nephritic and young MRL/lpr as well as NZB/W F1 mice and CD4-depleted, CD8-depleted or CD4- and CD8-depleted populations from lymph nodes were subjected to RT-PCR–SSCP. (a) Comparison of T cell clonality in different tissues between nephritic and young MRL/lpr mice. (b) Comparison of T cell clonality in different tissues between nephritic and young NZB/W F1 mice. Black triangles indicate the identical TCR Vβ clonotypes expanded in different organs. Lanes representative of different organs are indicated as arrows. Three individual mice were analysed in each group, and all mice yielded essentially the same results within each group.
To further determine the phenotype of clonally expanded and accumulated T cells in different tissues of nephritic mice, isolated pLNs were separated into CD4-depleted, CD8-depleted or CD4- and CD8-depleted populations using the magnetic cell sorting method (MACS), and were then subjected to RT-PCR–SSCP analysis. A number of these commonly expanded T cell clonotypes were of the CD4+ subset both in the NZB/W F1 and MRL/lpr mice, as many of these clonotypes disappeared upon CD4 or CD4·CD8 depletion. This is consistent with the FACS results, in which the percentages of CD4+ T cells in organs were found to be about two to three times higher than the percentages of CD8+ T cells (data not shown). In Fig. 1a, some bands of CD4·CD8 depletion exhibited a pattern similar as after CD8 depletion. Since we have confirmed the depletion of each of CD4, CD8 or CD4·8 depletion by FACS analysis, the residual bands in CD4·8 depletion seem to be derived from CD4–CD8–CD3+ cells accumulated in nephritic MRL/lpr mice [12]. Many of the residual bands in CD4·8 depletion did not showed commonly expansion in organs. These results indicate that the commonly expanded T cell clones exist in different tissues in the nephritic mice, and that a number of these identical clonotypes belong to the CD4+ subset.
TCR Vβ CDR3 sequence analysis
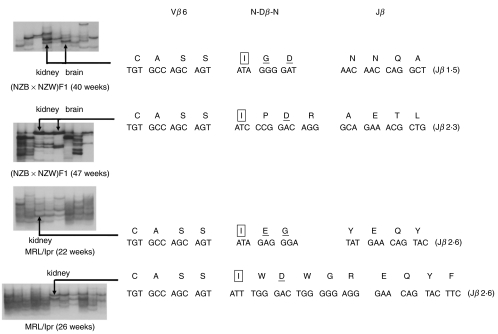
We next tried to examine whether these commonly expanded CD4+ T cell clonotypes in various organs of NZB/W F1 and MRL/lpr mice share some characteristics of the CDR3 sequences with the TCR Vβ chains. RT-PCR products of TCR Vβ6 genes of the kidney and brain from nephritic mice of these two strains were cloned and sequenced. Then the representative PCR products of plasmid DNA with different CDR3 sequences were subjected to SSCP electrophoresis with the former TCR Vβ6 PCR product. We determined which of the T cell clonotypes with unique CDR3 sequences migrate to the same position on gel as the designated identical bands in the analyses of various organs. The CDR3 sequence of these identical TCR Vβ6 clonotypes was thus determined. As shown in Fig. 2, the indicated bands with the same migration from the left kidney and brain in the same individual exhibited an identical CDR3 sequence. These TCR Vβ6 chains displayed a common amino acid residue motif of IXD in CDR3 loops from two different individuals of NZB/W F1 mice. Similarly, IXD was also found in one MRl/lpr mouse, and I and E was found in another. The amino acid of G was shown in three individuals of these two strains. These N(D)N regions encoding isoleucine gave rise to the sequence CASSI in TCR Vβ6 gene. These data strongly suggest an antigen-driven response in these commonly expanded CD4+ T cells.
Fig. 2.
The CDR3 sequence analysis of the TCR Vβ6 chain expressed by the identical clonotype accumulated in different organs from nephritic mice. Nucleotides and corresponding deduced amino acid sequences of the V(D)J junctional region of the identical TCR Vβ6 clones accumulated in the kidney and brain of nephritic NZB/W F1 and MRL/lpr mice are shown. Amino acid sequences are displayed in single letter code above the nucleotide sequences. Arrows indicate the identical bands at different sites that were sequenced. The indicated bands with the same migration from the left kidney and brain exhibited entirely identical CDR3 sequences. The common amino acid motif of ‘I’, ‘D’ or ‘E’ and ‘G’ found in these identical clonotypes from different individuals of the two strains were boxed, enlarged and underlined or simply underlined, respectively.
Phenotypic analysis of CD4+ T cells in nephritic NZB/W F1 mice
To demonstrate whether the CD4+ T cells that accumulated in different tissues were activated, we performed a FACS analysis of some activation markers expressed on CD4+ T cells in the spleen, kidney, lung and small intestine of nephritic NZB/W F1 mice. As can be seen in Table 1, the existence of CD69+CD4+ and CD62Llow CD4+ T cell populations were found in CD4+ T cells derived from the spleen, kidney, lung and small intestine. These findings indicated that the CD4+ T cells that accumulated in nephritic NZB/W F1 mice were activated.
Table 1.
The phenotype of T cells derived from nephritic NZB/W F1 mice
| Spleen | Kidney | Lung | Small intestine | |||||
|---|---|---|---|---|---|---|---|---|
| CD69+ | CD62Llow | CD69+ | CD62Llow | CD69+ | CD62Llow | CD69+ | CD62Llow | |
| Whole organ cells | 6·35 | 18·87 | 3·11 | 6·92 | 4·82 | 15·77 | 1·69 | 6·37 |
| CD4+ | 16·68 | 48·55 | 23·19 | 58·94 | 21·37 | 73·25 | 23·34 | 89·09 |
Numbers shown are percentages of CD69+ or CD62Llow cells either in the whole organ cell population or in the CD4+ population. Experiments were conducted at least three times using different mice, and the data were not significantly different. Representative data are shown.
Phenotype of T cell clone commonly expanded in various organs
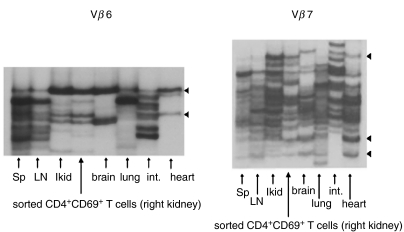
To investigate which phenotypes of the activated CD4+ T cells were involved in the common expanding in various organs, we purified CD4+CD69+ cells from the right kidney of a 47-week-old NZB/W F1 mouse by cell sorting on a FACSVantage. Total RNA of the sorted cells was extracted, reverse transcribed and amplified with primers specific for Vβ6 and Vβ7. Subsequent SSCP analysis was then carried out to compare the expanded clonotype of the sorted CD4+CD69+ population with that of the whole T cell populations in other tissues, including the left kidney. As presented in Fig. 3, the CD4+CD69+ cell population displayed expanded bands identical to those of the left kidney and other different tissues with respect either to Vβ6 or Vβ7. The strong identity of the SSCP electrophoresis pattern of the left and right kidneys (Fig. 1a and b) and left kidney and CD4+CD69+ sorted cells from the right kidney (Fig. 3) suggest the reproducibility of our SSCP analysis. These findings revealed that the commonly expanded T cell clonotypes in various organs in this lupus model have an activated phenotype.
Fig. 3.
Analysis of T cell clonality of FACS-sorted CD4+CD69+ from the right kidney of a NZB/W F1 nephritic mice at 47 weeks by RT-PCR–SSCP. FACS-sorted CD4+CD69+ cells from the right kidney and the whole T cell populations of other organs were subjected to RT-PCR–SSCP analysis to compare the expanded clonotypes of the sorted CD4+CD69+ population with those of the whole T cell populations in other organs, including the left kidney. Black triangles indicate identical expanded bands shared by the CD4+CD69+ population and the whole T cell populations in other organs. Lanes representative of different organs or sorted cells are indicated by arrows.
DISCUSSION
RT-PCR–SSCP analysis is thought to be one of the most sensitive techniques for detecting expanded and accumulated T cell clones in vivo and in vitro[13–15]. In the present study, we employed this method to investigate the pathogenic role of CD4+ T cells in SLE by comparing T cell clonality in various organs obtained from young and nephritic mice of the MRL/lpr and NZB/W F1 strains. Our results demonstrated that although young mice of these two strains showed a few distinct expanded T cell clones, no identical clonotypes were found among the different organs. In contrast, some identical T cell clonotypes with limited TCR Vβ usage showed expansion and accumulation in different organs taken from nephritic mice. These observations indicate the systemic nature of this disease and the possible pathogenic T cell clonotypes involved in these two strains as well. Moreover, these identical clonotypes were mainly CD4+ T cells, suggesting that there is selective expansion and accumulation of the CD4+ subset at different sites, and that these clonally expanded CD4+ T cells may play a dominant role in the disease development in these lupus mice.
Previous studies have revealed that in contrast to CD8+ and CD4–CD8– T cells, a number of CD4+ T cells showed a large disease-related increase in NZB/W F1 mice [1]. Also, in addition to a massive increase in aberrant double-negative (CD4–CD8–) T cells (DN T cells), MRL/lpr mice demonstrated expansions in the CD4+ population as well [2]. Although the lymphadenopathy in MRL/lpr mice is due primarily to the DN T cell accumulation, autoimmunities such as autoantibodies, vasculitis, arthritis and Ig-induced nephritis in this strain depend largely on CD4+ T cells [3,4]. Similar results were also obtained from SLE patients in that the expanded CD4+ T cell clonotypes were not restricted to the kidney but were also found in the peripheral blood and other lesions, such as pleural and pericardial effusions of patients with lupus serositis, and these expansions correlated with the disease activities [16–18]. Our findings are consistent with the above studies, and to our knowledge this is the first report of the existence of identical CD4+ T cell clonotypes having expanded in many organs of lupus mice. Because the CD4+ T cell is important in the pathogenesis of lupus [3–7], our results suggest that the clonally expanded CD4+ T cells in MRL/lpr and NZB/W F1 mice play an important role in the development of SLE. Our observation of restricted Vβ usage in commonly expanded clones suggests that these CD4+ T cells were likely to be activated by a particular autoantigen(s). The restricted use of TCR Vβ genes in aged diseased mice has been reported in MRL/lpr [19,20] and NZB/W F1 [21,22] mice.
We further addressed whether these identical expanded CD4+ T cell clones are activated by specific antigen(s) by performing CDR3 sequence analysis of the TCR Vβ6 chain expressed on these T cell subsets in nephritic MRL/lpr and NZB/W F1 mice. We found that these TCR Vβ6 chains displayed a common amino acid residue motif of IXD or E and G in the CDR3 loops either in the brain or kidney in these two strains. These N(D)N regions encoding isoleucine gave rise to the sequence CASSI in TCR Vβ6 gene. These findings suggest that CD4+ T cells in these strains may recognize restricted epitopes on autoantigens, thus leading to the specific antigen-driven activation and resulting in clonal expansion and accumulation in different sites of diseased mice. Previous studies showed that CDR3 of the TCR Vβ chain expressed by pathogenic T cell clones in lupus mice bears a recurrent motif of anionic residues such as D or E, suggesting that it is specific for autoantigens with cationic residue [23–27]. Among several autoantigens predicted to be possibly involved in SLE to date, nucleosomes that carry cationic residues are believed to be primary immunogens that initiate the cognate interaction between the pathogenic T and B cells of SLE and induce the pathogenic anti-DNA autoantibodies [26,27]. The existence of a D motif in the CDR3 region found in our experiments is in accordance with those studies. Previous reports suggest the associtation of Vβ6 with autoreactivity by the increased expression of Vβ6 in MRL/lpr mice compared to control mice [28], and by the existence of autoreactive T cell lines with Vβ6 from lupus-prone SNF1 mice [29]. Although these reports may be consistent with our results, the actual evaluation of autoreactivity of expanded T cells should be clarified by the cloning of TCR of these T cells.
SSCP-bands could result from co-migration of the cDNA molecules with different sequences. Although definitive proof of clonal T cells requires further actual cloning and sequencing, we found co-migration to be a relatively rare phenomenon in SSCP analysis, and each band we analysed was clearly derived from a single sequence (Fig. 2).
Our FACS analysis showed that the activated CD4+ T cells such as the CD4+CD69+ or CD4+CD62Llow cell population were detected in the spleen, kidney, lung and small intestine in nephritic NZB/W F1 mice. This indicates that CD4+ T cells infiltrating into different sites of diseased lupus mice were activated. It has been demonstrated that the CD4+CD69+ population shared the same clonotypes as those commonly expanded clonotypes of whole T cell populations in other organs, including the left kidney. This observation revealed that the commonly expanded and accumulated T cell clonotypes in various organs in this lupus model contained CD4+ T cell of activated phenotype. Therefore, we concluded that some CD4+ T cells in nephritic lupus mice were activated, and it is the CD4+CD69+ phenotype that clonally expanded and accumulated in various sites. Thus these cells might be involved in the disease pathogenesis.
CD69 is known to be an early T cell activation marker that is involved in signal transduction, cell proliferation and cytokine secretion [30]. Previous reports have shown that the CD4+CD69+ subset of peripheral blood T cells was larger in SLE patients than in normal controls [31] and the percentage of CD4+CD69+ T cells correlated with the disease activity in SLE patients [32]. The expression of this subset was also found to be significantly increased in peripheral lymphoid tissues and inflammatory infiltrates in the kidney and lung of aged NZB/W F1 mice without skewing to a particular TCR Vβ usage [33]. Our results showed agreement with the above studies, suggesting that the clonally expanded CD4+CD69+ phenotypes in various organs may play an important role in SLE pathogenesis via signal transduction, cell proliferation and cytokine secretion.
As is well known, MRL/lpr mice which develop severe, progressive, systemic autoimmune disease spontaneously often exhibit a high incidence of inflammatory central nervous system (CNS) disease [34–36]. Previous studies showed the dominant infiltration of CD4+ T cells in brains from this strain [35], and anti-CD4 therapy effectively prevented the development of CNS lesions [37]. Our SSCP study revealed that the systemically expanding CD4+ T cell clone also existed in the CNS of aged MRL/lpr mice. These results suggest that CD4+ T cells activated by systemic autoantigen migrate to the brain across the blood–brain barrier. The hypothesis in some degree may elucidate why the onset of CNS lesions in human SLE are often associated with disease severity and disease duration.
In summary, combining RT-PCR–SSCP analysis with the CDR3 sequence analysis of the TCR Vβ chain family expressed by T cell clones has allowed us to demonstrate that some CD4+ T cells are activated, clonally expanded and commonly accumulated in different tissues in nephritic MRL/lpr and NZB/W F1 lupus mice. These CD4+ T cell clonotypes might recognize restricted T cell epitopes on autoantigens and are involved in specific immune responses of SLE, thus playing a pathogenic role in these lupus mice.
Acknowledgments
This study was supported by grants from the Ministry of Health, Labour and Welfare and the Ministry of Education, Culture, Sports, Science and Technology of Japan. We are grateful to Kazumi Abe for her excellent technical assistance and Professor Bi Li-qi for her continuous encouragement.
References
- 1.Rozzo SJ, Drake CG, Chiang BL, et al. Evidence for polyclonal T cell activation in murine models of systemic lupus erythematosus. J Immunol. 1994;153:1340–51. [PubMed] [Google Scholar]
- 2.Giese T, Davidson WF. Evidence for early onset, polyclonal activation of T cell subsets in mice homozygous for lpr. J Immunol. 1992;149:3097–106. [PubMed] [Google Scholar]
- 3.Koh DR, Ho A, Rahemtulla A, et al. Murine lupus in MRL/lpr mice lacking CD4 or CD8 T cells. Eur J Immunol. 1995;25:2558–62. doi: 10.1002/eji.1830250923. [DOI] [PubMed] [Google Scholar]
- 4.Jevnikar AM, Grusby MJ, Glimcher LH. Prevention of nephritis in major histocompatibility complex class II-deficient MRL-lpr mice. J Exp Med. 1994;179:1137–43. doi: 10.1084/jem.179.4.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jabs DA, Burek CL, Hu Q, et al. Anti-CD4 monoclonal antibody therapy suppresses autoimmune disease in MRL/Mp-lpr/lpr mice. Cell Immunol. 1992;141:496–507. doi: 10.1016/0008-8749(92)90166-m. [DOI] [PubMed] [Google Scholar]
- 6.Santoro TJ, Portanova JP, Kotzin BL. The contribution of L3T4+ T cells to lymphoproliferation and autoantibody production in MRL-lpr/lpr mice. J Exp Med. 1988;167:1713–8. doi: 10.1084/jem.167.5.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wofsy D, Seaman WE. Successful treatment of autoimmunity in NZB/NZW F1 mice with monoclonal antibody to L3T4. J Exp Med. 1985;161:378–91. doi: 10.1084/jem.161.2.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sekigawa I, Okada T, Noguchi K, et al. Class-specific regulation of anti-DNA antibody synthesis and the age-associated changes in (NZB × NZW) F1 hybrid mice. J Immunol. 1987;138:2890–5. [PubMed] [Google Scholar]
- 9.Chesnutt MS, Finck BK, Killeen N, et al. Enhanced lymphoproliferation and diminished autoimmunity in CD4-deficient MRL/lpr mice. Clin Immunol Immunopathol. 1998;87:23–32. doi: 10.1006/clin.1997.4492. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto K, Masuko K, Takahashi S, et al. Accumulation of distinct T cell clonotypes in human solid tumors. J Immunol. 1995;154:1804–9. [PubMed] [Google Scholar]
- 11.Yamamoto K, Masuko-Hongo K, Tanaka A, et al. Establishment and application of a novel T cell clonality analysis using single-strand conformation polymorphism of T cell receptor messenger signals. Hum Immunol. 1996;48:23–31. doi: 10.1016/0198-8859(96)00080-8. [DOI] [PubMed] [Google Scholar]
- 12.Mixter PF, Russell JQ, Morrissette GJ, et al. A model for the origin of TCR-alphabeta+ CD4−CD8− B220+ cells based on high affinity TCR signals. J Immunol. 1999;162:5747–56. [PubMed] [Google Scholar]
- 13.Yamamoto K, Sakoda H, Nakajima T, et al. Accumulation of multiple T cell clonotypes in the synovial lesions of patients with rheumatoid arthritis revealed by a novel clonality analysis. Int Immunol. 1992;4:1219–23. doi: 10.1093/intimm/4.11.1219. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda Y, Masuko K, Nakai Y, et al. High frequencies of identical T cell clonotypes in synovial tissues of rheumatoid arthritis patients suggest the occurrence of common antigen-driven immune responses. Arthritis Rheum. 1996;39:446–53. doi: 10.1002/art.1780390312. [DOI] [PubMed] [Google Scholar]
- 15.Komagata Y, Masuko K, Tashiro F, et al. Clonal prevalence of T cells infiltrating into the pancreas of prediabetic non-obese diabetic mice. Int Immunol. 1996;8:807–14. doi: 10.1093/intimm/8.6.807. [DOI] [PubMed] [Google Scholar]
- 16.Mato T, Masuko K, Misaki Y, et al. Correlation of clonal T cell expansion with disease activity in systemic lupus erythematosus. Int Immunol. 1997;9:547–54. doi: 10.1093/intimm/9.4.547. [DOI] [PubMed] [Google Scholar]
- 17.Kolowos W, Gaipl US, Voll RE, et al. CD4 positive peripheral T cells from patients with systemic lupus erythematosus (SLE) are clonally expanded. Lupus. 2001;10:321–31. doi: 10.1191/096120301671176280. [DOI] [PubMed] [Google Scholar]
- 18.Kato T, Kurokawa M, Sasakawa H, et al. Analysis of accumulated T cell clonotypes in patients with systemic lupus erythematosus. Arthritis Rheum. 2000;43:2712–21. doi: 10.1002/1529-0131(200012)43:12<2712::AID-ANR11>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 19.Singer PA, Theofilopoulos AN. T-cell receptor V beta repertoire expression in murine models of SLE. Immunol Rev. 1990;118:103–27. doi: 10.1111/j.1600-065x.1990.tb00814.x. [DOI] [PubMed] [Google Scholar]
- 20.Singer PA, McEvilly RJ, Noonan DJ, et al. Clonal diversity and T-cell receptor beta-chain variable gene expression in enlarged lymph nodes of MRL-lpr/lpr lupus mice. Proc Natl Acad Sci USA. 1986;83:7018–22. doi: 10.1073/pnas.83.18.7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirose S, Tokushige K, Kinoshita K, et al. Contribution of the gene linked to the T cell receptor beta chain gene complex of NZW mice to the autoimmunity of (NZB × NZW) F1 mice. Eur J Immunol. 1991;21:823–6. doi: 10.1002/eji.1830210343. [DOI] [PubMed] [Google Scholar]
- 22.Yanagi Y, Hirose S, Nagasawa R, et al. Does the deletion within T cell receptor beta-chain gene of NZW mice contribute to autoimmunity in (NZB × NZW) F1 mice? Eur J Immunol. 1986;16:1179–82. doi: 10.1002/eji.1830160925. [DOI] [PubMed] [Google Scholar]
- 23.Datta SK, Patel H, Berry D. Induction of a cationic shift in IgG anti-DNA autoantibodies. Role of T helper cells with classical and novel phenotypes in three murine models of lupus nephritis. J Exp Med. 1987;165:1252–68. doi: 10.1084/jem.165.5.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suzuki N, Harada T, Mizushima Y, et al. Possible pathogenic role of cationic anti-DNA autoantibodies in the development of nephritis in patients with systemic lupus erythematosus. J Immunol. 1993;151:1128–36. [PubMed] [Google Scholar]
- 25.Desai-Mehta A, Mao C, Rajagopalan S, et al. Structure and specificity of T cell receptors expressed by potentially pathogenic anti-DNA autoantibody-inducing T cells in human lupus. J Clin Invest. 1995;95:531–41. doi: 10.1172/JCI117695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohan C, Adams S, Stanik V, et al. Nucleosome: a major immunogen for pathogenic autoantibody-inducing T cells of lupus. J Exp Med. 1993;177:1367–81. doi: 10.1084/jem.177.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams S, Leblanc P, Datta SK. Junctional region sequences of T-cell receptor beta-chain genes expressed by pathogenic anti-DNA autoantibody-inducing helper T cells from lupus mice: possible selection by cationic autoantigens. Proc Natl Acad Sci USA. 1991;88:11271–5. doi: 10.1073/pnas.88.24.11271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mountz JD, Smith TM, Toth KS. Altered expression of self-reactive T cell receptor V beta regions in autoimmune mice. J Immunol. 1990;144:2159–66. [PubMed] [Google Scholar]
- 29.Adams S, Zordan T, Sainis K, Datta S. T cell receptor V beta genes expressed by IgG anti-DNA autoantibody-inducing T cells in lupus nephritis: forbidden receptors and double-negative T cells. Eur J Immunol. 1990;20:1435–43. doi: 10.1002/eji.1830200705. [DOI] [PubMed] [Google Scholar]
- 30.Cebrian M, Yague E, Rincon M, et al. Triggering of T cell proliferation through AIM, an activation inducer molecule expressed on activated human lymphocytes. J Exp Med. 1988;168:1621–37. doi: 10.1084/jem.168.5.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crispin JC, Martinez A, de Pablo P, et al. Participation of the CD69 antigen in the T-cell activation process of patients with systemic lupus erythematosus. Scand J Immunol. 1998;48:196–200. doi: 10.1046/j.1365-3083.1998.00366.x. [DOI] [PubMed] [Google Scholar]
- 32.Portales-Perez D, Gonzalez-Amaro R, Abud-Mendoza C, et al. Abnormalities in CD69 expression, cytosolic pH and Ca2+ during activation of lymphocytes from patients with systemic lupus erythematosus. Lupus. 1997;6:48–56. doi: 10.1177/096120339700600107. [DOI] [PubMed] [Google Scholar]
- 33.Ishikawa S, Akakura S, Abe M, et al. A subset of CD4+ T cells expressing early activation antigen CD69 in murine lupus: possible abnormal regulatory role for cytokine imbalance. J Immunol. 1998;161:1267–73. [PubMed] [Google Scholar]
- 34.Alexander EL, Murphy ED, Roths JB, et al. Congenic autoimmune murine models of central nervous system disease in connective tissue disorders. Ann Neurol. 1983;14:242–8. doi: 10.1002/ana.410140211. [DOI] [PubMed] [Google Scholar]
- 35.Vogelweid CM, Johnson GC, Besch-Williford CL, et al. Inflammatory central nervous system disease in lupus-prone MRL/lpr mice: comparative histologic and immunohistochemical findings. J Neuroimmunol. 1991;35:89–99. doi: 10.1016/0165-5728(91)90164-3. [DOI] [PubMed] [Google Scholar]
- 36.Hess DC, Taormina M, Thompson J, et al. Cognitive and neurologic deficits in the MRL/lpr mouse: a clinicopathologic study. J Rheumatol. 1993;20:610–7. [PubMed] [Google Scholar]
- 37.O'Sullivan FX, Vogelweid CM, Besch-Williford CL, et al. Differential effects of CD4+ T cell depletion on inflammatory central nervous system disease, arthritis and sialadenitis in MRL/lpr mice. J Autoimmun. 1995;8:163–75. doi: 10.1006/jaut.1995.0013. [DOI] [PubMed] [Google Scholar]