Abstract
Cytokine production may be regulated by both genotypic (single nucleotide or tandem repeat polymorphisms) and non-genotypic factors relating to the environment and inherent biology (i.e. gender). Interleukin (IL)-1 is one of the body's most highly proinflammatory cytokines and is implicated in the pathophysiology of numerous diseases, but also in the maintenance of homeostasis in a number of tissues. The cytokine IL-1 receptor antagonist (IL-1Ra) is the competitive inhibitor of the IL-1 agonists IL-1α and IL-1β. In vivo IL-1Ra was measured in a cohort of 200 + blood donors and the effect of the IL-1 gene polymorphisms, environmental and biological factors assessed. In this study, we observed that possession of particular alleles of 5 IL-1 gene polymorphisms (IL1A-889, IL1Α VNTR, IL1B -511, IL1B +3953 and the IL1RN VNTR) did not correlate with higher plasma IL-1Ra levels. Environmental factors such as smoking and non-steroidal anti-inflammatory drug ingestion were associated with higher in vivo IL-1Ra levels (P = 0·015 and 0·022, respectively), but biological factors such as gender, age and menstruation status did not have any impact upon in vivo IL-1Ra levels. Genotypic associations of IL-1 gene family polymorphisms with disease features may reflect characteristics of stressed rather than normal control circuits for cytokine production.
Keywords: environmental, genotypic factors, interleukin-1
INTRODUCTION
Various factors have been observed to influence the production of cytokines, whether they are pro- or anti-inflammatory in nature. The regulation of cytokine production is influenced by genetic factors such as cytokine gene polymorphisms (CGPs) [1], and environmental factors such as nutritional status, smoking, therapy with steroids [2] and non-steroidal anti-inflammatory drugs (NSAID), and other inherent biological factors such as gender [3,4], age [3,5] and circadian rhythms [6]. A single or multiple of these factors may be responsible for variation in cytokine production between individuals.
A number of studies have shown a positive correlation between plasma and serum cytokine levels (in vivo) [7] and those produced in tissue culture systems (in vitro), where peripheral blood mononuclear cells are purely cultured and cytokine production analysed or stimulated with mitogens such as lipopolysaccharide (LPS), concavalin A (Con A) or phytohaemaglutinin (PHA) and the cytokine production analysed. Vamvakopoulos et al. have elegantly demonstrated the role of interleukin (IL)-1 (IL-1RN VNTR and IL-1B+3953) polymorphisms on stimulated IL-1β production [8] without formal estimation of haplotype frequencies in their study population. However, there are also a number of studies where the association of cytokine levels does not correlate with CGPs. A number of studies, in which polymorphisms in certain cytokine genes have been described previously to influence cytokine production, are analysed in various disease cohorts [1]. In these studies the cytokine in question is often implicated in the pathophysiology of the disease, most usually in the inflammatory process.
The IL-1 family of cytokines has been implicated in the pathology of a number of diseases with a proinflammatory element. The IL-1 receptor antagonist (IL-1Ra) competitively inhibits the binding of the two IL-1 agonists (IL-1α and -β, two of the body's most highly proinflammatory cytokines) to the type 1 IL-1 receptor, which is responsible for signal transduction and the resultant biological activities.
IL-1 has a wide role in immunobiology, denoted by the variety of cells that are capable of expressing and reacting to IL-1 binding. Monocytes are the main source of IL-1 production, where expression is predominantly IL-1β. In contrast, skin keratinocytes produce large quantities of IL-1α[9]. It has been shown that in the transition of monocyte to tissue macrophage there is also a transition between the production of IL-1β to IL-1α [10]. Expression of IL-1 is seen in macrophages of different sources (alveolar, spleen and peritoneal macrophages and Kupffer cells), peripheral neutrophil granulocytes, endothelial cells, fibroblasts, smooth muscle and Langerhans cells, osteoclasts, astrocytes, neural cell, T, B and natural killer (NK) cells [11].
Polymorphisms in the IL-1 gene cluster (IL-1α, IL-1β and IL-1Ra) located on chromosome 2q have been implicated in the regulation of IL-1 production [8,12], but have also been associated with many diseases [13–25] containing an inflammatory element. There have also been a number of studies where there was no association of genotype with disease, or where initial disease association reports have not been confirmed.
The aim of this study was to assess the impact of IL-1 gene polymorphisms and non-genetic factors, both environmental and biological (assessed by questionnaire), on the in vivo levels of IL-1Ra. Briefly, the study was unable to confirm previous findings [7] that a combination of IL-1β and IL-1Ra genotypes influenced in vivo levels. The ingestion of NSAIDs influenced the level of IL-1Ra, and those individuals who smoked also had a different level of IL-1Ra compared to those who did not smoke. A weak trend was observed (after multivariate modelling) between gender and IL-1Ra levels.
MATERIALS AND METHODS
Study populations
The Joint Local Ethics Committee (JLEC) for the Newcastle region approved the collection of blood samples (both EDTA-anticoagulated and serum) from consenting laboratory staff (n = 24) for a small pilot study to confirm the effect of clot formation on cytokine levels.
A total of 229 blood donors gave informed consent to participate voluntarily in the study, with ethics granted under the auspices of the National Blood Authority (NBA). This also meant that only one sample could be obtained from each subject. Therefore, an EDTA blood sample was taken at the end of the donation from the intravenous line after the completion of blood donation for the retrieval of both plasma for IL-1Ra measurement and leucocytes for DNA analysis of the IL-1 gene polymorphisms. Participation in the study also involved answering a questionnaire, which remained anonymous. Data were obtained on the donors’ age, gender, whether they smoked or were taking prescribed or over-the-counter medicines. Female donors were asked to complete further questions regarding their menstrual cycle: did they have regular menstrual bleeding, were oral contraceptives used, and if so did they contain oestrogen and/or progesterone? If they were not menstruating regularly, did they class themselves as pre- or postmenopausal? Those women who were menopausal were asked if they were taking hormone replacement therapy (HRT). The demographics of the blood donor population can be viewed in Table 1. The results analysed and discussed are centred wholly on the data generated from the samples and questionnaire answers from the NBA blood donors.
Table 1.
Demographics of the NBA study population
| NBA study population | |||
|---|---|---|---|
| Demographics | All n (%) | Male n (%) | Female n (%) |
| No. of volunteers | 229 | 99 | 130 |
| Age (years) | |||
| Mean | 41 | 44 | 39 |
| Median | 41 | 45 | 39 |
| Range | 17–67 | 17–67 | 17–67 |
| Smokers | 42 (18) | 15 (15) | 27 (21) |
| NSAID ingestion | 19 (8) | 4 (4) | 15 (12) |
| Regular menstruation | 93 (41) | 93 (72) | |
| Postmenopausal | 26 (11) | 26 (20) | |
| Hormone-based contraception | 26 (11) | 26 (20) | |
n: number of individuals; %: percentage of individuals of the total.
Sample preparation
All sample separation was performed under sterile conditions. Plasma was obtained from 4·5 ml EDTA blood Vacutainer samples after centrifugation at 380 g for 5–10 min. The plasma was removed and frozen in aliquots at −80°C until required. Plasma samples from the blood donors were separated within 3 h of collection. Data from our laboratory (not shown) have observed no significant difference between levels of plasma cytokines from samples separated within 30 min or 6 h of collection.
Peripheral blood leucocytes were obtained from the buffy coat of the separated EDTA whole blood samples. The cells were washed in TNE (Tris sodium EDTA) buffer and then frozen below −20°C in residual TNE buffer until DNA extraction was performed.
DNA extraction
Genomic DNA was obtained by standard sodium dodecyl sulphate (SDS) proteinase K digestion followed by phenol and separate chloroform steps. DNA was precipitated out of solution by the addition of ammonium acetate (to 2·5 m) and spooled out following the addition of ethanol (to 70%). DNAs were dissolved in TE buffer (10 mm Tris/HCl, 0·5 mm EDTA).
IL1A (−889) SNP polymorphism (LocusLink ID 3552)
Genomic DNA was analysed by polymerase chain reaction (PCR) essentially as described previously [13]. The PCR amplicon was digested with NcoI and was separated by 10% polyacrylamide gel electrophoresis (PAGE) and visualized by silver staining. Limited digestion to the assigned genotype was confirmed by double digestion of PCR with the same enzyme, and spot samples were also analysed by direct DNA sequencing. Briefly, a new set of forward and reverse primers were designed (MacVector, Accelrys, Inc., San Diego, CA, USA), IL1AF (5′-GAG ATG GGG GCT TCA CTA TG-3′) and IL1AR (5′-CGG GAG GTA TGC GTA AGG-3′), with universal M13 forward and reverse sequences attached, so that the new amplicon spanned the region of interest. PCR amplicons were purified and then cycle sequencing was performed by a CEQ 8000 (Beckman Coulter, High Wycombe, UK) using the manufacturer's standard method. The sequence data were analysed by CEQ software (Beckman Coulter) and the sequence was verified against the region of interest in the IL-1α gene (NCBI Blast search file X03883) [26].
IL1A (VNTR) polymorphism (LocusLink ID 3552)
Genomic DNA was amplified by PCR using primers designed by the MacVector software program (Accelrys, Inc.), IL-1α 3 (5′-TCT TCT GAT CCT TGG AGC TGT CC-3′) and IL-1α 4 (5′-GGA GGA TAT GGA AAA CAG ACA ACC C-3′), instead of those described previously [12]. The final concentrations in the PCR were: 1 × NH4 containing buffer (Bioline, London, UK), 1 mm MgCl2 (Bioline), 250 µm dNTPs (Roche Diagnostics, Lewes, UK), 0·2 µm of each primer and 0·5 U Taq polymerase (Bioline) per 20 µl reaction volume. PCR cycle conditions were: 94°C for 30 s, 50°C for 1 min, 72°C for 1 min for 30 cycles including a 5-s autoextension per cycle, with a final 10-min extension step at 72°C. Electrophoresis of PCR products was carried out using 1% agarose (0·9% GTG high resolution agarose (Microsieve, Flowgen, Ashby de la Zouch, UK) and 0·1% of LE low resolution agarose (SeaKem, Flowgen UK), stained with ethidium bromide (0·5 µg/ml) and visualized by UV transillumination. The PCR product was sequenced after cloning into the TA cloning vector (Invitrogen, Paisley, UK) and transformed into XL Blue Competent Escherichia coli (Stratagene, Cambridge, UK); sequence identity was verified against the region of interest in the IL-1α gene (NCBI Blast search file X03883) [26]. The fragment size of each allele was calculated from the flanking region (including forward and reverse primer sequence) either side of the VNTR repeat and the respective number of repeats in specific alleles as described previously [12]. The size of each of the alleles was as follows: allele 1, 638 base pairs (bp); allele 2, 1052 bp; allele 3, 592 bp; allele 4, 776 bp; allele 5, 454 bp; allele 6, 914 bp; and allele 7, 500 bp.
IL1B (−511) polymorphism (LocusLink ID 3553)
The DNA samples were analysed by PCR and restriction enzyme digestion as described [27]. AvaI digestion products were separated by 2% agarose gel electrophoresis (AGE), stained with ethidium bromide and visualized by UV transillumination.
IL1B (+3954) polymorphism (LocusLink ID 3553)
DNA was subjected to PCR and restriction enzyme digestion as described [27]; products were separated by 2% AGE, stained with ethidium bromide and visualized by UV transillumination.
IL1RN (VNTR) polymorphism (LocusLink ID 3557)
This polymorphism, located in intron 2 of the IL-1Ra gene, was analysed as described [28]. PCR products were separated by 2% AGE, stained with ethidium bromide and visualized under UV transillumination.
Plasma and serum IL-1Ra quantification
IL-1Ra levels were measured using Quantikine® IL-1Ra enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems Europe Ltd., Abingdon, UK). The method was performed as per the manufacturer's instructions, including a standard curve double-diluted from 3000 pg/ml to 46·1 pg/ml. The microtitre plates were scanned at 450 nm using an ELISA plate reader (Multiskan MCC/340, Labsystems, Ashford, UK), and the corresponding IL-Ra levels were calculated from the standard curve by the supplied software.
Statistical analysis
The Hardy–Weinberg equilibrium (χ2 test) was used to determine co-dominant inheritance of alleles in each polymorphism. Haplotype frequencies for pairs of alleles at different loci in the NBA normal population were estimated using the Estimating Haplotype frequencies (EH) software program as described (ftp://linkage.rockefeller.edu/software/eh). We have described previously the analysis of these predicted haplotypes between the five polymorphisms studied [16].
The data obtained were analysed by systat 9·0, spss10 and graphpad prism3 statistical software. P-values lower than 0·05 were considered significant. Plasma IL-1Ra levels from the blood donor samples were not normally distributed as assessed by the Kolmogorov–Smirnov Test (by Lilliefors). The plasma IL-1Ra data could not be transformed and were therefore analysed individually against genotypic and non-genotypic factors using non-parametric statistical tests (Mann–Whitney U-test, two-tailed). Allele 2 in each of the polymorphisms analysed was denoted as the allele previously associated with influence on production, disease incidence and/or severity. Genotypic and non-genotypic data were analysed with plasma IL-1Ra levels in univariate analysis by χ2 test, prior to analysis in a multivariate model using logistic regression. The criteria for inclusion were variables with P < 0·5. The final model comprised the non-genotypic factors: smoker, gender, age over 40 years and NSAID usage as independent variables and plasma IL-1Ra as the dependent variable (using a forward stepwise approach).
RESULTS
The effect of clot formation upon IL-1Ra levels was observed in our small pilot study of laboratory volunteers. Significantly higher results were present in the serum compared to plasma taken at the same time each individual (data not shown). Unfortunately, the study size was unable to demonstrate any possible difference between IL-1Ra levels with genotypic or non-genotypic factors as analysed in the NBA study population.
Allele frequency and linkage disequilibrium analysis
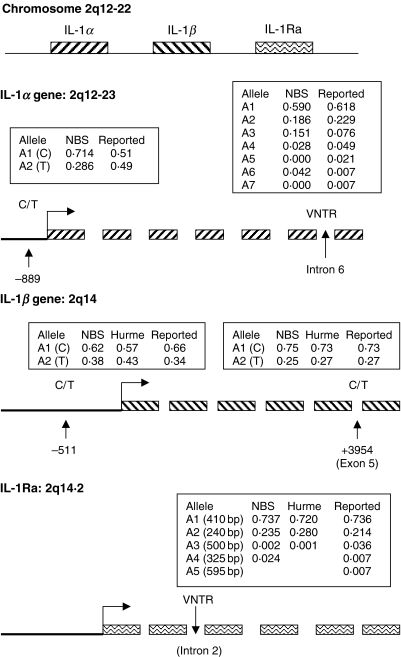
The allele frequencies of the five polymorphisms studied in the normal blood donor volunteers (see Fig. 1) have been reported in our previous studies in a normal control population for IL-1 gene polymorphism analysis [16,17]. The slight but not statistically significant difference (data not shown) in allele frequencies observed in our study compared to those reported previously could be explained by differences in study numbers or by possible ethnic differences between the populations studied.
Fig. 1.
Allele frequencies of the five IL-1 gene polymorphisms studied in the NBA normal population, a comparative study by Hurme et al. [7] and those in previously reported studies; IL1A -889 [13]. IL1A VNTR [54], IL1B -511 [55], IL1B +3954 [56] and the IL1RN VNTR polymorphism. Gene locations are archived in the National Center for Biotechnology Information (NCBI) database [26].
We have reported previously analysis of genotype frequencies, using the EH haplotype software to predict a number of conserved haplotypes in this population (Table 2) [17]. Some polymorphic loci were found to be in strong linkage disequilibrium. Allele 2 of the IL1A (−889) and allele 1 of the IL1A (VNTR) polymorphism being linked together in a commonly occurring conserved haplotype (see Table 2) (P < 0·0001). Haplotypes containing allele 2 at both these two loci are not common. In contrast, other loci show little evidence of linkage, alleles of the IL1A (−889) with the IL1B (−511) polymorphisms (P = 0·316) and the IL1A (−889) and the IL1RN (VNTR) polymorphism (P = 0·5124).
Table 2.
Estimated haplotype frequencies of the IL-1 genotypes from a normal control population
| Loci | Haplotype | |||||||
|---|---|---|---|---|---|---|---|---|
| 1–1 | 1–2 | 2–1 | 2–2 | 1–3,4,5,6 | 2–3,4,5,6 | χ2 | P | |
| IL1A−889/IL1A VNTR | 0·352 | 0·198 | 0·239 | 0·009 | 0·173 | 0·027 | 36·45 | <0·0001 |
| IL1A−889/IL1B−511 | 0·428 | 0·3 | 0·187 | 0·085 | 4·72 | 0·316 | ||
| IL1A−889/IL1B +3954 | 0·66 | 0·064 | 0·089 | 0·187 | 117·5 | <0·0001 | ||
| IL1A−889/IL1RN VNTR | 0·531 | 0·17 | 0·212 | 0·061 | 0·026 | <0·001 | 3·227 | 0·5214 |
| IL1B−511/IL1A VNTR | 0·472 | 0·073 | 0·120 | 0·134 | 0·077 | 0·125 | 66·6 | <0·0001 |
| IL1B−511/IL1B +3954 | 0·421 | 0·196 | 0·329 | 0·053 | 11·87 | 0·0183 | ||
| IL1B−511/IL1RN VNTR | 0·506 | 0·094 | 0·231 | 0·142 | 0·016 | 0·01 | 20·45 | 0·0004 |
| IL1B +3954/IL1A VNTR* | 0·35 | 0·201 | 0·245 | <0·001 | 0·192 | 0·012 | 55·69 | <0·0001 |
| IL1B +3954/IL1RN VNTR | 0·516 | 0·207 | 0·220 | 0·031 | 0·027 | <0·001 | 9·35 | 0·0529 |
| IL1A VNTR/IL1RN VNTR | 0·463 | 0·121 | 0·147 | 0·06 | 0·007 | |||
P-values indicate significance of association where loci can be demonstrated to be linked, the specific alleles that are linked together are illustrated by higher frequency values as described in the text.
These polymorphic loci are not listed in order across the locus to aid the clarity of the table.
The pattern of linkage across the IL-1 gene cluster is not uniform. The IL1A (−889) and IL1B (+3953) loci are in strong linkage disequilibrium with each other (P = 0·0001), yet show little linkage to other loci which physically map between them, namely the IL1A (−889) and IL1B (−511) (P = 0·316) polymorphisms. The strong linkage observed between allele 2 of the IL1A (−889) and the IL1B (+3953) polymorphisms meant that separate analysis of the IL1A (−889) polymorphism with plasma IL-1Ra levels was redundant.
Effect of IL-1 genotype on in vivo IL-1Ra levels
Genotype was compared with IL-1Ra levels, as described. There was no significant difference between possession or non-possession of allele 2 (as the production-associated allele) in any of the IL-1β and IL-1Ra polymorphisms studied when analysed separately with the level of IL-1Ra. We also compared the presence or absence of allele 2 in the IL1RN (VNTR) with presence or absence of allele 2 in the IL1B (−511) or IL1B (+3953) polymorphisms [7]. The plasma levels of IL-1Ra with the four-haplotype combinations when comparing IL-1Ra VNTR and each IL-1β polymorphism were not statistically different when analysed. Therefore, the association of IL1RN (VNTR) allele 2+/IL1B (−511) allele 2+, IL1RN (VNTR) allele 2+/IL1B (+3953) allele 2- and IL1RN (VNTR) allele 2+ alone with increased plasma IL-1Ra level described by Hurme et al. [7] could not be confirmed in our cohort.
The effect of non-genotypic factors on in vivo IL-1Ra levels
Univariate analysis.
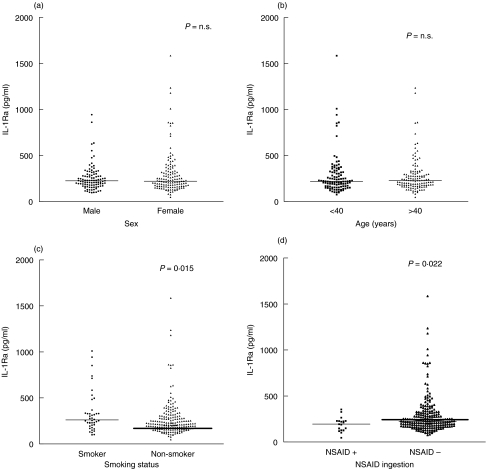
There was no significant difference in IL-1Ra plasma levels between male and female blood donors (see Fig. 2a). Female donors were asked a number of questions relating to hormonal influences that might affect IL-1 production. These included whether they had regular periods, were taking the oral contraceptive pill (and whether it was an oestrogen-containing pill or progesterone-only) and if they were menopausal (with or without the use of HRT). No differences in IL-1Ra production were seen relating to use of the combined oral contraceptive pill or in those women who had passed the menopause compared to women who had not (data not shown).
Fig. 2.
The effect of non-genotypic factors on plasma IL-1Ra levels. (a) Plasma IL-1Ra levels, comparing subjects for gender. There was no difference in plasma IL-1Ra levels between male and female blood donor volunteers. (b) Plasma IL-1Ra levels, comparing subjects for age. There was no difference in plasma IL-1Ra levels between blood donors > or <40 years of age. (c) Plasma IL-1Ra levels, comparing smokers and non-smokers. Smokers had higher plasma levels than non-smokers (P = 0·015). (d) Plasma IL-1Ra levels, examining the effect of NSAIDs. Those donors who had taken NSAIDs in the 2 weeks prior to donation also had higher plasma IL-1Ra levels (P = 0·022).
Age was categorized into three groups: those aged 17–29 years, 30–39 years and over 40 years. There was no statistically significant difference of IL-1Ra levels between the three age categories analysed (see Fig. 2b for the difference between donors under and above 40 years).
The volunteer donors that smoked had significantly higher plasma IL-1Ra levels than non-smokers (P = 0·015) (see Fig. 2c). Those donors that had taken NSAIDs in the 2 weeks prior to their blood donation (n = 18) had significantly lower plasma IL-1Ra levels compared to those donors (n = 230) who had not taken NSAIDs in the same time period (P = 0·022) (see Fig. 2d).
In order to account for multiple comparisons the P-values obtained for univariate analysis could be corrected (Bonferroni). In the case of the effect of smoking the data obtained would indicate a trend (P = 0·075) on the sample size. In this study, however, utilizing a stepwise approach in the multivariate regression model performed reduces the rate of false discovery [29].
Multivariate analysis.
The only factor that achieved significance in the model was whether the blood donor smoked (P = 0·04415). Both gender and NSAID drug usage displayed a weak trend (P = 0·17856 and 0·17170, respectively).
DISCUSSION
Our study was comparable in design to the study described by Hurme et al. [7]. However, we failed to confirm the significant finding observed by Hurme and co-workers between the possession of allele 2 in the IL-1β-511 or the absence of allele 2 in the IL-1β + 3953 polymorphisms, with possession of allele 2 in the IL-1Ra VNTR polymorphisms with increased plasma IL-1Ra levels. The number of blood donor volunteers in our study was similar to that described by Hurme et al. [7], with comparable allele frequencies to that of the Finnish population. The estimated haplotypes of the 5 IL-1 gene polymorphisms we have studied indicates that there could be co-ordinated regulation of IL-1 production in individuals; however, this could not be confirmed in vivo in our study.
The IL-1Ra VNTR genotype has been shown in a few studies to be implicated in the pathology of a number of diseases, including severity of acute graft-versus-host disease (GVHD) and incidence of chronic GVHD [17,25], skin diseases [30], sepsis [31], osteoporotic fractures [32] and lung function decline (IL-1 haplotypes) [33,34]. These associations may reflect the biology of individuals under immune stress or peripheral leucocytes stimulated with bacterial endotoxin.
The roles of non-genotypic factors are also important in the regulation of proinflammatory cytokine production. It has been shown that age can influence the levels of IL-1β in unstimulated and stimulated lipopolysaccharide (LPS) monocyte-enriched cultures [5]. However, the low level of IL-1β in plasma compared to serum made its measurement in the NBA samples impractical and subsequent analysis with the genotypic and non-genotypic factors difficult. The measurement of plasma IL-1Ra levels was therefore carried out due to the suggested co-ordinated regulation of agonist and antagonist [7,27]. Interindividual variation in IL-1 production between males and females has been described in in vitro studies with stimulated peripheral blood mononuclear cell (PBMC) cultures [4], and with differences in tumour necrosis factor (TNF)-α production between males and postmenopausal women but not premenopausal women [35]. The weak effect of gender on in vivo IL-1Ra levels observed in our study (when corrected for genotypic and non-genotypic factors) in a multivariate model illustrates a possible effect of gender on IL-1 production.
There is increasing evidence that the gender effect may be due to sex hormones, and that oestrogens and their receptors may have a particular influence on the regulation of cytokines. The effect of oestrogen in the development of osteoporosis has been studied widely [35–37], where oestrogen deficiency postmenopause has been associated with increased bone turnover, loss and resorption due to the increasing osteoclast activity by decreasing the levels of proinflammatory cytokines [36]. Oestrogen replacement therapy in postmenopausal women with osteoporosis, compared to controls, reduced the in vitro production of both IL-1β and IL-1Ra from PBMC cultures. Unfortunately, our study could not illustrate the effect of oestrogen on IL-1Ra levels by the ingestion of oral contraceptives or pre- versus postmenopausal differences, but this may have been due purely to sample size. The role of oestrogen in the modulation of the immune system could also explain the increased frequency and severity of autoimmune disorders in women [38].
Increases in in vivo levels of IL-1β and IL-1Ra (alongside other proinflammatory cytokines) proteins have been observed in inflammatory diseases such as asthma [39,40] and periodontal disease [41]. Unfortunately, such studies have not always been able to demonstrate the link between increases in proinflammatory cytokines (TNF-α, IL-1 and IL-6) and disease severity in patients who smoke, suggesting that this might be a confounding variable [33,34,41]. In our study of normal individuals we observed a correlation between smoking and raised plasma IL-1Ra levels. The total number of circulating leucocytes has been correlated with smoking activity with varying increases between the leucocyte subpopulations [42], which may be caused by an inflammatory response to particles in cigarette smoke [43]. This may account for the observed increased plasma levels in the sample subjects who smoke; however, a total leucocyte count was not performed on the samples collected to either confirm or refute this as a possible factor. Multivariant analysis in our study determined that smoking affected IL-1Ra levels independently, without being influenced by IL-1 genotype.
Modulation of cytokine expression by glucocorticoids binding to negative regulatory elements present in gene promoter regions has been well described [6,44,45]. The body naturally produces cortisol in a diurnal rhythm, with endogenous release and exogenous administration promoting an anti-inflammatory response. The severity of symptoms in autoimmune diseases such as rheumatoid arthritis or asthma can be exacerbated at night or early morning [46,47], when proinflammatory cytokines such as TNF-1 and IL-1, widely accepted in the disease pathology daily [48,49], are at their peak [6].
The metabolites of arachidonic acid, prostaglandins and leukotrienes, are also endogenous regulators of IL-1, produced through the lipoxygenase and cyclo-oxygenase pathways [50]. Inhibition of these pathways by drugs such as aspirin and ibuprofen (NSAIDs) results in a decrease in the inflammatory response. These drugs have been shown to increase the levels of IL-1β by reduction of prostaglandin E2 (PGE2), which is responsible for reducing the expression of IL-1 at post-transcriptional levels by the induction of elevated cyclic AMP [2]. This may result in increased IL-1Ra levels by the co-ordinated regulation of IL-1β and IL-1Ra [7]. This effect was illustrated in the increase in plasma IL-1Ra levels with NSAID intake observed in our study. In a wider context the ingestion of NSAIDs may also regulate the production of other anti-inflammatory cytokines such as IL-10, leading to an overall anti-inflammatory response.
Comparison of studies that are based upon in vivo, in vitro or genetic associations with disease phenotypes are problematic. A wide variety of studies rely upon the exogenous stimulation of separated peripheral leucocytes by mitogens such as LPS. Although these studies provide evidence for cellular response to stress, temporal resting cytokine production can only be measured accurately in plasma or serum samples. This itself can also introduce variables between the types of sample obtained for analysis (as we have observed in our small pilot study between plasma and serum samples from laboratory volunteers). The response to stress as modelled by in vitro studies is an important illustrator of the possible responses of the immune system in disease pathophysiology. The advances in genetics, where it is thought that variation in the genome sequences (be they SNPs or VNTRs) might underlie the difference in individuals to susceptibility or protection from diseases [51,52], has led to numerous studies that associate specific singular polymorphisms of particular haplotypes with a disease [1,53]. These studies often do not examine the functional affect of such diversity. Identification of the functional or immunological effects of the designated ‘high-risk or protective alleles’ from a significant disease association observation have to be compared with appropriate in vivo or in vitro measurement. The advent of the human genome project may provide greater knowledge of the true interplay between nature and nurture, as the genome does not work alone − the environment also plays a role [53].
In summary, our study illustrates the role which non-genetic factors may have on the production of the cytokine IL-1Ra. Although the possession of allele 2 in the IL-1Ra VNTR polymorphism was not found to have a significant impact on the plasma level of IL-1Ra, as shown in a comparative study, numerous studies have observed significant association with the severity of disease. The difference in IL-1Ra levels between smokers and non-smokers illustrate further the ‘stress’ that smoking places upon the immune system.
Acknowledgments
The authors would like to thank the blood donors of the Newcastle region for their co-operation in this study, Dr Cath Chapman and the staff of the Newcastle region National Blood Authority who took the research samples after blood donation. The authors would also like to thank Dr Rob Taylor and Geoff Taylor (Mitochondrial and Neurological Research Group) for performing the direct M13 sequencing of PCR amplicons and Dr Chris Redfern who advised on statistical analysis of the data. This study was funded by a Tyneside Leukaemia Research Association (TLRA) PhD Studentship grant and a European Commission FP5 grant (QLRT2000-00010) EUROBANK, contract no. QLRT 2000-00010.
REFERENCES
- 1.Bidwell JL, Keen LJ, Gallagher G, et al. Cytokine gene polymorphism in human disease: on-line databases. Genes Immun. 1999;1:3–19. doi: 10.1038/sj.gene.6363645. [DOI] [PubMed] [Google Scholar]
- 2.Knudsen PJ, Dinarello CA, Strom TB. Glucocorticoids inhibit transcriptional and post-transcriptional expression of interleukin-1 in U937 cells. J Immunol. 1987;139:4129–34. [PubMed] [Google Scholar]
- 3.Daun JM, Ball RW, Cannon JG. Glucocorticoid sensitivity of interleukin-1 agonist and antagonist secretion: the effects of age and gender. Am J Physiol. 2000;278:R855–62. doi: 10.1152/ajpregu.2000.278.4.R855. [DOI] [PubMed] [Google Scholar]
- 4.Lynch EA, Dinarello CA, Cannon JG. Gender differences in IL-1a, IL-1b, and IL-1 receptor antagonist secretion from mononuclear cells and urinary excretion. J Immunol. 1994;153:300–6. [PubMed] [Google Scholar]
- 5.Riancho JA, Zarrabeitia MT, Amado JA, Olmos JM, Gonzalez-Macias J. Age-related difference in cytokine secretion. Geronotology. 1994;40:8–12. doi: 10.1159/000213568. [DOI] [PubMed] [Google Scholar]
- 6.Petrovsky N, McNair P, Harrison LC. Diurnal rhythms of pro-inflammatory cytokine: regulation by plasma cortisol and therapeutic implications. Cytokine. 1998;10:307–12. doi: 10.1006/cyto.1997.0289. [DOI] [PubMed] [Google Scholar]
- 7.Hurme M, Santtila S. IL-1 receptor antagonist (IL-1Ra) plasma levels are co-ordinately regulated by both IL-1Ra and IL-1beta genes. Eur J Immunol. 1998;28:2598–602. doi: 10.1002/(SICI)1521-4141(199808)28:08<2598::AID-IMMU2598>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 8.Vamvakopoulos J, Green C, Metcalfe S. Genetic control of IL-1b bioactivity through differential regulation of the IL-1 receptor antagonist. Eur J Immunol. 2002;32:2988–96. doi: 10.1002/1521-4141(2002010)32:10<2988::AID-IMMU2988>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 9.Kristensen M, Deleuran B, Eedy DJ, Feldmann M, Breathnach SM, Brennan FM. Distribution of interleukin 1 receptor antagonist protein (IRAP), interleukin 1 receptor, and interleukin α in normal and psoriatic skin. Decreased expression of IRAP in psoriatic lesional epidermis. Br J Dermatol. 1992;127:305–11. doi: 10.1111/j.1365-2133.1992.tb00446.x. [DOI] [PubMed] [Google Scholar]
- 10.Beuscher HU, Rausch U-P, Otterness IG, Rollonghoff M. Transition from interleukin 1β (IL-1β) to IL-1α production during maturation of inflammatory macrophages in vivo. J Exp Med. 1992;175:1793–7. doi: 10.1084/jem.175.6.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinarello CA. The interleukin-1 family: 10 years of discovery. FASEB J. 1994;8:1314–25. [PubMed] [Google Scholar]
- 12.Bailly S, Israel N, Fay M, Gougerot-Pocidalo MA, Duff GW. An intronic polymorphic repeat sequence modulates interleukin-1 alpha gene regulation. Mol Immunol. 1996;33:999–1006. doi: 10.1016/s0161-5890(96)00042-9. [DOI] [PubMed] [Google Scholar]
- 13.McDowell TL, Symons JA, Ploski R, Forre O, Duff GD. Genetic association between juvenile rheumatoid arthritis and a novel interleukin-1α polymorphism. Arthritis Rheum. 1995;38:221–8. doi: 10.1002/art.1780380210. [DOI] [PubMed] [Google Scholar]
- 14.Blakemore AI, Tarlow JK, Cork MJ, Gordon C, Emery P, Duff GW. Interleukin-1 receptor antagonist gene polymorphism as a disease severity factor in systemic lupus erythematosus. Arthritis Rheum. 1994;37:1380–5. doi: 10.1002/art.1780370917. [DOI] [PubMed] [Google Scholar]
- 15.Clay FE, Cork MJ, Tarlow JK. Interleukin-1 receptor antagonist gene polymorphism association with lichen sclerosus. Hum Genet. 1994;94:407–10. doi: 10.1007/BF00201602. [DOI] [PubMed] [Google Scholar]
- 16.Cullup H, Dickinson AM, Cavet J, Jackson G, Taylor PRA, Middleton PG. Donor interleukin-1 receptor antagonist genotype associated with acute graft-versus-host disease in human leucocyte antigen-matched sibling allogeneic transplants. Br J Haematol. 2001;113:807–13. doi: 10.1046/j.1365-2141.2001.02811.x. [DOI] [PubMed] [Google Scholar]
- 17.Cullup H, Dickinson AM, Cavet J, Jackson GH, Middleton PG. Polymorphisms of IL-1α constitute independent risk factors for chronic graft versus host disease following allogeneic bone marrow transplantation. Br J Haematol. 2003;122:778–87. doi: 10.1046/j.1365-2141.2003.04510.x. [DOI] [PubMed] [Google Scholar]
- 18.De Caterina R. Polymorphisms related to inflammation in the interleukin-1 system genes: a step forward in the search for predisposing conditions to ischemic heart disease. Cardiologia. 1999;44:831–4. [Editorial; Comment] [PubMed] [Google Scholar]
- 19.Diehl SR, Wang Y, Brooks CN, et al. Linkage disequilibrium of interleukin-1 genetic polymorphisms with early-onset periodontitis. J Periodontol. 1999;70:418–30. doi: 10.1902/jop.1999.70.4.418. [DOI] [PubMed] [Google Scholar]
- 20.El-Omar EM, Carrington M, Chow W-H, et al. Interleukin-1 polymorphisms associated with increased risk of gastric cancer. Nature. 2000;404:398–402. doi: 10.1038/35006081. [DOI] [PubMed] [Google Scholar]
- 21.Ehmke B, Kress W, Karch H, Grimm T, Klaiber B, Flemmig TF. Interleukin-1 haplotype and periodontal disease progression following therapy. J Clin Periodontol. 1999;26:810–3. doi: 10.1111/j.1600-051x.1999.tb02525.x. [DOI] [PubMed] [Google Scholar]
- 22.Gore EA, Sanders JJ, Pandey JP, Palesch Y, Galbraith GM. Interleukin-1beta+3953 allele 2: association with disease status in adult periodontitis. J Clin Periodontol. 1998;25:781–5. doi: 10.1111/j.1600-051x.1998.tb02370.x. [DOI] [PubMed] [Google Scholar]
- 23.Keen RW, Woodford-Richens KL, Lanchbury JS, Spector TD. Allelic variation at the interleukin-1 receptor antagonist gene is associated with early postmenopausal bone loss at the spine. Bone. 1998;23:367–71. doi: 10.1016/s8756-3282(98)00109-4. [DOI] [PubMed] [Google Scholar]
- 24.Parkhill JM, Hennig BJW, Chapple ILC, Heasman PA, Taylor JJ. Association of interleukin-1 gene polymorphisms with early-onset periodontitis. J Clin Periodontol. 2000;27:682–9. doi: 10.1034/j.1600-051x.2000.027009682.x. [DOI] [PubMed] [Google Scholar]
- 25.Rocha V, Franco RF, Porcher R, et al. Host defense and inflammatory gene polymorphisms are associated with outcomes after HLA-identical sibling bone marrow transplantation. Blood. 2002;100:3908–18. doi: 10.1182/blood-2002-04-1033. [DOI] [PubMed] [Google Scholar]
- 26.National Center for Biotechnology Information (NCBI) Locus link search page URL. http://www.ncbi.nim.nih.gov/locuslink.
- 27.Santtila S, Savinainen K, Hurme M. Presence of the IL-1RA allele 2 (IL1RN*2) is associated with enhanced IL-1beta production in vitro. Scand J Immunol. 1998;47:195–8. doi: 10.1046/j.1365-3083.1998.00300.x. [DOI] [PubMed] [Google Scholar]
- 28.Tarlow JK, Blakemore AIF, Lennard A, et al. Polymorphism in human IL-1 receptor antagonist gene intron 2 is caused by variable numbers of an 86-bp tandem repeat. Hum Genet. 1993;91:403–4. doi: 10.1007/BF00217368. [DOI] [PubMed] [Google Scholar]
- 29.Edwin JCG, Sullivan PF. False discoveries and models for gene discovery. Trends Genet. 2003;19:537–42. doi: 10.1016/j.tig.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Corke MJ, Tarlow JK, Clay FE. An allele of the interleukin-1 receptor antagonist as a genetic severity factor in alopecia areata. J Invest Dermatol. 1995;104:S15–16. doi: 10.1038/jid.1995.37. [DOI] [PubMed] [Google Scholar]
- 31.Fang XM, Schroder S, Hoeft A, Stuber F. Comparison of two polymorphisms of the interleukin-1 gene family: interleukin-1 receptor antagonist polymorphism contributes to susceptibility to severe sepsis. Crit Care Med. 1999;27:1330–4. doi: 10.1097/00003246-199907000-00024. [DOI] [PubMed] [Google Scholar]
- 32.Langdahl BL, Lokke E, Carstens M, Stenkjaer LL, Eriksen EF. Osteoporotic fractures are associated with an 86-base pair repeat polymorphism in the interleukin-1-receptor antagonist gene but not with polymorphisms in the interleukin-1beta gene. J Bone Miner Res. 2000;15:402–14. doi: 10.1359/jbmr.2000.15.3.402. [DOI] [PubMed] [Google Scholar]
- 33.Joos L, McIntyre JL, Ruan J, et al. Association of IL-1beta and IL-1 receptor antagonist haplotypes with rate of decline in lung function in smokers. Thorax. 2001;56:863–6. doi: 10.1136/thorax.56.11.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karjalainen J, Hulkkonen J, Hurme MI. L-1 haplotypes and lung function decline. Thorax. 2002;57:561–2. doi: 10.1136/thorax.57.6.561-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Danis VA, Millington M, Hyland VJ, Grennan D. Cytokine production by normal human monocytes: inter-subject variation and relationship to an IL-1 receptor antagonist (IL-1 Ra) gene polymorphism. Clin Exp Immunol. 1995;99:303–10. doi: 10.1111/j.1365-2249.1995.tb05549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kassem M, Khosla S, Spelsberg TC, Riggs BL. Cytokine production in the bone marrow microenvironment: failure to demonstrate estrogen regulation in early postmenopausal women. J Clin Endocrinol Metab. 1996;81:513–18. doi: 10.1210/jcem.81.2.8636260. [DOI] [PubMed] [Google Scholar]
- 37.Pacifici R, Vannice JL, Rifas L KR. Monocytic secretion of interleukin-1 receptor antagonist in normal and osteoporotic women. Effects of menopause and estrogen/progesterone therapy. J Clin Endocrinol Metab. 1993;77:1135–41. doi: 10.1210/jcem.77.5.8077304. [DOI] [PubMed] [Google Scholar]
- 38.Ruh MF, Bi Y, D’Alonzo R, Bellone CJ. Effect of environmental estrogens on IL-1beta promoter activity in a macrophage cell line. J Steroid Biochem Mol Biol. 1998;66:203–10. doi: 10.1016/s0960-0760(98)00042-9. [DOI] [PubMed] [Google Scholar]
- 39.Sousa AR, Lane SJ, Nakhosteen JA. Expression of interleukin-1beta and interleukin-1 receptor antagonist (IL-1Ra) on asthmatic bronchial epithelium. Am J Respir Crit Care Med. 1996;154:1061–6. doi: 10.1164/ajrccm.154.4.8887608. [DOI] [PubMed] [Google Scholar]
- 40.Pujol JL, Cosso B, Daures JP. Interleukin-1 release by alveolar macrophages in asthmatic patients and healthy subjects. Int Arch Allergy Appl Immunol. 1990;91:207–10. doi: 10.1159/000235117. [DOI] [PubMed] [Google Scholar]
- 41.Bostrom L, Linder LE, Bergstrom J. Smoking and GCF levels of IL-1b and IL-1Ra in periodontal disease. J Clin Periodontol. 2000;27:250–5. doi: 10.1034/j.1600-051x.2000.027004250.x. [DOI] [PubMed] [Google Scholar]
- 42.Van Tiel ED, Peeter PHM, Smit HA, et al. Quitting smoking may restore hematological characteristics within five years. Ann Epidemiol. 2002;12:378–88. doi: 10.1016/s1047-2797(01)00282-4. [DOI] [PubMed] [Google Scholar]
- 43.Schwartz J, Weiss ST. Cigarette smoking and peripheral blood leukocyte differentials. Ann Epidemiol. 1994;4:236–42. doi: 10.1016/1047-2797(94)90102-3. [DOI] [PubMed] [Google Scholar]
- 44.de Rijk R, Michelson D, Karp B, et al. Exercise and circadian rhythm-induced variations in plasma cortisol differentially regulate interleukin-1β (IL-1β), IL-6 and tumour necrosis factor-α (TNFα) production in humans: high sensitivity of TNFα and resistance of IL-6. J Clin Endocrinol Metab. 1997;82:2182–91. doi: 10.1210/jcem.82.7.4041. [DOI] [PubMed] [Google Scholar]
- 45.Schmidt M, Pauels HG, Lufering N, Domschke W, Kucharzik T. Glucocortocoids induce apotosis in human monocytes: potential role of IL-1 beta. J Immunol. 1999;163:3484–90. [PubMed] [Google Scholar]
- 46.Harkness JA, Richter MB, Panayi GS, et al. Diurnal variation in disease activity of rheumatoid arthritis. Br Med J Clin Res. 1982;284:551–4. doi: 10.1136/bmj.284.6315.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin JR, Cicutto LC, Smith HR, Ballard RD, Szefler SJ. Airways inflammation in nocturnal asthma. Am J Respir Dis. 1991;143:351–7. doi: 10.1164/ajrccm/143.2.351. [DOI] [PubMed] [Google Scholar]
- 48.Laurincova B. Interleukin-1 family: from genes to human disease. Acta Univ Palacki Olomuc Fac Med. 2000;143:19–29. [PubMed] [Google Scholar]
- 49.Hurme M, Lahdenpohja N, Santtila S. Gene polymorphisms of interleukins 1 and 10 in infectious and autoimmune diseases. Ann Med. 1998;30:469–73. doi: 10.3109/07853899809002488. [DOI] [PubMed] [Google Scholar]
- 50.Endres S, Cannon JG, Ghorbani R, et al. In vitro production of IL1β, IL1α, TNF and IL2 in healthy subjects: distribution, effect of cyclooxygenase inhibition and evidence of independent gene regulation. Eur J Immunol. 1989;19:2327–33. doi: 10.1002/eji.1830191222. [DOI] [PubMed] [Google Scholar]
- 51.Fahrer AM, Bazan JF, Papathanasiou P, Nelms KA, Goodnow CC. A genomic view of immunology. Nature. 2001;409:836–8. doi: 10.1038/35057020. [DOI] [PubMed] [Google Scholar]
- 52.Group TISMW. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–33. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 53.Chakravarti A. To a future of genetic medicine. Nature. 2001;409:822–3. doi: 10.1038/35057281. [DOI] [PubMed] [Google Scholar]
- 54.Bailly S, di Giovine FS, Duff GW. Polymorphic tandem repeat region in interleukin-1 alpha intron 6. Hum Genet. 1993;91:85–6. doi: 10.1007/BF00230231. [DOI] [PubMed] [Google Scholar]
- 55.Di Giovine FS, Takhsh E, Blakemore AIF, Duff GW. Single base polymorphism at -511 in the human interleukin-1β gene. Hum Mol Genet. 1992;1:450. doi: 10.1093/hmg/1.6.450. [DOI] [PubMed] [Google Scholar]
- 56.Pociot F, Molvig J, Wogensen L, Worsaae H, Nerup J. A Taq1 polymorphism in human interleukin-1β (IL-1β) gene correlates with IL-1β secretion in vitro. Eur J Clin Invest. 1992;22:396–402. doi: 10.1111/j.1365-2362.1992.tb01480.x. [DOI] [PubMed] [Google Scholar]