Abstract
mRNA expression of two recently described human β-defensins (hBD-3 and hBD-4) in epithelial cells of normal small and large intestine and the impact of chronic intestinal inflammation on their expression levels was investigated. Intestinal specimens from patients with ulcerative colitis (UC), Crohn's disease (CD) and controls with no history of inflammatory bowel disease were studied. hBD-3 and hBD-4 mRNAs were determined in freshly isolated epithelial cells by real-time quantitative reverse transcription–polymerase chain reaction (QRT-PCR) and by in situ hybridization. The effect of proinflammatory cytokines on hBD-3 and hBD-4 mRNA expression in colon carcinoma cells was also investigated. Purified epithelial cells of normal small and large intestine expressed both hBD-3 and hBD-4 mRNA, with higher expression levels of hBD-3 mRNA. In situ hybridization revealed higher levels of mRNA expression in the crypt- compared to the villus/luminal-compartment. Interferon (IFN)-γ, but not tumour necrosis factor (TNF)-α or IL-1β, augmented hBD-3 mRNA expression. None of these agents stimulated hBD-4 expression. Colonic epithelial cells from patients with UC displayed a significant increase in hBD-3 and hBD-4 mRNA compared to epithelial cells of controls. In contrast, small intestinal epithelial cells from CD patients did not show increased expression levels compared to the corresponding control cells. Moreover, Crohn's colitis did not show increased expression of hBD-4 mRNA, while the data are inconclusive for hBD-3 mRNA. We conclude that the chronic inflammatory reaction induced in the colon of UC patients enhances hBD-3 and hBD-4 mRNA expression in the epithelium, whereas in CD this is less evident.
Keywords: Crohn's disease, cytokines, human, inflammatory bowel disease, qRT-PCR
INTRODUCTION
Human survival requires constant co-ordinated activity of the immune system. The ability to prevent infection depends on both innate and adaptive immune mechanisms. A particularly vulnerable area for microbial attack is the intestine, because of constant exposure to abrasive food components and the abundance of microorganisms in its lower part. Chronic inflammatory bowel disease (IBD), in particular ulcerative colitis (UC), may be regarded as a consequence of failing adaptive and/or innate immune system to cope with the onslaught of pathogenic, opportunistic or commensal microorganisms. An important aspect of the innate defence is the release of antimicrobial peptides/proteins at the luminal surface of the epithelium. Present knowledge about innate immune functions in IBD is scarce. It has been shown that expression of human β-defensin-2 (hΒD-2) is up-regulated in colonic enterocytes in patients with UC and that human α-defensins 5 and 6 (HD-5, HD-6) and lysozyme are over-expressed, due mainly to metaplastic Paneth-cell differentiation in UC colon [1–5]. A recent study indicates that hBD-3 is increased in UC colon [6]. Similarly, human neutrophil α-defensins 1–3 were shown recently to be expressed in epithelial cells in active IBD [7].
The defensins comprise a family of small, cationic antimicrobial peptides [8–10]. They are classified into α- and β-defensins. Six α-defensins have been identified in humans; two (HD-5 and HD-6) are expressed by Paneth-cells of the small intestine and in the female reproductive tract [11,12] and four [human neutrophil peptide (HNP)-1–4] are expressed mainly by neutrophils. Based on in silico analysis a total of 28 β-defensins have been predicted in the human genome [13].The genes are located in five different clusters, of which the cluster on chromosome 8p22–23 is the largest and contains all eight β-defensins known to be expressed (i.e. hBD-1 to hBD-4, DEF105 to DEF108) [14–18].
hBD-1 [17] and hBD-2 [14] are expressed in various epithelial tissues including intestinal epithelial cells (IEC) [2,3,19], whereas hBD-1 is expressed constitutively in most tissues; hBD-2 is up-regulated by certain proinflammatory cytokines and/or bacteria [2,14,20–23]. hBD-3 was initially cloned [24,25] and isolated from epidermal keratinocytes of patients with psoriasis [15] and was reported to be induced by interferon (IFN)-γ in lung epithelial cells. hBD-4 was first demonstrated in lung cDNA[16] and is up-regulated by Gram-negative and Gram-positive bacteria in respiratory cells.
hBD-1 and hBD-2 display antibacterial activity against Gram-negative bacteria [14,26,27], whereas hBD-3 is in addition active against Gram-positive bacteria and Saccharomyces cerevisiae[15,24]. hBD-3 is salt-resistant, much more basic than hBD-1 and -2, and is a dimer in solution [28], properties which may explain its broader antimicrobial activity. Growth inhibition of Gram-negative and Gram-positive bacteria as well as Saccharomyces cerevisiae[16] has been reported for hBD-4. Interestingly, both hBD-3 and hBD-4 are chemotactic to monocytes [16,24].
The aims of this study were twofold: to investigate the expression of hBD-3 and hBD-4 in the epithelium at different levels of the normal human intestine and to determine whether their expression levels are affected by chronic inflammation.
MATERIALS AND METHODS
Patients
Surgical IBD specimens were obtained from patients suffering from UC (n = 19, mean age 37 ± 13 years) and CD (n = 11, mean age 32 ± 14 years). The diagnosis of UC and colonic CD was established by the combination of clinical symptoms, endoscopic findings and histology. For small intestinal CD additional investigations with small bowel X-ray and/or leucocyte scintigraphy were performed. Eight of the UC samples were from patients with active disease, 10 were from patients with moderately active disease and one was from a patient with inactive disease. Four UC patients had no drug treatment, whereas 11 were on corticosteroids, in two combined with azatioprin, and in one with 5-ASA. Two patients were treated with azatioprin alone and two with 5-ASA alone. Six samples from CD patients were from ileum (all with active disease), one from jejunum (active disease) and five from colon (four with active disease and one with moderately active disease). Eight CD patients were treated with local or systemic corticosteroids, of whom two also had 5-ASA. Azatioprin were administered to four patients either alone or combined with 5-ASA.
Control colon specimens were from seven male and 17 female patients (mean age 66 ± 11 years) with colorectal cancer (n = 21) or non-inflammatory benign conditions (n = 3). Control ileum specimens were from eight male and five female patients (65 ± 12 years) with colon cancer (n = 11), Meckels diverticuli (n = 1) or colon polyps (n = 1). Control jejunum specimens were from eight male and seven female patients (69 ± 13 years) with gastric, colonic or pancreatic cancer (n = 13) or gastric ulcer (n = 1). None of the control patients had been subjected to radio- or chemotherapy, long-standing antibiotic medication or steroid treatment. The control samples were taken distant to macroscopically detectable lesions. All patients received a single intravenous dose of antibiotics 2 h prior to surgery according to preoperative standard procedure. The ethics committee at the Faculty of Medicine and Odontology of Umeå University Hospital approved the study and the patients gave their informed consent.
Isolation of intestinal epithelial cells
IEC were isolated from surgical specimens as previously described [2, 29,30]. The procedure yields two epithelial cell populations. One from the villous/luminal compartment (referred to as l/v-IEC) and one from the crypt compartment (c-IEC). The procedure includes depletion of leucocytes by treatment with anti-CD45 charged paramagnetic beads. Isolated cells were washed in RNase-free phosphate buffered saline (PBS) and kept at −80°C until RNA extraction.
Cell lines
Four human colon carcinoma cell lines were used; moderately differentiated HT-29 cells and well-differentiated LS174T cells were maintained in Parker199 medium (SBL Vaccin, Stockholm, Sweden) supplemented with 8% fetal calf serum (FCS). Well-differentiated T-84 cells were maintained in 1 : 1 Dulbecco/Vogt modified Eagle's minimal essential medium: F12-HAM (Invitrogen), supplemented with 15 mm HEPES buffer and 8% FC. Well-differentiated HCT-8 cells were maintained in 1 : 1 Parker199: F12-HAM and 8% FCS. The fetal small intestinal cell line INT407 was maintained in Dulbecco/Vogt modified Eagle's minimal essential medium supplemented with 15% FCS. The lung epithelial cell line A549 and the keratinocytic cell line HaCaT were maintained in Dulbecco/Vogt modified Eagle's minimal essential medium supplemented with 10% FCS and 20 mm or 10 mm HEPES buffer, respectively. Antibiotics and extra l-glutamine were added to all culture media. Cells were maintained at 37°C in humidified air with 5% CO2 and trypsinized weekly.
Stimulation of cell lines
Cells were trypsinized and washed in complete tissue culture medium. Thereafter, 1·5–2 × 106 cells were seeded in 3 ml of the appropriate medium in six-well tissue culture plates and incubated for 2–24 h at 37°C with 50 ng/ml of recombinant human interleukin-1β (rhIL-1β; Endogen, Woburn, MA, USA), 20 ng/ml of rh-tumour necrosis factor-α (rhTNF-α; Endogen) or 100 or 200 U/ml of rh-interferon-γ (rhIFN-γ; Promega, Madison, WI, USA). Cytokine was added at the time of seeding. After incubation, the cells were trypsinized, washed in RNase-free PBS and kept at −80°C until RNA extraction.
RNA copy standard preparation and real-time quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR)
Total RNA was extracted from IEC and cell lines as described [2]. Total RNA from HaCaT cells and cDNA from testis (Stratagene, La Jolla, CA, USA) were used as starting material for cloning of hBD-3 and hBD-4 cDNA, respectively. The primers used for RT-PCR were hBD-3 forward: 5′-TCTCAGCGTGGGGTGAAGC-3′ and hBD-3 reversed: 5′-CGGCCGCCTCTGACTCTG-3′; hBD-4 forward: 5′-AGCCCCAGCATTATGCAGAGA-3′and hBD-4 reversed: 5′-GCGACTCTAGGGACCAGCACTAC-3′. The PCR products, which include the respective sequences amplified in the real-time quantitative RT-PCR assays, were cloned, sequenced and used for RNA copy standard preparation and optimization of qRT-PCR assays. The RNA copy standards were prepared by in vitro transcription with T7 polymerase/RiboProbe® In Vitro Transcription Systems (Promega) according to the manufacturer's instructions (for experimental details see reference [2]).
Real-time qRT-PCR assays for hBD-3 and hBD-4 mRNAs were constructed using the TaqMan EZ® technology (Applied Biosystems, Foster City, CA). The hBD-3 system consisted of hBD-3 forward primer: 5′-TGAAGCCTAGCAGCTATGAG GATC-3′, hBD-3 reverse primer: 5′-CCGCCTCTGACTCTG CAATAA-3′ and a dual-labelled fluorescent hBD-3 probe: 5′-(FAM) TTGGTGCCTGTTCCAGGTCATGGAG (TAMRA)-3′. The hBD-4 system consisted of hBD-4 forward primer: 5′-CCCAGCATTATGCAGAGACTT-3′, hBD-4 reverse primer: 5′-ACCACATATTCTGTCCAATTCAAAT-3′ and hBD-4 probe: 5′-(FAM) TGCTGCTATTAGCCGTTTCTCTTCTACTCTAT CAA (TAMRA)-3′. The primers were chosen to hybridize to different exons. Because of the high similarity between the ends of exon 1 and the intron of hBD-3, the possibility to amplify DNA or consumption of primers by contaminating nuclear DNA was eliminated by treatment of the RNA samples with DNaseI (Invitrogen) according to the manufacturer's advice. Emission from released reporter dye (FAM) was monitored by the ABI Prism® 7700 Sequence Detection System (Applied Biosystems). The assays gave a linear relation between log concentrations of standard RNA and PCR cycles over a range of at least five logs. Determinations were carried out in triplicates and expressed as copies of mRNA/µl using the external copy standard. The concentration of 18S rRNA (Applied Biosystems) was determined in each sample and the result expressed as mRNA copies of defensin per unit of 18S rRNA.
Immunoflow cytometry
Purity of the isolated IEC fractions was estimated by two-colour immunoflow cytometry using phycoerythrin conjugated anti-CD45 monoclonal antibodies (MoAbs) and fluorescein isothiocyanate (FITC)-conjugated anti-epithelial antigen MoAb BerEp4 (DakoCytomation, Glostrup, Denmark), as described [2]. Conjugated irrelevant concentration and isotype-matched mAbs (DakoCytomation) served as controls. Cells were analysed on a FACScan flowcytometer (Becton-Dickinson, Mountain-View, CA, USA) using the cellquest program.
In situ hybridization
In situ hybridization was performed on 10 µm cryosections fixed with 4% paraformaldehyde using digoxigenin-labelled RNA probes and alkaline phosphatase-labelled Fab-fragments of sheep antidigoxigenin (Roche Diagnostics, Basel, Switzerland) according to Panoskaltsis-Mortari and Bucy with minor modifications, as described previously [31,32]. Cells with hybridized probe were visualized by incubation with nitroblue tetrazolium, 5-bromo-4-chloro-3-indolyl phosphate and levamisole in 100 mm Tris-HCl (pH 9·5), 100 mm NaCl, 50 mm MgCl2.
Statistical analyses
The significance of differences in antimicrobial peptide mRNA expression levels between groups and the fold increase in stimulated cell lines was evaluated by two-tailed Mann–Whitney's rank sum test. Expression levels in cell lines are given as mean (± standard error) and fold increase after stimulation as geometric mean (± geometric standard error).
RESULTS
hBD-3 and hBD-4 mRNAs are expressed in normal intestinal epithelium
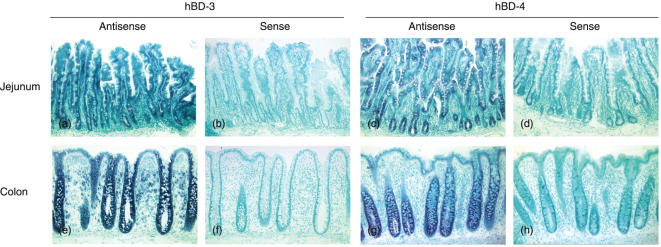
Tissue sections from normal jejunum (n = 4), ileum (n = 3) and colon (n = 7) were subjected to in situ hybridization with hBD-3 and hBD-4 antisense and sense probes, respectively. The epithelium in all three intestinal compartments gave positive signals with the antisense probes and no signal with the sense probes (Fig. 1a). The strongest signal intensity was observed in cryptal cells, both in large and small intestine. Differences between individual samples were not seen.
Fig. 1.
Expression of hBD-3 and hBD-4 mRNA in human intestinal mucosa as determined by in situ hybridization. (a) jejunum, hBD-3 antisense probe; (b) jejunum, hBD-3 sense probe; (c) jejunum, hBD-4 antisense probe; (d) jejunum, hBD-4 sense probe; (e) colon, hBD-3 antisense probe; (f) colon, hBD-3 sense probe; (g) colon, hBD-4 antisense probe; (h) colon, hBD-4 sense probe. Original magnification: × 55.
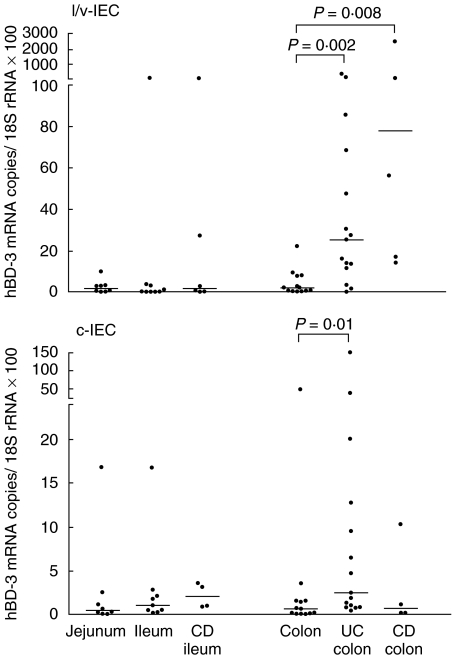
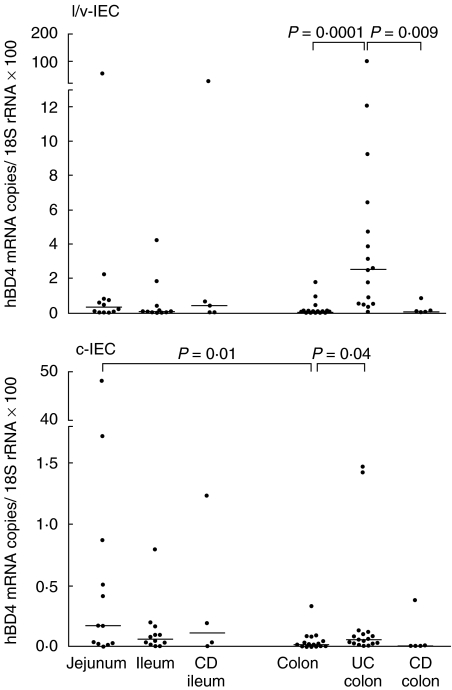
To quantify and compare the levels of hBD-3 and hBD-4 mRNA in the epithelial cells we developed specific real-time qRT-PCR assays with copy standards for the two β-defensin mRNA species. IEC were isolated from surgical specimens of jejunum, ileum and colon by a procedure yielding one cell population enriched in villous/luminal IEC (l/v-IEC) and one population enriched in cryptal IEC (c-IEC). Less than 2% of the cells in the IEC preparations were leucocytes as determined by immunoflow cytometry. Figures 2 and 3 show the results of qRT-PCR analysis of hBD-3 and hBD-4 mRNAs in l/v-IEC and c-IEC isolated from normal intestine. Both defensin mRNAs were present at all three levels of the intestine with hBD-3 mRNA being expressed at about 10 times higher copy numbers/18S rRNA unit than hBD-4 mRNA. hBD-3 mRNA was detected at similar levels along the intestinal axis. hBD-4 mRNA, in contrast, showed a tendency to be expressed at higher levels in the small intestine, the difference reaching statistical significance between jejunum c-IEC and colon c-IEC (P = 0·01). To verify the specificity of the qRT-PCR assays the qRT-PCR products from several epithelial cell samples were sequenced. All products had the expected sequences.
Fig. 2.
Expression levels of hBD-3 mRNA in freshly isolated l/v-IEC and c-IEC from jejunal, ileal and colonic mucosa of controls with no history of intestinal inflammation, colonic mucosa of UC and CD patients and ileal mucosa of CD patients as determined by qRT-PCR. mRNA levels are expresses as copies of hBD-3 mRNA/18S rRNA unit. Statistically significant differences are indicated. Horizontal bars indicate median values.
Fig. 3.
Expression levels of hBD-4 mRNA in freshly isolated l/v-IEC and c-IEC from jejunal, ileal and colonic mucosa of controls with no history of intestinal inflammation, colonic mucosa of UC and CD patients and ileal mucosa of CD patients as determined by qRT-PCR. mRNA levels are expresses as copies of hBD-4 mRNA/18S rRNA unit. Statistically significant differences are indicated. Horizontal bars indicate median values.
IEC from UC and CD patients express different levels of defensins
The expression levels of hBD-3 and hBD-4 mRNA in isolated IEC from patients with UC or CD were studied (Figs 2 and 3). The UC colon IEC fractions contained significantly higher mRNA levels of the two defensins than the corresponding fractions of normal colon. Both mRNA species were expressed at higher levels in the l/v-IEC fraction than in the c-IEC fraction.
Colon IEC from patients with CD located to colon, i.e. Crohn's colitis, or small intestinal IEC from CD patients with the disease located in the small intestine were analysed with respect to hBD-3 and hBD-4 mRNA expression and compared with the corresponding values in normal intestine (Figs 2 and 3). With the exception of the hBD-3 mRNA levels in the colonic l/v-IEC fraction of Crohn's colitis patients there was no difference between patients and controls. Thus, increased concentrations of defensin mRNA in epithelial cells seem to be a property of UC not seen in CD.
About half the number of patients had received corticosteroid treatment prior to operation. We therefore investigated whether this treatment affected defensin mRNA expression. There was a tendency to increased expression in the group of patients that did not receive steroid treatment. For hBD-4 in the c-IEC fraction the difference was statistically significant (P-value of 0·02). We also investigated whether there was a correlation between hBD-3 and hBD-4 mRNA levels in individual UC patients. No such correlation was found.
hBD-3 mRNA levels in colon carcinoma cell lines is up-regulated by IFN-γ
Three colon adenocarcinoma cell lines and a fetal small intestinal cell line expressed significant levels of hBD-3 and hBD-4 mRNA before addition of any stimulatory cytokine (Table 1). T-84 showed the highest expression levels of hBD-3 mRNA and HT-29 the lowest level. hBD-4 mRNA was expressed at 6–18 times lower levels than hBD-3 mRNA.
Table 1.
Expression levels of hBD-3 and hBD-4 mRNA in the three colon carcinoma cell lines LS174T, T-84 and HT-29 and the fetal small intestinal cell line INT407
| Cell line | hBD-3 | hBD-4 | n* |
|---|---|---|---|
| LS174T | 0·65 ± 0·30** | 0·028 ± 0·0087 | 15 |
| T-84 | 3·9 ± 0·96 | 0·20 ± 0·053 | 12 |
| HT-29 | 0·068 ± 0·029 | 0·011 ± 0·0047 | 6 |
| INT407 | 0·26 ± 0·027 | 0·075 ± 0·023 | 3 |
n = number of determinations.
mRNA copies per 18S rRNA unit × 100. Mean value (± standard error).
To investigate whether proinflammatory cytokines could increase hBD-3 and hBD-4 mRNA expression in IEC, the cell lines were stimulated with rhIL-1β, rhIFN-γ and rhTNF-α for 2–24 h. Maximal induction was seen after 4–6 h. The results for LS174T and T-84 cells at 4 and 6 h are shown in Table 2. IFN-γ induced increased hBD-3 mRNA expression in LS174T cells but not in T-84 cells. The greatest increase was sixfold, which was statistically significant (P = 0·03). TNF-α showed only a weak stimulatory effect and IL-1β did not have any effect on hBD-3 mRNA expression levels (Table 2). To ascertain that IL-1β was biologically active we also analysed hBD-2 mRNA expression. hBD-2 mRNA was clearly up-regulated already after 4 h of stimulation with IL-1β (data not shown). There was a tendency to increased hBD-4 expression with TNF-α. The difference did not, however, reach statistical significance.
Table 2.
Effect of proinflammatory cytokines on the expression levels of hBD-3 and hBD-4 mRNA in two colon carcinoma cell lines
| hBD-3 | hBD-4 | ||||||
|---|---|---|---|---|---|---|---|
| Cell line | Time of stimulation | IFN-γ | TNF-α | IL-1β | IFN-γ | TNF-α | IL-1β |
| LS174T | 4 h | 6·0 ± 0·15† | 2·3 ± 0·29 | 1·6 ± 0·098 | 4·4 ± 0·61 | 13 ± 0·34 | 2·6 ± 0·48 |
| 6 h | 4·0 ± 0·18 | 1·3 ± 0·42 | 1·1 ± 0·098 | 0·85 ± 0·25 | 0·99 ± 0·22 | 0·88 ± 0·28 | |
| T-84 | 4 h | 1·0 ± 0·27 | 1·0 ± 0·36 | 1·1 ± 0·18 | 0·78 ± 0·17 | 0·72 ± 0·34 | 1·4 ± 0·43 |
| 6 h | 1·2 ± 0·25) | 0·95 ± 0·097 | 0·49 ± 0·18 | 0·70 ± 0·11 | 1·0 ± 0·090 | 0·50 ± 0·19 | |
Data expressed as fold increase compared to unstimulated cells (= 1·0, see Table 1 for absolute values). Geometric mean value (± standard error of the geometric mean) of four experiments.
DISCUSSION
This is the first unequivocal demonstration of hBD-3 and hBD-4 expression in the intestinal epithelium of human small and large intestine. Previous studies have indicated that hBD-3 mRNA is present in whole tissue extracts of the small and large intestine [6,15], while hBD-4 mRNA was not previously found in the intestine [16]. In situ hybridization revealed that mRNA for the two defensins was expressed most abundantly in the crypt epithelium, particularly in the lower parts of the crypts. Lack of specific antibodies precluded analysis of hBD-3 and hBD-4 peptide expression and processing. In this study we used all commercially available antibodies against hBD-3, including purified polyclonal rabbit IgG claimed to react with residues 23–33 (GIINTLQKYYC) of hBD-3 (Koma Biotech, Korea), but found them to be either non-reactive or unspecifically positive in immunohistochemistry and immunoflow cytometry.
In normal intestinal epithelium hBD-3 mRNA was expressed at approximately 10 times higher concentrations than hBD-4 mRNA. For hBD-3 mRNA there was no significant difference in expression levels between epithelial cells in normal small and large intestine while for hBD-4 mRNA the small intestine expressed higher levels than the large intestine.
The most interesting finding in this study was that IEC of UC colon expressed significantly higher levels of both hBD-3 and hBD-4 mRNAs than control colon IEC. This was true both for the l/v-IEC and c-IEC fractions. Thus, these two recently discovered β-defensins show a similar up-regulation in UC colon IEC as hBD-2 [2,4]. This is in contrast to hBD-1, which shows a slightly decreased expression level in UC colon compared to controls [2,4]. Up-regulation of hBD-3 mRNA in UC colon was recently demonstrated by Wehkamp and coworkers [6] in RNA extracts of colon biopsies. Our study confirms and extends their results. By using isolated epithelial cells depleted in leucocytes and in situ hybridization we identify the epithelium as a major source of these defensins. hBD-4 m-RNA levels in UC colon has not been analysed previously.
Our data on IEC from CD are limited. Nevertheless, they indicate that there may be a difference in β-defensin mRNA expression in IEC of CD patients compared to IEC of UC patients. Thus, small intestinal IEC from CD patients did not show increased expression of hBD-3 or hBD-4 mRNA. Interestingly, similar results were previously obtained for hBD-2 [2]. When colon samples from Crohn's colitis were studied the result was less clear-cut. There was a statistically significant increase in expression of hBD-3 mRNA in the l/v-IEC colon fraction compared to the l/v-IEC of control colon. However, the c-IEC fractions did not show this difference. hBD-4 mRNA was unchanged compared to controls and low compared to UC colon IEC. Wehkamp and coworkers recently found the same type of difference between CD and UC in expression of hBD-2 and hBD-3 mRNA [6]. The authors suggest that there is a defect in β-defensin expression in CD [33]. Whether this is indeed the explanation is currently unclear. The microbial load is vastly different in the healthy small intestine compared to the healthy large intestine. As we have no information about the degree of microbial infiltration in the small intestine of CD patients with active disease, this will remain an open question.
Why do colonic epithelial cells in UC express increased levels of mRNA for hBD-2, -3 and -4 but not for hBD-1? Two, not mutually exclusive, explanations seem likely. hBD-2, -3 and -4 but not hBD-1 are induced by microbial pathogens gaining access to the epithelial surface where they interact with Toll-like receptors, which in turn activate defensin expression. Indeed, increased hBD-2 and hBD-3 but not hBD-1 expression have been observed by exposing epithelial cells to different bacteria [2,3,14,15,23]. The other possibility would be that up-regulation of β-defensins are the consequence of increased expression of proinflammatory cytokines released from immune cells in the intraepithelial compartment or in lamina propria. The finding that hBD-2 is induced by TNF-α and IL-1β[2,3,21,23] and hBD-3 by IFN-γ (this study) in epithelial cells is compatible with the latter idea. The capacity of IFN-γ to up-regulate hBD-3 has also been demonstrated in the keratinocyte cell line HaCaT and in differentiated airway epithelia [24]. Analysis of the hBD-3 gene reveals IFN-γ response elements, several consensus sequences for activator protein-1 (AP-1) response elements, GM-CSF response elements and NF-IL-6 response elements [25] but no nuclear transcription factor κB (NFκB) consensus elements. Different types of inflammatory stimuli probably regulate BD-2 compared to hBD-3 gene expression.
In this study we were not able to induce increased hBD-4 expression by IL-1β, IFN-γ or TNF-α in the intestinal epithelial cell lines. Similarly, hBD-4 mRNA was not affected by IL-1α, IL-6, IFN-γ or TNF-α, whereas phorbol ester and bacteria were potent inducers in a lung epithelial cell line [16]. Moreover, the hBD-4 promoter region revealed absence of NFκB or STAT binding sites [16].
In UC there is a strong augmentation of the local adaptive immune response with heavy infiltration of immune cells in lamina propria [34] and a distorted T cell cytokine profile [35] indicative of a T-reg type of response. There was only weak production of IFN-γ or TNF-α in lamina propria T cells of UC colon. It seems unlikely, therefore, that these cytokines are responsible for up-regulation of hBD-2, -3 -4 in UC. Consequently, increased adherence of bacteria to the epithelium in UC colon [36,37] is the more probable explanation.
So far all well-characterized human β-defensins have been found in IEC. While the ‘epithelial’α-defensins HD-5 and HD-6 are confined to Paneth cells in the small intestine in normal physiology, the β-defensins seem to be expressed in the whole intestine. This indicates that they are important in the innate defence of the intestinal mucosa. Because exposure of the epithelium to pathogenic microorganisms will occur more or less regularly, there must be a fast and reliable first line of defence to protect from infection. Because hBD-1–4 are reactive against a broad range of microorganisms, it is reasonable to assume that an important role of the defensins is to kill microorganisms. Antimicrobial peptides may, however, have multiple roles. They may in addition act as mediators of inflammation. hBD-3 and hBD-4 are chemotactic to monocytes in vitro[16,24], hBD-1 and hBD-2 attract memory T cells and dendritic cells [38]. Thus, an equally important role for the β-defensins of IEC in UC may be to attract more immune cells to the site of inflammation.
Acknowledgments
This project was supported by grants from the ‘Network for Inflammation Research’ funded by the Swedish Foundation for Strategic Research (S. H.), the Swedish Cancer Society (S. H.), the Swedish Research Council, Medicine (Å. D. and S. H.), and the Swedish Research Council, Natural Sciences and Technology (M.-L. H.). We thank Dr Åke Öberg and colleagues for the surgical samples, Marianne Sjöstedt for skilful technical assistance, Anne Israelsson and Anna Bas for advice on different technical matters.
REFERENCES
- 1.Cunliffe RN, Rose FR, Keyte J, Abberley L, Chan WC, Mahida YR. Human defensin 5 is stored in precursor form in normal Paneth cells and is expressed by some villous epithelial cells and by metaplastic Paneth cells in the colon in inflammatory bowel disease. Gut. 2001;48:176–85. doi: 10.1136/gut.48.2.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fahlgren A, Hammarström S, Danielsson Å, Hammarström ML. Increased expression of antimicrobial peptides and lysozyme in colonic epithelial cells of patients with ulcerative colitis. Clin Exp Immunol. 2003;131:90–101. doi: 10.1046/j.1365-2249.2003.02035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Neil DA, Porter EM, Elewaut D, et al. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J Immunol. 1999;163:6718–24. [PubMed] [Google Scholar]
- 4.Wehkamp J, Fellermann K, Herrlinger KR, et al. Human beta-defensin 2 but not beta-defensin 1 is expressed preferentially in colonic mucosa of inflammatory bowel disease. Eur J Gastroenterol Hepatol. 2002;14:745–52. doi: 10.1097/00042737-200207000-00006. [DOI] [PubMed] [Google Scholar]
- 5.Wehkamp J, Schwind B, Herrlinger KR, et al. Innate immunity and colonic inflammation: enhanced expression of epithelial alpha-defensins. Dig Dis Sci. 2002;47:1349–55. doi: 10.1023/a:1015334917273. [DOI] [PubMed] [Google Scholar]
- 6.Wehkamp J, Harder J, Weichenthal M, et al. Inducible and constitutive beta-defensins are differentially expressed in Crohn's disease and ulcerative colitis. Inflamm Bowel Dis. 2003;9:215–23. doi: 10.1097/00054725-200307000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Cunliffe RN, Kamal M, Rose FR, James PD, Mahida YR. Expression of antimicrobial neutrophil defensins in epithelial cells of active inflammatory bowel disease mucosa. J Clin Pathol. 2002;55:298–304. doi: 10.1136/jcp.55.4.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lehrer RI, Ganz T. Defensins of vertebrate animals. Curr Opin Immunol. 2002;14:96–102. doi: 10.1016/s0952-7915(01)00303-x. [DOI] [PubMed] [Google Scholar]
- 9.Yang D, Biragyn A, Kwak LW, Oppenheim JJ. Mammalian defensins in immunity: more than just microbicidal. Trends Immunol. 2002;23:291–6. doi: 10.1016/s1471-4906(02)02246-9. [DOI] [PubMed] [Google Scholar]
- 10.Raj PA, Dentino AR. Current status of defensins and their role in innate and adaptive immunity. FEMS Microbiol Lett. 2002;206:9–18. doi: 10.1111/j.1574-6968.2002.tb10979.x. [DOI] [PubMed] [Google Scholar]
- 11.Ouellette AJ, Bevins CL. Paneth cell defensins and innate immunity of the small bowel. Inflamm Bowel Dis. 2001;7:43–50. doi: 10.1097/00054725-200102000-00007. [DOI] [PubMed] [Google Scholar]
- 12.Quayle AJ, Porter EM, Nussbaum AA, et al. Gene expression, immunolocalization, and secretion of human defensin-5 in human female reproductive tract. Am J Pathol. 1998;152:1247–58. [PMC free article] [PubMed] [Google Scholar]
- 13.Schutte BC, Mitros JP, Bartlett JA, et al. Discovery of five conserved beta-defensin gene clusters using a computational search strategy. Proc Natl Acad Sci USA. 2002;99:2129–33. doi: 10.1073/pnas.042692699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harder J, Bartels J, Christophers E, Schroder JM. A peptide antibiotic from human skin. Nature. 1997;387:861. doi: 10.1038/43088. [DOI] [PubMed] [Google Scholar]
- 15.Harder J, Bartels J, Christophers E, Schroder JM. Isolation and characterization of human beta-defensin-3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276:5707–13. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 16.Garcia JR, Krause A, Schulz S, et al. Human beta-defensin 4: a novel inducible peptide with a specific salt-sensitive spectrum of antimicrobial activity. FASEB J. 2001;15:1819–21. [PubMed] [Google Scholar]
- 17.Bensch KW, Raida M, Magert HJ, Schulz-Knappe P, Forssmann WG. hBD-1: a novel beta-defensin from human plasma. FEBS Lett. 1995;368:331–5. doi: 10.1016/0014-5793(95)00687-5. [DOI] [PubMed] [Google Scholar]
- 18.Semple CA, Rolfe M, Dorin JR. Duplication and selection in the evolution of primate beta-defensin genes. Genome Biol. 2003;4:R31. doi: 10.1186/gb-2003-4-5-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frye M, Bargon J, Lembcke B, Wagner TO, Gropp R. Differential expression of human alpha- and beta-defensins mRNA in gastrointestinal epithelia. Eur J Clin Invest. 2000;30:695–701. doi: 10.1046/j.1365-2362.2000.00696.x. [DOI] [PubMed] [Google Scholar]
- 20.Singh PK, Jia HP, Wiles K, et al. Production of beta-defensins by human airway epithelia. Proc Natl Acad Sci USA. 1998;95:14961–6. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Neil DA, Cole SP, Martin-Porter E, et al. Regulation of human beta-defensins by gastric epithelial cells in response to infection with Helicobacter pylori or stimulation with interleukin-1. Infect Immun. 2000;68:5412–5. doi: 10.1128/iai.68.9.5412-5415.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehmann J, Retz M, Harder J, et al. Expression of human beta-defensins 1 and 2 in kidneys with chronic bacterial infection. BMC Infect Dis. 2002;2:20. doi: 10.1186/1471-2334-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harder J, Meyer-Hoffert U, Teran LM, et al. Mucoid Pseudomonas aeruginosa, TNF-alpha, and IL-1beta, but not IL-6, induce human beta-defensin-2 in respiratory epithelia. Am J Respir Cell Mol Biol. 2000;22:714–21. doi: 10.1165/ajrcmb.22.6.4023. [DOI] [PubMed] [Google Scholar]
- 24.Garcia JR, Jaumann F, Schulz S, et al. Identification of a novel, multifunctional beta-defensin (human beta-defensin 3) with specific antimicrobial activity. Its interaction with plasma membranes of Xenopus oocytes and the induction of macrophage chemoattraction. Cell Tissue Res. 2001;306:257–64. doi: 10.1007/s004410100433. [DOI] [PubMed] [Google Scholar]
- 25.Jia HP, Schutte BC, Schudy A, et al. Discovery of new human beta-defensins using a genomics-based approach. Gene. 2001;263:211–8. doi: 10.1016/s0378-1119(00)00569-2. [DOI] [PubMed] [Google Scholar]
- 26.Valore EV, Park CH, Quayle AJ, Wiles KR, McCray PB, Jr, Ganz T. Human beta-defensin-1: an antimicrobial peptide of urogenital tissues. J Clin Invest. 1998;101:1633–42. doi: 10.1172/JCI1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldman MJ, Anderson GM, Stolzenberg ED, Kari UP, Zasloff M, Wilson JM. Human beta-defensin-1 is a salt-sensitive antibiotic in lung that is inactivated in cystic fibrosis. Cell. 1997;88:553–60. doi: 10.1016/s0092-8674(00)81895-4. [DOI] [PubMed] [Google Scholar]
- 28.Schibli DJ, Hunter HN, Aseyev V, et al. The solution structures of the human beta-defensins lead to a better understanding of the potent bactericidal activity of HBD3 against Staphylococcus aureus. J Biol Chem. 2002;277:8279–89. doi: 10.1074/jbc.M108830200. [DOI] [PubMed] [Google Scholar]
- 29.Lundqvist C, Melgar S, Yeung MMW, Hammarström S, Hammarström ML. Intraepithelial lymphocytes in human gut have lytic potential and a cytokine profile that suggest T helper 1 and cytotoxic functions. J Immunol. 1996;157:1926–34. [PubMed] [Google Scholar]
- 30.Lundqvist C, Hammarström ML, Athlin L, Hammarström S. Isolation of functionally active intraepithelial lymphocytes and enterocytes from human small and large intestine. J Immunol Meth. 1992;152:253–63. doi: 10.1016/0022-1759(92)90147-l. [DOI] [PubMed] [Google Scholar]
- 31.Panoskaltsis-Mortari A, Bucy RP. In situ hybridization with digoxigenin-labeled RNA probes: facts and artifacts. Biotechniques. 1995;18:300–7. [PubMed] [Google Scholar]
- 32.Bas A, Hammarström SG, Hammarström ML. Extrathymic TCR gene rearrangement in human small intestine. Identification of new splice forms of recombination activating gene-1 mRNA with selective tissue expression. J Immunol. 2003;171:3359–71. doi: 10.4049/jimmunol.171.7.3359. [DOI] [PubMed] [Google Scholar]
- 33.Fellermann K, Wehkamp J, Herrlinger KR, Stange EF. Crohn's disease: a defensin deficiency syndrome? Eur J Gastroenterol Hepatol. 2003;15:627–34. doi: 10.1097/00042737-200306000-00008. [DOI] [PubMed] [Google Scholar]
- 34.Yeung MMW, Melgar S, Baranov V, et al. Characterisation of mucosal lymphoid aggregates in ulcerative colitis: immune cell phenotype and TcR-gammadelta expression. Gut. 2000;47:215–27. doi: 10.1136/gut.47.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melgar S, Yeung MMW, Bas A, et al. Over-expression of interleukin 10 in mucosal T cells of patients with active ulcerative colitis. Clin Exp Immunol. 2003;134:127–37. doi: 10.1046/j.1365-2249.2003.02268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swidsinski A, Ladhoff A, Pernthaler A, et al. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 37.Schultsz C, Van Den Berg FM, Ten Kate FW, Tytgat GN, Dankert J. The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology. 1999;117:1089–97. doi: 10.1016/s0016-5085(99)70393-8. [DOI] [PubMed] [Google Scholar]
- 38.Yang D, Chertov O, Bykovskaia SN, et al. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science. 1999;286:525–8. doi: 10.1126/science.286.5439.525. [DOI] [PubMed] [Google Scholar]