Abstract
Umbilical cord blood has emerged as an alternative source of haematopoietic CD34+ cells for allogeneic stem cell transplantation. Although bacteraemia induced by Escherichia coli is considered one of the complications of transplantation, expression of proinflammatory cytokines is poorly understood. In this study, we report the altered expression of proinflammatory cytokines in CD34+ cells and their in vitro cultured cells following E. coli infection. CD34+ stem cells and their cultured cells up-regulated expression of proinflammatory cytokines such as interleukin (IL)-1α, IL-6, IL-8 and tumour necrosis factor (TNF)-α after infection with E. coli. Expression of the proinflammatory cytokines was generated mainly by the granulocyte-macrophage lineages. E. coli infection activated the signals of p50/p50 nuclear factor-kappaB (NF-κB) homodimers and IκB kinase. Furthermore, inhibition of NF-κB activation lowered the up-regulated expression of the proinflammatory cytokines. These results suggest that CD34+ cells and their cultured cells infected with E. coli induce the expression of proinflammatory cytokines via the NF-κB pathway.
Keywords: CD34, cytokine, Escherichia coli, NF-κB
INTRODUCTION
Umbilical cord blood contains a significantly higher number of CD34+ stem cells than adult peripheral blood [1]. Moreover, the number of colony forming unit–granulocyte-macrophages (CFU–GM) is much higher in umbilical cord blood than peripheral blood [2,3]. Therefore, umbilical cord blood is proposed as an ideal alternative to bone marrow and peripheral blood for haematopoietic stem cell transplantation. However, sepsis induced by bacterial infection is known to be one of the complications after cord blood transplantation. For example, early transplant-related mortality after cord blood transplantation is close to 50%, due mainly to infectious complications such as bacteraemia [4]. In addition, a report demonstrated that the sepsis induced by Escherichia coli infection has been considered as the one of the complications upon transplantation treatment [5].
Proinflammatory cytokines, including tumour necrosis factor (TNF)-α, interleukin (IL)-1, IL-6 and IL-8, are synthesized and secreted by numerous cell types and tissues in response to pathogenic infection [6,7]. TNF-α, IL-1α and IL-6 are involved in the induction of acute phase proteins, generation of fever and shock. This phenomenon is known to be observed during the sepsis. In addition, many neutrophils are found in the blood in septic patients. The migration and activation of neutrophils are induced by chemokine IL-8 [8,9]. In this regard, cytokine-induced inflammatory responses are highly characteristic of the septic insult produced by Gram-negative bacterial infection. Considering cytokine-induced inflammatory responses in sepsis, there is a possibility that proinflammatory cytokines may be expressed in CD34+ cells or their cultured cells when bacterial infection occurs during the period of cord blood transplantation. However, the expression of proinflammatory cytokines in the CD34+ stem cells is poorly understood.
Many of the genes that are activated after bacterial infection are target genes of the transcription nuclear factor-kappa B (NF-κB) [10–14]. NF-κB is a dimeric transcription factor composed of homodimers or heterodimers of Rel protein, of which there are five family members in mammalian cells [i.e. RelA (p65), c-Rel, Rel B, NF-κB1 (p50) and NF-κB2 (p52)] [15–17]. NF-κB dimers are held in the cytoplasm in an inactive state by inhibitory proteins, the IκBs. Stimulation of cells with cytokine or bacterial infection activates a signalling cascade that culminates in the phosphorylation of IκBs [11,13]. Two isoforms of IκB kinases, IκB kinase (IKK)α and IKKα, phosphorylate IκBs directly on serine residues and are components of a high molecular weight cytoplasmic IKK complex. Phosphorylation of IκBs on conserved serine residues targets IκBs for subsequent ubiquitination and degradation. However, the role of NF-κB in E. coli-induced signal transduction of CD34+ cells has not been clarified.
The present study thus tested whether CD34+ cells or their cultured cells could use an NF-κB signal transduction pathway to activate the proinflammatory cytokines in response to E. coli infection. We report here that E. coli infection can induce NF-κB signals in CD34+ cells and their derivatives and that inhibition of NF-κB showed the suppression of the proinflammatory cytokine expression. These results suggest that activated NF-κB signals may be a major regulator of the activation of proinflammatory cytokine genes in CD34+ cells and their cultivated cells in response to E. coli infection.
MATERIALS AND METHODS
Isolation of CD34+ cells in umbilical cord blood
Normal umbilical cord blood scheduled for discarding after delivery was obtained with a maternal consent. Low-density mononuclear cells were isolated on Ficoll-Paque (1·077 g/ml) (Amersham Pharmacia Biotech, Piscataway, NJ, USA), washed twice in a phosphate-buffered saline (PBS) containing 2 mm of EDTA, and centrifuged for 10 min at 200 g at 20°C. The cell pellet was resuspended in a final volume of 300 µl buffer/108 total cells. CD34+ cells were incubated with a monoclonal antihuman CD34+ antibody (QBEND/10), followed by a positive selection of immunomagnetic beads and MACS separators according to the manufacturer's recommendations (Miltenyl Biotech, Auburn, CA, USA) [18]. The purity of CD34+ cells routinely exceeded ∼98%, as determined by fluorescence-activated cell sorting (FACS) analysis (Becton Dickinson, San Jose, CA, USA) (Fig. 1).
Fig. 1.
Flow cytometric analysis of CD34+ cells on cord blood cells. CD34+ cells were isolated from cord blood cells using MACS separation. The isolated CD34+ cells were stained with CD34-fluorescein isothiocyanate (FITC, solid line) or isotype control-FITC (dot line) for 15 min and fixed with 2% paraformaldehyde solution. Fixed cells were suspended in PBS and analysed by flow cytometry. Data for 10 000 cells were analysed by using a VellQuest version 3·11. software (Becton Dickinson). The area marked as M to the right peak contains the CD34+ cells (98·6%).
To differentiate CD34+ cells in vitro, CD34+ cells (1 × 105/ml) were seeded at Dulbecco's minimum essential medium (DMEM) with 10% heat-inactivated fetal bovine serum (FBS) containing FLT3 ligand (FL, 50 ng/ml, Chemicon, Temecula, CA, USA), thrombopoietin (TPO, 10 ng/ml, Kirin Brewery, Gunma, Japan) and stem cell factor (SCF, 50 ng/ml, Kirin Brewery) in 35 mm dishes at 37°C in a humidified atmosphere of 5% CO2 in air. Every 3–4 days, a half of the culture volume was removed from the wells to be replaced with fresh medium and growth factors. The cells were harvested at various intervals (days 7 and 14). Viability of the cells was assessed using the trypan blue exclusion assay. Under this culture condition, CD34+ fraction was decreased to 21–30% at day 7 and was only 5–6% at day 14, as assessed by FACS analyses. When 1 × 105 isolated CD34+ cells were incubated with FL + TPO + SCF at day 0, the viable cells were increased to (1·6 ± 0·7) × 107 cells after 7 days’ culture and (8·6 ± 1·5) × 108 cells after 14 days culture (mean ± s.d. of five separate experiments). The percentage of living cells was 96·2 ± 2·6% (day 7) and 86·8 ± 3·6% (day 14) of total cells (mean ± s.d. of five separate experiments).
To isolate CFU–GM, BFU–E (burst-forming unit–erythroid) and CFU–GEMM (colony forming unit–granulocyte, erythrocyte, macrophage, megakaryocyte), CD34+ cells were cultured in complete MethoCultTM medium (1% methylcellulose in DMEM, 30% FBS, 1% bovine serum albumin, 3 U/ml erythropoietin, 10−4m 2-mercaptoethanol, 2 mM l-glutamine, 50 ng/ml stem cell factor, 10 ng/ml GM–CSF, 10 ng/ml IL-3; Stem Cell Technologies, Vancouver, Canada). After 14 days of CD34+ cell culture in the complete MethoCultTM medium according to the manufacturer's protocol, the fractions of CFU–GM, BFU–E and CFU–GEMM were 68%, 29% and 3%, respectively (mean value of three separate experiments). Under the optical inverted microscopy, CFU–GM or BFU–E was collected.
Infection protocol
E. coli O29 was obtained from the American Type Culture Collection (ATCC; no. 23892). Bacteria were grown to late log phase at 37°C in tryptic soy broth before infection. CD34+ stem cells (day 0) or their cultured cells (days 7 or 14) in the six-well tissue culture plate were infected with E. coli for 1, 3, 6, 9 or 12 h in electrophoretic mobility shift assays; for 6, 12 or 18 h in reverse transcription–polymerase chain reaction (RT-PCR); for 18 h in cytokine production experiments [enzyme-linked immunosorbent assay (ELISA)]. The ratio of E. coli to the cells was adjusted to 10 : 1. In some experiments, CD34+ cells or their cultured cells were treated with an inhibitor of NF-κB activation, calpain-1 inhibitor (25 µm, Calbiochem, La Jolla, CA, USA), for 1 h before exposure to E. coli and during the incubation period of the experiment. A similar protocol was used for the experiments using NF-κB essential modifier (NEMO)-binding domain (NBD) peptide (200 µm, Peptron, Daejeon, Korea). An NBD peptide can block association of NEMO with the IKK complex and inhibit NF-κB activation [19]. Sequences of the wild-type and mutant peptides are drqikiwfqnrrmkwkkTALDWSWLQTE (wild-type) and drqikiwfqnrrmkwkkTALDASALQT E (mutant). Positions of the WeA mutations are underlined [19].
Quantitative RT-PCR analysis
CD34+ cells and their cultured cells were incubated with E. coli for the indicated times, after which total cellular RNA was extracted from the cells using Trizol reagent (Gibco BRL, Life Technologies, Palo Alto, CA, USA). Quantitative RT-PCR using internal standards was used to quantify cytokine mRNA levels, as described previously [20,21]. Synthetic standard RNA was kindly provided by Dr Kagnoff of the University of California, San Diego. PCR amplification consisted of 35 cycles of 1-min denaturation at 95°C, 2·5-min annealing and extension at either 60°C (IL-1α, IL-6, IL-8 and eotaxin), 65°C [TNF-α, MCP-1 and normal T cell expressed and secreted (RANTES)] or 72°C (β-actin). Cytokine mRNA levels of 5 × 103 molecules/µg of total RNA were considered positive. Although lower levels could be detected and quantified, they were considered unlikely to be biologically meaningful as they would, on average, reflect < 1 mRNA transcript/20 cells [22].
Cytokine ELISA in culture supernatants
Before the measurement of cytokines, the supernatants of E. coli-infected cell culture media were filtered through a 0·22-µm filter to remove any contaminants. The levels of human IL-1α, IL-6, IL-8, MCP-1 and TNF-α were determined by Quantikine immunoassay kits (R&D Systems, Minneapolis, MN, USA). Each sample was tested in triplicate. The detection limit was 15 pg/ml for the five cytokines.
Electrophoretic mobility shift assays (EMSA) and supershift EMSA
Cells were harvested, and nuclear extracts were prepared as described [13]. The concentrations of proteins in the extracts were determined by the Bradford assay (Bio-Rad, Hercules, CA, USA). Electrophoretic mobility shift assays (EMSA) were performed according to the protocol of the manufacturer (Promega, Madison, WI, USA). Five µg of nuclear extracts (obtained from ∼3 × 107 cultured cells derived from CD34+) or 0·5 µg of nuclear extracts (obtained from ∼ 3 × 106 CD34+ cells) was used. In brief, nuclear extracts were incubated for 30 min at room temperature with γ32P-labelled oligonucleotide probe corresponding to a consensus NF-κB binding site. After incubation, bound and free DNAs were resolved on 5% native polyacrylamide gels as described previously [11,13].
Supershift assays were used to identify the specific members of the NF-κB family which could be activated by infection with E. coli. EMSA was performed as described above except that rabbit antibodies (1 µg/reaction) against that NF-κB proteins p50, p52, p65, c-Rel and Rel B (Santa Cruz Biotechnology, Santa Cruz, CA, USA) were added during the binding reaction period [13].
Immunoblots
Cells were washed with ice-cold PBS and lysed in a 0·5 ml/well lysis buffer (150 mm NaCl, 20 mm Tris, pH 7·5, 0·1% Triton X-100, 1 mm PMSF, 10 µg/ml aprotonin), as described previously [13]. Protein concentrations in the lysates were determined by the Bradford method (Bio-Rad). Five to 15 µg protein/lane was size-fractionated on a denaturing, non-reducing 6% polyacrylamide minigel (Mini-PROTEIN II; Bio-Rad) and electrophoretically transferred to a nitrocellulose membrane (0·1-µm pore size). Specific proteins were detected using mouse antihuman IκBα (Santa Cruz Biotechnology), phospho-IκB kinase (IKK)α (Cell Signalling Technology, Beverly, MA, USA) or actin (Santa Cruz Biotechnology) as a primary antibody and peroxidase-conjugated antimouse or antirabbit IgG (Transduction Laboratories, Lexington, KY, USA) as a secondary antibody. Specifically bound peroxidase was detected by enhanced chemiluminescence (ECL system; Amersham Life Science, Bucks, UK) and exposure to X-ray film (XAR5; Eastman Kodak Company, Rochester, NY, USA) for 10–30 s.
Recombinant retrovirus and retrovirus infection
Dominant-negative IκBα (S32A, S36A) [23] was amplified with sense (5′-aacc ATGGCATACCCATACGACGTCCCA GACTACGCTttccaggcggccgagcgcccccaggag-3′) and antisense (5′-aaaaGGATCCtcataacgtcagacgctggcct-3′) primers using high fidelity Taq polymerase (Gibco BRL). The capital letters represent nucleotides encoding a HA tag. The PCR products were digested with Nco-1 and BamH1 restriction enzymes. These enzymes were incubated with the buffer [150 mm NaCl, 10 mm Tris-HCl, 10 mm MgCl2, 1 mm dithiothreitol (pH 7·9) and 100 µg/ml bovine serum albumin] at 37°C for 2 h. The digested PCR products were cloned into the corresponding sites in MFG retroviral vector by replacing the GFP sequence of MFG.GFP.IRES.puro [24]. The retroviral plasmids obtained were introduced into the 293 gpg retrovirus packaging cell line [25] by transient transfection. Briefly, the plasmid DNA (4 µg) was mixed with Lipofectamine (Gibco BRL) in optiMEM (Gibco BRL) at room temperature for 30 min. These mixtures were overlaid to the 293 gpg retrovirus packaging cell line (3 × 106 cells/60 mm culture dish) and incubated for 8 h at 37°C. After 8 h, DMEM supplemented with 10% FBS and puromycin (2 µg/ml, Sigma, St Louis, MO, USA) were added. At 24 and 48 h later, the culture media were change to new media. After 72 h the supernatants were harvested and used for retroviral infection. The virus titres, measured in NIH3T3 cell line by puromycin-resistant colony formation, were between 105 and 5 × 105/ml (retrovirus-IκBα-AA). The infection and selection of target cells with puromycin was performed as described previously [24].
Transfection of plasmids and luciferase assay
pIL8-luciferase and pRSV-β-galactosidase transcriptional reporters were kindly donated by Dr Kagnoff of the University of California, San Diego [11]. The cultured cells derived from CD34+ (day 14) were transfected with 1·5 µg of plasmid DNA using GenePorter transfection reagent (Gene Therapy Systems, San Diego, CA, USA), according to the manufacturer's instructions. The transfected cells were incubated for 48 h at 37°C in a 5% CO2 incubator. Cells were incubated with E. coli for the indicated periods. Luciferase activity was determined and normalized relative to β-galactosidase expression, in accordance with the manufacturer's instructions (Tropix Inc., Bedford, MA, USA). Light release was quantified for 10 s using a luminometer (MicroLumat Plus, Berthold GmbH KG, Bad Wildbad, Germany), as described previously [11,13].
Statistical analysis
Data are presented as the mean ± standard deviation (s.d.) for quantitative RT-PCR, and the mean ± standard error of the means (SEM) for the ELISA and luciferase assays. Wilcoxon's rank sum test was used for statistical analysis. A P-value less than 0·05 was considered statistically significant.
RESULTS
E. coli infection up-regulates expression of IL-1α, IL-6, IL-8 and TNF-α in the CD34+ cells or their cultured cells
IL-1α, IL-6, IL-8, MCP-1 and TNF-α are proinflammatory cytokines that are involved in the inflammatory process. We assessed the gene expression of these cytokines in CD34+ cells following E. coli infection. CD34+ cells constitutively expressed low levels of IL-1α, IL-6, IL-8 and TNF-α mRNA expression, but the expression of these proinflammatory cytokines increased after E. coli infection (Fig. 2). In addition, up-regulated proinflammatory cytokine mRNA expression such as IL-1α, IL-6, IL-8 and TNF-α were also noted in E. coli-infected cultured cells derived from CD34+ (days 7 and 14) (Fig. 2). Quantification of mRNA using synthetic standard RNA, as shown in Table 1, showed that IL-1α, IL-6, IL-8 and TNF-α mRNA expression by E. coli infection was 2–19 times greater than control in CD34+ cells. The cultured cells derived from CD34+ stem cells also increased the numbers of proinflammatory cytokine mRNA transcripts in response to E. coli infection (Table 2). However, mRNA expression of MCP-1, RANTES and eotaxin was not increased significantly in E. coli-infected CD34+ cells (day 0) and their cultured cells (days 7 and 14) (MCP-1, ∼8·5 × 104 transcripts/µg total RNA; RANTES, <5 × 103 transcripts/µg total RNA; eotaxin, <5 × 103 transcripts/µg total RNA; β-actin, ∼1 × 106 transcripts/µg total RNA).
Fig. 2.
RT-PCR analysis of proinflammatory cytokine mRNA in E. coli-infected CD34+ cells and their cultured cells. The cells (1 × 106) in the six-well tissue culture plate were infected with E. coli for the indicated hours. The ratio of E. coli to the cells was adjusted to 10 : 1. The expression of mRNA for each cytokine and β-actin was assessed by RT-PCR using specific primers. The data are representative of more than five separate experiments. (a) Freshly isolated CD34+ haematopoietic stem cells; (b) or (c), cultured cells which were obtained from CD34+ stem cells incubated with FL + TPO + SCF for 7 days or 14 days, respectively.
Table 1.
Proinflammatory cytokine mRNA expression in CD34+ stem cells infected with E. coli*
| Time after infection (h) | ||||
|---|---|---|---|---|
| 0 | 6 | 12 | 18 | |
| IL-1α | 3·8 ± 4·6† | 35·6 ± 20·8 (9) | 45·6 ± 29·0 (12) | 41·8 ± 28·9 (11) |
| IL-6 | 8·2 ± 9·8 | 124 ± 67 (15) | 145 ± 49 (18) | 118 ± 34 (14) |
| IL-8 | 3·7 ± 3·3 | 7·8 ± 2·7 (2) | 19·8 ± 7·3 (5) | 32·6 ± 11·3 (9) |
| TNF-α | 0·6 ± 0·8 | 11·4 ± 6·2 (19) | 9·2 ± 3·3 (16) | 8·2 ± 3·4 (14) |
| β-actin | 106 ± 61 | 195 ± 125 (1·8) | 104 ± 56 (1·0) | 123 ± 50 (1·2) |
CD34+ cells (purity > 98%, 1 × 106 cells) in six-well plates were incubated with E. coli for the indicated hours. The ratio of E. coli to CD34+ cells was adjusted to 10 : 1. For quantification of the expressed transcripts, total RNA was reverse-transcribed using an oligo(dT) primer and synthetic internal RNA standards, and amplified by PCR.
Mean numbers ± s.d. of mRNA transcripts (104)/µg RNA (n = 5). Parentheses are mean fold-induction compared with uninfected control.
Table 2.
Proinflammatory cytokine mRNA expression in E. coli-infected cultured cells derived from CD34 + stem cells*
| Time after infection (h) | ||||
|---|---|---|---|---|
| 0 | 6 | 12 | 18 | |
| 7 days’ culture | ||||
| IL-1α | 8·5 ± 6·2† | 46·2 ± 18·0 (5) | 57·2 ± 30·8 (7) | 46·6 ± 27·4 (5) |
| IL-6 | 6·8 ± 3·3 | 10·1 ± 3·5 (1·5) | 23·4 ± 11·0 (3·4) | 9·7 ± 3·4 (1·4) |
| IL-8 | 7·2 ± 3·6 | 36·4 ± 15·6 (5) | 88·4 ± 25·0 (12) | 129 ± 46 (18) |
| TNF-α | 0·8 ± 0·3 | 11·4 ± 4·2 (14) | 10·3 ± 3·8 (12) | 6·9 ± 3·2 (8) |
| β-actin | 112 ± 49 | 125 ± 75 (1·1) | 256 ± 143 (2·3) | 190 ± 123 (1·7) |
| 14 days’ culture | ||||
| IL-1α | 9·2 ± 5·2 | 120 ± 110 (13) | 148 ± 106 (16) | 133 ± 73 (14) |
| IL-6 | 9·2 ± 3·8 | 468 ± 267 (51) | 436 ± 254 (47) | 450 ± 258 (49) |
| IL-8 | 8·7 ± 3·5 | 40·6 ± 17·0 (5) | 278 ± 154 (32) | 678 ± 355 (78) |
| TNF-α | 0·3 ± 0·2 | 20·1 ± 12·8 (63) | 17·3 ± 10·0 (54) | 16·4 ± 12·2 (51) |
| β-actin | 231 ± 119 | 175 ± 95 (0·8) | 151 ± 60 (0·7) | 174 ± 55 (0·8) |
CD34+ stem cells (purity > 98%, 1 × 105 cells) were incubated with FLT3 ligand (50 ng/ml) + thrombopoietin (10 ng/ml) + stem cell factor (50 ng/ml) for 7 or 14 days. Every 3–4 days, half the culture volume was removed from the wells to be replaced with fresh medium and growth factors. The cultured cells (1 × 106 cells) in six-well plates were incubated with E. coli for the indicated hours. The ratio of E. coli to the cultured cells was adjusted to 10 : 1. For quantification of the expressed transcripts, total RNA was reverse-transcribed using an oligo(dT) primer and synthetic internal RNA standards, and amplified by PCR.
Mean numbers ± s.d. of mRNA transcripts (104)/µg RNA (n = 5). Parentheses are mean fold-induction compared with uninfected control.
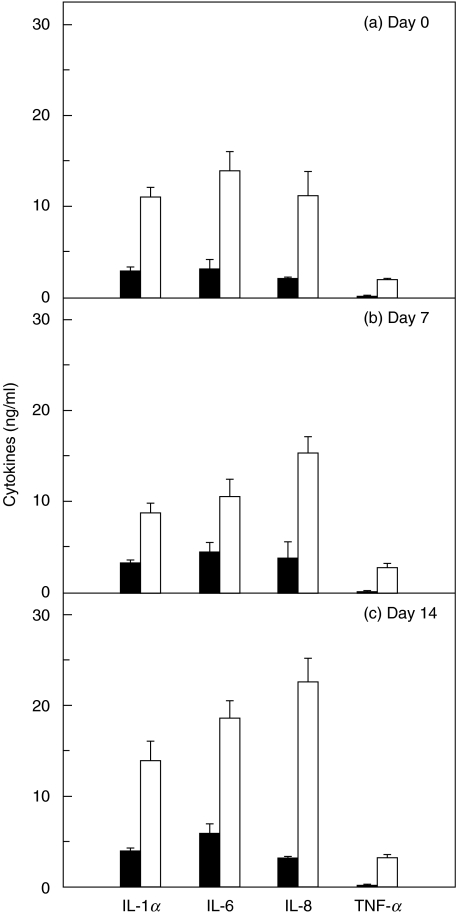
To determine whether increased cytokine mRNA levels were accompanied by increased protein secretion, we measured the amount of cytokine proteins in culture supernatants. The secretions of each cytokine were paralleled by each mRNA expression (Fig. 3). For example, the cultured cells derived from CD34+ (day 14) infected with E. coli produced 3·6-fold higher amounts of IL-1α compared with uninfected controls. However, MCP-1 production in infected and uninfected cells remained relatively constant (∼ 18·5 ng/ml). These data suggest that the increased proinflammatory cytokine secretion in response to E. coli infection may be due in large part to pretranslational events.
Fig. 3.
Proinflammatory cytokine secretion by CD34+ cells or their cultured cells infected with E. coli. CD34+ cells (day 0, a) or their cultured cells [day 7 (b) and 14 (c)] in six-well plates were incubated with E. coli for 18 h and protein levels of each cytokine were determined by ELISA. Data are the mean ± s.e.m. of seven separate experiments. White bar, E. coli-infected; black bar, non-infected controls.
The magnitude of the cytokine response was dependent on the number of infected E. coli per CD34+ cells. Infection of the cultured cells derived from CD34+ (day 14, 1 × 106 cells) with increasing numbers of E. coli for 6 h was paralleled by increased IL-1α mRNA expression. At 6 h after infection of the cultured cells with 104, 105, 106, 107 and 108 CFU of E. coli, IL-1α mRNA transcripts increased 5·4 ± 2·5, 8·2 ± 1·4, 13·2 ± 3·1, 19·0 ± 3·6 and 17·3 ± 3·2-fold, respectively, relative to those of uninfected control (mean fold-increase ± s.d., n = 3). In this experiment, the mean number of IL-1α mRNA transcripts of the uninfected control was 1·2 × 105 transcripts/µg RNA.
To test the subpopulation of the cultured cells derived from CD34+ involved in the induction of proinflammatory cytokines in response to E. coli infection, the proinflammatory cytokine expression levels were compared between CFU–GM and BFU–E. As shown in Fig. 4, up-regulation of the proinflammatory cytokines by E. coli infection was attributed mainly to CFU–GM in the cultured cells derived from CD34+.
Fig. 4.
Secretion of proinflammatory cytokines from CFU–GM or BFU-E in response to E. coli infection. CFU–GM or BFU–E were isolated from the cultured cells derived from CD34+ cells. The freshly isolated CD34+ cells were cultured in complete MethoCultTM medium for 14 days. Isolated CFU–GM or BFU–E (1 × 104) were incubated with E. coli (1 × 105 CFU) for 18 h and protein levels of each cytokine were determined by ELISA. Data are the mean ± s.e.m. of five separate experiments. Asterisks indicate statistical significance with P < 0·05 in comparison with control.
E. coli infection induces p50 homodimeric NF-κB activation and IκBα degradation in CD34+ cells and their cultured cells
To determine whether E. coli activates NF-κB in human CD34+ cells and their cultured cells, DNA binding studies were performed using cell extracts after infection of the cultured cells derived from CD34+ cells (day 14) with E. coli. Following infection, these cells increased the DNA binding activity of NF-κB, as shown by EMSA (Fig. 5a). In addition, degradation of IκBα was observed in E. coli-infected cultured cells derived from CD34+, as determined by immunoblot analysis. Notably, an IκBα signal completely disappeared at 1 h after infection, but then continuously recovered (Fig. 5a). Similar results were obtained in the CD34+ cells (day 0) (Fig. 5b).
Fig. 5.
NF-κB activation and IκBα degradation in the CD34+ cells or their cultured cells infected with E. coli. (a) The cultured cells (day 14) were incubated with E. coli. The ratio of E. coli to the cells was adjusted to 10 : 1. NF-κB DNA binding activity was assessed by EMSA at the indicated times. Immunoblots for concurrent IκBα and actin under the same condition are provided beneath each EMSA time point. The results are representative of five repeated experiments. (+) Represents a positive control whereby the cultured cells derived from CD34+ were treated with TNF-α (20 ng/ml) for 3 h (–) represents negative control. (b) Similar results of NF-κB activation and IκBα degradation were observed in the CD34+ stem cells (day 0). (c) Activation of specific NF-κB subunits in the cultured cells (c, day 14) infected with E. coli. Supershift assays were performed using antibodies to p50, p52, p65, c-Rel and Rel B. The antibody to p50 shifts the entire NF-κB signals. Anti-p52, p65, c-Rel and Rel B did not show the shifts. The results are representative of three repeated experiments. (d) Similar results of supershift assay were obtained from the CD34+ cells (day 0).
NF-κB exist as dimeric complexes, either homo- or heterodimers [17]. To identify the specific NF-κB subunits that comprise the NF-κB signal detected by EMSAs in E. coli-infected CD34+ cells or their cultured cells, a supershift assay was performed. Specific antibodies to p50, p52, p65, c-Rel and Rel B were used for these experiments. Supershift studies demonstrated that the antibody to p50 shifted the entire signal. However, antip52, antip65, antic-Rel or anti-Rel B antibodies did not shift the NF-κB signals (Fig. 5c,d). These results indicate that NF-κB activation by E. coli infection is mediated predominantly by homodimers of p50.
Inhibition of NF-κB activity down-regulates the proinflammatory cytokine expression in CD34+ cells infected with E. coli
Based on E. coli infection activating the NF-κB signals in CD34+ cells or their cultured cells (Fig. 5), we next determined whether increased proinflammatory cytokine expression after E. coli infection was associated with NF-κB pathways. Addition of calphain-1 inhibitor to E. coli-infected cultured cells derived from CD34+ cells (day 14) significantly decreased the proinflammatory cytokine secretion (Table 2). Consistent with this, the expression of up-regulated IL-1α mRNA transcripts was also decreased by the treatment with calpain-1 inhibitor [E. coli-infected, 37 ± 26; E. coli+ calpain-1 inhibitor, 6·9 ± 1·3; non-infected control, 1·5 ± 1·8; mean ± s.d. (× 105) mRNA transcripts/µg total RNA; 6 h post-infection, n = 3].
One of the major pathways of NF-κB activation involves the phosphorylation of IκBα, which is followed in turn by IκB degradation and the subsequent migration of NF-κB dimers from the cytoplasm to the nucleus [15,16]. We assayed the expression of proinflammatory cytokines in CD34+ cells with suppressed NF-κB activity, which had been transfected with the retrovirus-IκBα-AA. To confirm that the transfection with retrovirus-IκBα-AA was related to a decrease in an NF-κB signal, EMSA was performed. Activation of NF-κB signals was inhibited in the cultured cells derived from CD34+ (day 14) transfected with retrovirus-IκBα-AA (Fig. 6a). Consistent with this, mRNA expression of IL-1, IL-6, IL-8 and TNF-α in response to E. coli infection was also decreased in NF-κB-suppressed cultured cells derived from CD34+ (Fig. 6b).
Fig. 6.
Proinflammatory cytokine expression in E. coli-infected cultured cells transfected with retrovirus containing the IκBα super-repressor. (a) The cultured cells (day 14) were transfected with either retrovirus containing IκBα super-repressor (retrovirus-IκBα-AA) or control virus (retrovirus-GFP). At 48 h after transfection, the cells were incubated with E. coli for 3 h. NF-κB binding activity was assayed by EMSA. The results are representative of three repeated experiments. (b) Transfected cultured cells (1 × 108 cells) were incubated with E. coli (1 × 109 CFU) for 18 h. For quantification of the expressed transcripts, total RNA was reverse-transcribed using an oligo(dT) primer and synthetic internal RNA standards, and amplified by PCR. Data are presented as numbers of mRNA transcripts/µg of total RNA (mean ± SD, n = 5). Asterisks indicate statistical significance with P < 0·05 in comparison with control virus-transfected cells infected with E. coli. □, control; ▪, E. coli-infected;  , E. coli-infected cells transfected with retrovirus-IκBα-AA;
, E. coli-infected cells transfected with retrovirus-IκBα-AA;  , E. coli-infected cells transfected with control virus (retrovirus-GFP)
, E. coli-infected cells transfected with control virus (retrovirus-GFP)
E. coli infection increases phosphorylated IKK signals in the cultured cells derived from CD34+
Major pathways for NF-κB activation are involved in the activation of IKK, which is followed by IκB degradation [16]. In this study, we found that E. coli infection increased the signals of phosphorylated IKKα in the cultured cells derived from CD34+ (day 14) (Fig. 7a). These cultured cells were obtained from CD34+ cells incubated with FL + TPO + SCF for 14 days. In order to study whether the activation of IKK was one of the major pathways that culminated in the expression of proinflammatory cytokines following E. coli infection, cultured cells derived from CD34+ (day 14) were treated with an NBD peptide which can block the association of NEMO with the IKK complex [19]. Secretion of proinflammatory cytokines was inhibited by treatment with an NBD peptide, but not by treatment with the mutant type of NBD peptide (Table 3). Similar results were obtained from the CD34+ cells (day 0) infected with E. coli[IL-1α, control, 1·78 ± 0·66; E. coli-infected, 9·33 ± 2·22; E. coli+ NBD (wild-type), 3·62 ± 1·01; E. coli+ NBD (mutant), 10·20 ± 1·51; mean ± s.e.m. (ng/ml) of three separate experiments], [TNF-α, control, 0·03 ± 0·02; E. coli-infected, 1·91 ± 0·15; E. coli+ NBD (wild-type), 0·73 ± 0·13; E. coli+ NBD (mutant), 1·87 ± 0·27; mean ± s.e.m. (ng/ml) of three separate experiments]. To confirm that E. coli-induced IKK activation was directly associated with reporter gene activation, luciferase assays were also performed. Addition of NBD peptide decreased the activation of IL-8 reporter genes in the cultured cells infected with E. coli (Fig. 7b). These cultured cells were obtained from CD34+ cells incubated with FL + TPO + SCF for day 14. These results demonstrate that the activation of IKK are involved as crucial steps for NF-κB activation in CD34+ cells or their cultured cells following E. coli infection.
Fig. 7.
Phosphorylation of IKK and IL-8 reporter gene activation in E. coli-infected cultured cells derived from CD34+. (a) The cultured cells (day 14) were incubated with E. coli for the indicated times. The ratio of E. coli to the cells was adjusted to 10 : 1. Phosphorylation and protein expression of IKKα and actin were assessed by immunoblot. (b) Cultured cells (day 14) were transfected with pIL-8-luciferase transcriptional reporters; 48 h later the transfected cells were incubated with E. coli (black bar) or TNF-α (20 ng/ml, open bar) for 6 h. Data are expressed as the mean fold induction ± s.e.m. in luciferase activity relative to non-stimulated controls (n = 5).
Table 3.
Proinflammatory cytokine production in E. coli-infected cultured cells derived from CD34+ in the presence of NF-κB inhibitor or IKK inhibitor*
| E. coli-infected | |||||
|---|---|---|---|---|---|
| NBD peptide | |||||
| Cytokines | Control | E. coli-infected | Calpain-1 inhibitor | Wild-type | Mutant-type |
| IL-1α | 3·10 ± 0·47† | 12·23 ± 1·04 | 7·36 ± 0·89‡ | 6·71 ± 0·92‡ | 13·09 ± 1·48 |
| IL-6 | 5·92 ± 1·41 | 16·85 ± 2·08 | 9·63 ± 1·72‡ | 7·78 ± 1·43‡ | 15·85 ± 1·39 |
| IL-8 | 6·72 ± 0·93 | 22·94 ± 2·51 | 11·36 ± 1·46‡ | 12·18 ± 2·76‡ | 22·56 ± 3·28 |
| TNF-α | 0·04 ± 0·03 | 2·68 ± 0·55 | 1·14 ± 0·19‡ | 0·98 ± 0·13‡ | 2·35 ± 0·53 |
CD34+ stem cells (purity > 98%, 1 × 105 cells) were incubated with FLT3 ligand (50 ng/ml) + thrombopoietin (10 ng/ml) + stem cell factor (50 ng/ml) for day 14. These cultured cells (1 ×108 cells) were incubated with an NF-κB inhibitor, calpain-1 inhibitor (25 µm), or an IKK inhibitor, NBD peptides (200 µm) for 1 h followed by infection with E. coli for 18 h. The ratio of E. coli to the cells was adjusted to 10 : 1. Protein levels of cytokines in culture supernatants were determined by ELISA.
Mean ± s.e.m. (ng/ml) of five separate experiments.
Significantly different from value for the E. coli-infected group (P < 0·05).
DISCUSSION
Bacteraemia or sepsis causes complication of stem cell transplantation [5]. We have shown that the infection of CD34+ cells or their cultured cells with E. coli up-regulated expression of the proinflammatory cytokines such as IL-1α, IL-6, IL-8 and TNF-α. Furthermore, we demonstrated that the NF-κB activation pathway plays a crucial role in the expression of proinflammatory cytokines in response to the infection of CD34+ cells or their cultured cells with E. coli.
The release of proinflammatory cytokines can contribute to the inflammatory cell infiltration that accompanies bacteraemia. In addition, cytokines IL-1, IL-6 and TNF-α are known to be importantly involved in the systemic inflammatory process such as shock, fever and production of acute phase proteins and antibodies [26,27]. IL-8 is also known to be a chemoattractant and activator for neutrophils [28]. Our results showed that these cytokines were up-regulated in CD34+ cells infected with E. coli. Notably, the kinetics of IL-8 mRNA expression was delayed relative to the other cytokines. Therefore, there is a possibility that cytokines released from the cells during infection may mediate the IL-8 response to E. coli infection.
NF-κB has a key role in regulating the transcription of several members of a proinflammatory gene family that is induced in response to inflammation or infection with pathogens [10–14]. Activation of NF-κB in the cytoplasm involves the inducible phosphorylation of IκBs, which then undergoes ubiquitin-mediated proteolysis, thereby releasing NF-κB dimers to translocate to the nucleus [15–17]. In this study, E. coli infection activated NF-κB in CD34+ cells or their cultured cells as assayed by EMSA. In addition, degradation of IκBα was observed in E. coli-infected cells. Furthermore, blocking the NF-κB activation with calpain-1 inhibitor or retrovirus-IκBα-AA transfection significantly decreased expression of the proinflammatory cytokines in E. coli-infected cells. These results indicate that phosphorylation and degradation of the inhibitory protein IκBα and the subsequent dissocation of this protein from the NF-κB complex are necessary for the proinflammatory cytokine expression in response to E. coli infection. However, considering that the addition with calpain-1 inhibitor or transfection with retrovirus-IκBα-AA did not suppress completely proinflammatory cytokine expression, there is a possibility that other pathways may be involved in the expression of the proinflammatory cytokines induced by E. coli infection.
Distinct expression profiles for each of the five mammalian NF-κB/Rel members have been observed in developing tissues and organs. Thus, p50/p65 heterodimers are activated readily in most cell types while, in contrast, c-Rel complexes (e.g. p50/c-Rel heterodimers and c-Rel homodimers) are found predominantly in cells of haematopoietic lineage [29]. NF-κB/Rel is known not to affect the potency of haematopoietic stem cells to differentiate into common lymphoid and common myeloid cells. Nevertheless, p50/p65 is crucial for lymphopoiesis, whereas p65/c-Rel is complementary for myeloid development [29]. Our supershift studies showed that E. coli infection induced p50/p50 homodimers in CD34+ cells and their cultured cells. Until now, there has been little understanding of the induction of p50/p50 homodimers in CD34+ cells and their cultured cells. A recent study reported that p50/p65 was required for proper development of myeloid dendritic cells [30]. However, loss of p50/c-Rel or c-Rel alone did not perturb dendritic cell development, but rather affected the maturation and survival of dendritic cells [30,31]. These findings thus suggest that p50 is involved in dendritic cell development. Considering that CFU–GM was involved mainly in proinflammatory cytokine expression in E. coli-infected cultured cells derived from CD34+, the activation of p50/p50 homodimer may be due to myeloid lineages of CD34+ stem cells. Further study of the NF-κB subunits in the stage of myeloid development from CD34+ cells seems to be necessary.
In this study, E. coli infection increased the signals of phosphorylated IKKα in the cultured cells derived from CD34+. NEMO is required for the activation of IKK by inflammatory stimuli such as TNF-α [32,33]. Treatment with an NBD peptide which blocks the association of NEMO with the IKK complex decreased proinflammatory cytokine expression in E. coli-infected CD34+ cells and their cultured cells. These findings suggest that transcription of the proinflammatory cytokines in response to E. coli infection is regulated via IKK activation.
In conclusion, our data indicate that the NF-κB activation pathway is involved in E. coli-induced expression of IL-1α, IL-6, IL-8 and TNF-α from CD34+ cells. These proinflammatory cytokines may contribute to the inflammatory process following bacteraemia in transplantation treatment.
Acknowledgments
The experiments comply with the current laws of our country in which the experiments were performed. We thank Dr Martin F. Kagnoff for gifts of standard RNAs, Dr Hee-Young Chung for gifts of retrovirus-IκBα-AA and Joo Hyoung Lee, Su Jin Cho, Hye-Sook Lee, Shin-Jai Kang, Yun-Kyung Lee and Jin-Young Lee for their excellent technical help. This work was supported by a grant from the Korea Science and Engineering Foundation (KOSEF R01-2002-000-00024-0).
References
- 1.Broxmeyer HE, Gluckman E, Auerbach AD, et al. Human umbilical cord blood: a clinically useful source of transplantable hematopoietic stem/progenitor cells. Int J Cell Cloning. 1990;8:76–91. doi: 10.1002/stem.5530080708. [DOI] [PubMed] [Google Scholar]
- 2.Broxmeyer H, Hangoc G, Cooper S, et al. Growth characteristics and expansion of human umbilical cord blood and estimation of its potential for transplantation in adults. Proc Natl Acad Sci USA. 1992;89:4109–13. doi: 10.1073/pnas.89.9.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emerson SG, Sieff CA, Wang EA, et al. Purification of fetal hemopoietic progenitors and demonstration of recombinant multipotential colony-stimulating activity. J Clin Invest. 1985;76:1286–90. doi: 10.1172/JCI112087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saavedra S, Sanz GF, Jarque I, et al. Early infections in adult patients undergoing unrelated donor cord blood transplantation. Bone Marrow Transplant. 2002;30:937–43. doi: 10.1038/sj.bmt.1703764. [DOI] [PubMed] [Google Scholar]
- 5.Mullen CA, Nair J, Sandesh S, et al. Fever and neutropenia in pediatric hematopoietic stem cell transplant patients. Bone Marrow Transplant. 2000;25:59–65. doi: 10.1038/sj.bmt.1702109. [DOI] [PubMed] [Google Scholar]
- 6.Azuma I. Inducer of cytokines in vivo: overview of field and romurtide experience. Int J Immunopharmacol. 1992;14:487–96. doi: 10.1016/0192-0561(92)90180-s. [DOI] [PubMed] [Google Scholar]
- 7.Salazar-Mather TP, Hokeness KL. Calling in the troops: regulation of inflammatory cell trafficking through innate cytokine/chemokine networks. Viral Immunol. 2003;16:291–306. doi: 10.1089/088282403322396109. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki K, Nakaji S, Yamada M, et al. Systemic inflammatory response to exhaustive exercise. Cytokine kinetics. Exerc Immunol Rev. 2002;8:6–48. [PubMed] [Google Scholar]
- 9.Janeway CA, Jr, Travers P, Walport M, et al. Immunobiology. 8. Edinburgh: Churchill Livingstone; 2001. Innate immunity; pp. 35–91. [Google Scholar]
- 10.Arnalich F, Garcia-Palomero E, López J, et al. Predictive value of nuclear factor-κB activity and plasma cytokine levels in patients with sepsis. Infect Immun. 2000;68:1942–5. doi: 10.1128/iai.68.4.1942-1945.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elewaut D, DiDonato JA, Kim JM, et al. NF-kappa B is a central regulator of the intestinal epithelial cell innate immune response induced by infection with enteroinvasive bacteria. J Immunol. 1999;163:1457–66. [PubMed] [Google Scholar]
- 12.Hawiger J. Innate immunity and inflammation: a transcriptional paradigm. Immunol Res. 2001;23:99–109. doi: 10.1385/IR:23:2-3:099. [DOI] [PubMed] [Google Scholar]
- 13.Kim JM, Cho SJ, Oh YK, et al. Nuclear factor-kappa B activation pathway in intestinal epithelial cells is a major regulator of chemokine gene expression and neutrophil migration induced by Bacteroides fragilis enterotoxin. Clin Exp Immunol. 2002;130:59–66. doi: 10.1046/j.1365-2249.2002.01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tato CM, Hunter CA. Host–pathogen interactions: subversion and utilization of the NF-κB pathway during infection. Infect Immun. 2002;70:3311–7. doi: 10.1128/IAI.70.7.3311-3317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baeuerle PA, Henkel T. Function and activation of NF-κB in the immune system. Annu Rev Immunol. 1994;12:141–79. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 16.Baldwin AS. The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–83. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 17.Thanos D, Maniatis T. NF-κB: a lesson in family values. Cell. 1995;80:529–32. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 18.Lee HS, Jang MY, Choi JH, et al. Proliferation, apoptosis and telomerase activity in human cord blood CD34+ cells cultured with various cytokine combinations. J Microbiol Biotechnol. 2003;13:422–8. [Google Scholar]
- 19.May MJ, D’Acquisto F, Madge LA, et al. Selective inhibition of NF-kappaB activation by a peptide that blocks the interaction of NEMO with the IkappaB kinase complex. Science. 2000;289:1550–4. doi: 10.1126/science.289.5484.1550. [DOI] [PubMed] [Google Scholar]
- 20.Jung HC, Eckmann L, Yang SK, et al. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J Clin Invest. 1995;95:55–65. doi: 10.1172/JCI117676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim JM, Oh YK, Kim YJ, et al. Polarized secretion of CXC chemokines by human intestinal epithelial cells in response to Bacteroides fragilis enterotoxin: NF-κB plays a major role in the regulation of IL-8 expression. Clin Exp Immunol. 2001;123:421–7. doi: 10.1046/j.1365-2249.2001.01462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JM, Jung HC, Im K, et al. Synergy between Entamoeba histolytica and Escherichia coli in the induction of cytokine gene expression in human colon epithelial cells. Parasitol Res. 1998;84:509–12. doi: 10.1007/BF03356595. [DOI] [PubMed] [Google Scholar]
- 23.DiDonato J, Mercurio F, Rosette C, et al. Mapping of the inducible IkappaB phosphorylation sites that signal its ubiquitination and degradation. Mol Cell Biol. 1996;16:1295–304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JM, Kim JS, Jung HC, et al. Helicobacter pylori infection activates the NF-κB signaling pathway to induce iNOS and protect human gastric epithelial cells from apoptosis. Am J Physiol-Gastrointest Liver. 2003;285:G1171–80. doi: 10.1152/ajpgi.00502.2002. [DOI] [PubMed] [Google Scholar]
- 25.Ory DS, Neugeboren BA, Mulligan RC. A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc Natl Acad Sci USA. 1996;93:11400–6. doi: 10.1073/pnas.93.21.11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Van Amersfoort ES, Van Berkel TJ, Kuiper J. Receptors, mediators, and mechanisms involved in bacterial sepsis and septic shock. Clin Microbiol Rev. 2003;16:379–414. doi: 10.1128/CMR.16.3.379-414.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waage A, Steinshamn S. Cytokine mediators of septic infections in the normal and granulocytopenic host. Eur J Haematol. 1993;50:243–9. doi: 10.1111/j.1600-0609.1993.tb00156.x. [DOI] [PubMed] [Google Scholar]
- 28.Rollins BJ. Chemokines Blood. 1997;90:909–28. [PubMed] [Google Scholar]
- 29.Liou HC, Hsia CY. Distinctions between c-Rel and other NF-kappaB proteins in immunity and disease. Bioessays. 2003;25:767–80. doi: 10.1002/bies.10306. [DOI] [PubMed] [Google Scholar]
- 30.Ouaaz F, Arron J, Zheng Y, et al. Dendritic cell development and survival require distinct NF-kappaB subunits. Immunity. 2002;16:257–70. doi: 10.1016/s1074-7613(02)00272-8. [DOI] [PubMed] [Google Scholar]
- 31.Grumont R, Hochrein H, O'Keeffe M, et al. c-Rel regulates interleukin 12 p70 expression in CD8 (+) dendritic cells by specifically inducing p35 gene transcription. J Exp Med. 2001;194:1021–32. doi: 10.1084/jem.194.8.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang ED, Wang CY, Xiong Y, et al. A role for NF-kappaB essential modifier/IkappaB kinase-gamma (NEMO/IKKgamma) ubiquitination in the activation of the IkappaB kinase complex by tumor necrosis factor-alpha. J Biol Chem. 2003;278:37297–305. doi: 10.1074/jbc.M303389200. [DOI] [PubMed] [Google Scholar]
- 33.Carter RS, Pennington KN, Ungurait BJ, et al. In vivo identification of inducible phosphoacceptors in the IKKgamma/NEMO subunit of human IkappaB kinase. J Biol Chem. 2003;278:19642–8. doi: 10.1074/jbc.M301705200. [DOI] [PubMed] [Google Scholar]