Abstract
Nitric oxide (NO) produced by the inducible form of nitric oxide synthase (iNOS) has bactericidal and virocidal effects. Although NO synthesis and iNOS expression in macrophages affect several aspects of human immunodeficiency virus (HIV) type-1 pathogenesis, their role in HIV disease remains largely unknown. In humans, the expression of iNOS is influenced by a functional CCTTT-repeat polymorphism in the promoter region of the gene. We investigated the association of this polymorphism with HIV pathogenesis in naive HIV-infected patients before the initiation of antiretroviral therapy. The allele frequencies of the iNOS CCTTT-repeat polymorphism were assessed by PCR in 857 patients from the Swiss HIV Cohort Study, including rapid progressors and long-term nonprogressors, and in 240 healthy volunteers. In HIV-infected patients, the initial viral load and the decline in total CD4 cells was calculated to estimate disease progression. Allele frequencies of the iNOS CCTTT-repeat polymorphism were similar between the HIV-infected and noninfected blood donors. In treatment-naive HIV-positive patients, there was no association of the iNOS polymorphism with viral load or with the course of CD4 cells. Regulation of iNOS expression by the functional CCTTT-polymorphism does not modify HIV pathogenesis.
Keywords: HIV, inducible nitric oxide synthase, polymorphism, pathogenesis
INTRODUCTION
Nitric oxide (NO) is a labile intercellular messenger molecule and an effector molecule in macrophage cytotoxicity [1,2]. NO is produced by a family of enzymes, the NO synthases (NOS). Monocytes contain the constitutively expressed endothelial NOS and the inducible NOS (iNOS) [3]. Expression of iNOS is increased by a variety of stimuli, including microorganisms, pro-inflammatory cytokines and interferon-α and -γ [4]. Up-regulation of iNOS leads to increased NO production, which has bactericidal and virocidal effects in vitro and in vivo [5]. Compared to littermate controls, mice deficient in iNOS or pretreated with NOS inhibitors exhibited higher mortality, greater viral replication, and delayed clearance of virus when challenged with different viruses [6,7].
Because NO mediates other viral infections, it is a tempting potential target for therapeutic intervention in treatment of HIV disease. However, NO's role in HIV infection remains largely unknown. While most in vitro studies concluded that HIV [8] or HIV proteins [9,10] increase iNOS [11] and subsequently NO production, the ultimate effects of increased NO production on HIV replication are controversial. In some studies, NO increased HIV replication in monocyte-derived macrophages (MDM) [12] and caused apoptosis of cells near the HIV-infected MDM [13], potentially contributing to disease progression. In other studies, NO inactivated HIV protease [14], reverse transcriptase [15], and increased MIP-1α in MDM [16], suggesting a protective role for NO in HIV infection.
In vivo studies support the up-regulation of NO by HIV. Compared to noninfected controls, NO production substantially increased in HIV-infected peripheral blood monocytes [17] and sera from patients with advanced HIV infection. Serum levels of NO metabolites correlated with viral load and with activation of mononuclear phagocytes [18,19]. Furthermore, iNOS expression is associated with the development of the AIDS dementia complex [17,20–22] and with HIV-associated cardiomyopathy [23,24].
The gene for human iNOS has a functional CCTTT polymorphism within its promoter region that alters the transcriptional activity of the gene. Thirteen different alleles have been detected in Caucasians. Alleles with eight or nine CCTTT-repeats have lower levels of transcriptional activity and are less inducible by Il-1β than alleles with 12–15 repeats [25]. Thus, alleles with lower repeat numbers likely produce less iNOS and subsequently less NO than alleles with higher repeat numbers.
To investigate the role of NO in HIV pathogenesis, we examined the association of specific CCTTT alleles with HIV infection, progression rate and HIV RNA over time.
PATIENTS AND METHODS
Study participants
Written consent was obtained from all participants according to good clinical practice. We recruited 857 HIV-1-infected patients from the Genetic Core of the Swiss HIV Cohort Study (SHCS). Patients had to be at least 18 years of age. The decline in numbers of CD4 cells was calculated in untreated patients with at least two CD4 measurements separated by more than 1 year. Viral load was determined in untreated patients as the mean of log10 of HIV-1 RNA copies/ml with at least three independent measurements of HIV RNA levels (Roche Amplicor HIV-1 Ultrasensitive Monitor assay, version 1·5) over time before the start of treatment. Two CD4 cell count measurements separated by one year reflect quite precisely CD4 decline. In contrast HIV RNA is more inclined to variability and therefore, we claimed three independent measurements of HIV RNA to calculate the mean of log10 of HIV-RNA. The control group consisted of HIV-negative blood donors (n = 240).
iNOS Genotype
Blood samples supplemented with EDTA were extracted with the QIAmp DNA Mini Kit (QIAGEN AG, Basel, Switzerland). To amplify the region of the iNOS CCTTT repeat polymorphism, 50 ng DNA was added with primer U5553 ACCCCTGGAAGC CTACAACTGCAT and the fluorescently (Joe) labelled primer L5735 GCCACTGCACCCTAGCCTGTCTCA (Microsynth, Balgach, Switzerland) with the Amplitaq Gold System (Applied Biosytems, Foster City, CA, USA) in a total volume of 12·5 µl. PCR products were then analysed on an ABI310 genetic analyser with GeneScan 500 ROX as internal standard. Allele assignment was done with the ABI Genotyper software (all from Applied Biosystems).
Statistical analysis
Data were analysed with the statistical software packages R and SAS 8·1. Genotype frequencies between groups were compared with the chi–square test. Associations between CD4 decline or viral load and iNOS repeat number were analysed by linear regression. Differences between the presence and absence of a iNOS CCTTT 14 repeat allele were analysed using a t-test assuming equal variance. All P-values are two-sided, and P-values below 0·05 were considered statistically significant.
RESULTS
Allelic distribution of functional iNOS CCTTT repeat polymorphism
To investigate the role of the functional CCTTT repeat polymorphism in iNOS on HIV pathogenesis, we analysed the iNOS genotype of 857 HIV-positive patients and of 240 noninfected blood donors (Table 1). We detected 13 different CCTTT-alleles and an allele frequency similar to Caucasians reported in other studies [25,26]. No significant difference in iNOS allele frequencies was observed between the HIV-infected patients and noninfected blood donors.
Table 1.
iNOS Allele frequencies in the study
| iNOS alleles (repeats) | HIV Infected (n = 857) | Blood donors (n = 240) |
|---|---|---|
| 6 | 0·01 | |
| 8 | 0·08 | 0·004 |
| 9 | 0·041 | 0·037 |
| 10 | 0·128 | 0·123 |
| 11 | 0·185 | 0·19 |
| 12 | 0·325 | 0·335 |
| 13 | 0·172 | 0·165 |
| 14 | 0·087 | 0·071 |
| 15 | 0·039 | 0·056 |
| 16 | 0·011 | 0·013 |
| 17 | 0·002 | 0·006 |
| 18 | 0·001 | |
| 19 | 0·001 |
HIV RNA and iNOS CCTTT repeat polymorphism
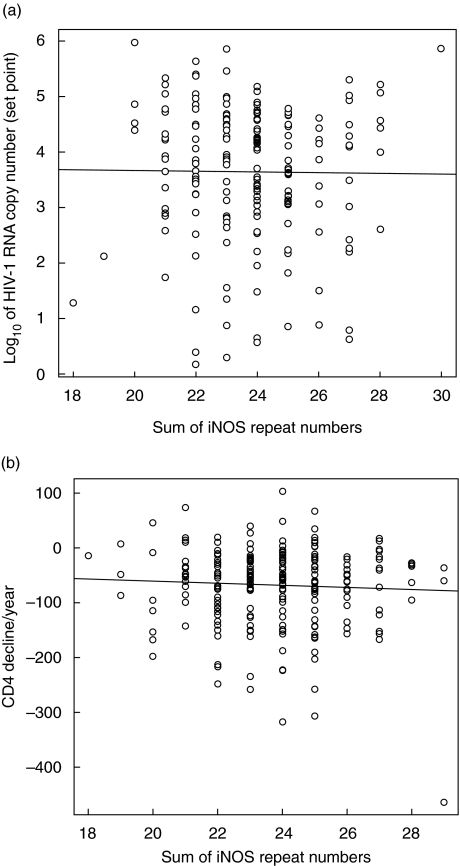
We next analysed the association of iNOS alleles with HIV RNA levels in HIV-positive patients (n = 183). Although there was a wide range in the means of HIV RNA levels in the study group, linear regression revealed no association of the iNOS CCTTT repeat polymorphism with the mean HIV RNA in a dominant (i.e. with the longer of the two alleles for each patient, P = 0·54, data not shown) or in a codominant model (i.e. the sum of the two alleles, P = 0·87, Fig. 1a). The power of the study to detect a difference of 0·2 log10 HIV-1 RNA copies in the dominant and codominant model was 82% and 99%, respectively, suggesting that this repeat polymorphism has no effect on HIV replication in vivo.
Fig. 1.
(a) Log10 of HIV-1 RNA copy numbers in naive patients, according to the iNOS CCTTT genotype in a codominant model (sum of the repeat numbers). (b) Decline of CD4 numbers per year in naive patients, according to the iNOS CCTTT genotype in a codominant model (sum of the repeat numbers).
CD4 decline and iNOS CCTTT repeat polymorphism
To assess the influence of the iNOS polymorphism on HIV disease progression, we analysed the association of the polymorphism with the decline in CD4 cells over time (CD4 decline) in 246 naive patients (Fig. 1b). Since the study included rapid progressors and long-term nonprogressors, a broad variation in CD4 decline over time was observed (−300 to + 100 cells/year). Nevertheless, there was no association between the iNOS CCTTT repeat number and the CD4 decline in a dominant (P = 0·10) or a codominant model (P = 0·44). The power of the study to detect a difference of 10 CD4 cells/µl in the dominant and codominant model was 82% and 99%, respectively. These results suggest that this polymorphism does not influence disease progression rate as defined by the CD4 decline in HIV-infected patients.
Since the iNOS 14 CCTTT repeat allele was previously associated with a variety of disease states, we analysed this allele for an association with HIV disease progression. 34 out of 246 patients included in the CD4 decline analysis had at least one iNOS 14 CCTTT repeat allele. However, no significant association with CD4 decline (P = 0·64) was detected. Similarly, there was no significant difference in viral load (P = 0·13) between the patients having at least one iNOS 14 CCTTT repeat allele (n = 30) and the rest (n = 153).
DISCUSSION
HIV progression rate is influenced by distinct host genetic factors, such as CCR5Δ32 and Rantes (reviewed in [27]). However, these factors explain progression in only a minority of patients. The effect of NO on HIV pathogenesis is controversial but iNOS-dependent NO production is assumed to be a pathogenic factor in malaria [28], dementia [29], and diabetic retinopathy [25]. This perception is based on an association of a CCTTT repeat polymorphism with the outcome in these diseases. Therefore, we hypothesized that the CCTTT repeat polymorphism in the iNOS gene may impact transmission or progression of HIV disease.
We detected all known CCTTT repeat alleles of the iNOS gene among the HIV-positive patients but the distribution of the alleles was similar to that of a control group of HIV-negative volunteers. This implies that HIV transmission is independent of this iNOS polymorphism. When we investigated the effect of the CCTTT repeat polymorphism on disease progression, we also detected no association with either the mean HIV RNA levels or the CD4 decline before treatment assuming a codominant or a dominant model. Thus, the CCTTT repeat polymorphism has also no impact on disease progression and no prognostic significance in HIV pathogenesis
Our findings may be explained in a couple of different ways. First, the lack of an association of this polymorphism with HIV disease progression may be due to variability in NO levels produced by the different iNOS alleles that are too small to be reflected in distinct HIV progression rates. Alternatively, NO may not influence HIV pathogenesis in man. However, this is less likely considering the manifold functions of NO in the immune system. It is also possible that another region of the iNOS gene is more critical for NO production and thus for HIV pathogenesis. Several other single nucleotide polymorphisms have recently been identified in the promoter and the coding region of the iNOS gene which revealed an association between certain iNOS haplotypes and the outcome of hepatitis C infection [30]. We focused on a polymorphism within the iNOS promoter which showed functional consequences in in vitro assays. Definition of transcriptional activity of the iNOS gene due to different iNOS polymorphisms and haplotypes will be critical for dissecting the genetic impact of iNOS on disease in the future.
Acknowledgments
We thank the patients for their commitment and participation. We acknowledge the excellent secretary assistance of Ms C. Vögtli. This work was supported by the Swiss HIV Cohort Study (♯368) and supported by the Swiss National Science Foundation (Grant 3345–062041 and 631–062898).
APPENDIX
The members of the Swiss HIV Cohort Study are:
S. Bachmann, M. Battegay, E. Bernasconi, H. Bucher, Ph. Bürgisser, S. Cattacin, M. Egger, P. Erb, W. Fierz, M. Fischer, M. Flepp (Chairman of the Clinical and Laboratory Committee), P. Francioli (President of the SHCS, Centre Hospitalier Universitaire Vaudois, CH-1011-Lausanne), H.J. Furrer, M. Gorgievski, H. Günthard, B. Hirschel, L. Kaiser, C. Kind, Th. Klimkait, B. Ledergerber, U. Lauper, M. Opravil, G. Pantaleo, L. Perrin, J.-C. Piffaretti, M. Rickenbach (Head of Data Center), C. Rudin (Chairman of the Mother & Child Substudy), J. Schüpbach, R. Speck, A. Telenti, A. Trkola, P. Vernazza (Chairman of the Scientific Board), R. Weber, S. Yerly.
References
- 1.Burgner D, Rockett K, Kwiatkowski D. Nitric oxide and infectious diseases. Arch Dis Child. 1999;81:185–8. doi: 10.1136/adc.81.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Symeonides S, Balk RA. Nitric oxide in the pathogenesis of sepsis. Infect Dis Clin North Am. 1999;13:449–63. doi: 10.1016/s0891-5520(05)70085-4. x. [DOI] [PubMed] [Google Scholar]
- 3.Reiling N, Ulmer AJ, Duchrow M, Ernst M, Flad HD, Hauschildt S. Nitric oxide synthase: mRNA expression of different isoforms in human monocytes/macrophages. Eur J Immunol. 1994;24:1941–4. doi: 10.1002/eji.1830240836. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg JB, Misukonis MA, Shami PJ, et al. Human mononuclear phagocyte inducible nitric oxide synthase (iNOS): analysis of iNOS mRNA, iNOS protein, biopterin, and nitric oxide production by blood monocytes and peritoneal macrophages. Blood. 1995;86:1184–95. [PubMed] [Google Scholar]
- 5.Benz D, Cadet P, Mantione K, Zhu W, Stefano G. Tonal nitric oxide and health. Anti-bacterial and – viral actions and implications for HIV. Med SciMonit. 2002;8:RA27–RA31. [PubMed] [Google Scholar]
- 6.MacLean A, Wei XQ, Huang FP, Al Alem UA, Chan WL, Liew FY. Mice lacking inducible nitric-oxide synthase are more susceptible to herpes simplex virus infection despite enhanced Th1 cell responses. J Generalvirol. 1998;79:825–30. doi: 10.1099/0022-1317-79-4-825. [DOI] [PubMed] [Google Scholar]
- 7.Noda S, Tanaka K, Sawamura S, et al. Role of nitric oxide synthase type 2 in acute infection with murine cytomegalovirus. J Immunol. 2001;166:3533–41. doi: 10.4049/jimmunol.166.5.3533. [DOI] [PubMed] [Google Scholar]
- 8.Hori K, Burd PR, Furuke K, Kutza J, Weih KA, Clouse KA. Human immunodeficiency virus-1-infected macrophages induce inducible nitric oxide synthase and nitric oxide (NO) production in astrocytes: astrocytic NO as a possible mediator of neural damage in acquired immunodeficiency syndrome. Blood. 1999;93:1843–50. [PubMed] [Google Scholar]
- 9.Mansur A, Chu CM, Karnik A, Frieri M. Nitric oxide production and apoptosis by GP120. Allergy Asthma Proc. 2000;21:145–9. doi: 10.2500/108854100778148936. [DOI] [PubMed] [Google Scholar]
- 10.Pietraforte D, Tritarelli E, Testa U, Minetti M. gp120 HIV envelope glycoprotein increases the production of nitric oxide in human monocyte-derived macrophages. J Leukoc Biol. 1994;55:175–82. doi: 10.1002/jlb.55.2.175. [DOI] [PubMed] [Google Scholar]
- 11.Polazzi E, Levi G, Minghetti L. Human immunodeficiency virus type 1 Tat protein stimulates inducible nitric oxide synthase expression and nitric oxide production in microglial cultures. J Neuropathol Exp Neurol. 1999;58:825–31. doi: 10.1097/00005072-199908000-00005. [DOI] [PubMed] [Google Scholar]
- 12.Blond D, Raoul H, Le Grand R, Dormont D. Nitric oxide synthesis enhances human immunodeficiency virus replication in primary human macrophages. J Virol. 2000;74:8904–12. doi: 10.1128/jvi.74.19.8904-8912.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hermann E, Idziorek T, Kusnierz JP, Mouton Y, Capron A, Bahr GM. Role of nitric oxide in the regulation of lymphocyte apoptosis and HIV- 1 replication. Int J Immunopharmacol. 1997;19:387–97. doi: 10.1016/s0192-0561(97)00060-x. [DOI] [PubMed] [Google Scholar]
- 14.Sehajpal PK, Basu A, Ogiste JS, Lander HM. Reversible S-nitrosation and inhibition of HIV-1 protease. Biochemistry. 1999;38:13407–13. doi: 10.1021/bi9912995. [DOI] [PubMed] [Google Scholar]
- 15.Persichini T, Colasanti M, Fraziano M, et al. Nitric oxide inhibits the HIV-1 reverse transcriptase activity. Biochem Biophys Res Commun. 1999;258:624–7. doi: 10.1006/bbrc.1999.0581. [DOI] [PubMed] [Google Scholar]
- 16.Sherry B, Schmidtmayerova H, Zybarth G, Dubrovsky L, Raabe T, Bukrinsky M. Nitric oxide regulates MIP-1alpha expression in primary macrophages and T lymphocytes: implications for anti-HIV-1 response. Mol Med. 2000;6:542–9. [PMC free article] [PubMed] [Google Scholar]
- 17.Bukrinsky MI, Nottet HS, Schmidtmayerova H, et al. Regulation of nitric oxide synthase activity in human immunodeficiency virus type 1 (HIV-1) -infected monocytes: implications for HIV- associated neurological disease. J Exp Med. 1995;181:735–45. doi: 10.1084/jem.181.2.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zangerle R, Fuchs D, Reibnegger G, et al. Serum nitrite plus nitrate in infection with human immunodeficiency virus type-1. Immunobiology. 1995;193:59–70. doi: 10.1016/S0171-2985(11)80155-5. [DOI] [PubMed] [Google Scholar]
- 19.Groeneveld PH, Kroon FP, Nibbering PH, Bruisten SM, van Swieten P, van Furth R. Increased production of nitric oxide correlates with viral load and activation of mononuclear phagocytes in HIV-infected patients. Scand J Infect Dis. 1996;28:341–5. doi: 10.3109/00365549609037916. [DOI] [PubMed] [Google Scholar]
- 20.Adamson DC, Wildemann B, Sasaki M, et al. Immunologic NO synthase: elevation in severe AIDS dementia and induction by HIV-1 gp41. Science. 1996;274:1917–21. doi: 10.1126/science.274.5294.1917. [DOI] [PubMed] [Google Scholar]
- 21.Adamson DC, McArthur JC, Dawson TM, Dawson VL. Rate and severity of HIV-associated dementia (HAD): correlations with Gp41 and iNOS. Mol Med. 1999;5:98–109. [PMC free article] [PubMed] [Google Scholar]
- 22.Rostasy K, Monti L, Yiannoutsos C, et al. NFκB activation, TNF-alpha expression, and apoptosis in the AIDS-Dementia-Complex. J Neurovirol. 2000;6:537–43. doi: 10.3109/13550280009091954. [DOI] [PubMed] [Google Scholar]
- 23.Barbaro G, Di Lorenzo G, Soldini M, et al. Clinical course of cardiomyopathy in HIV-infected patients with or without encephalopathy related to the myocardial expression of tumour necrosis factor-alpha and nitric oxide synthase. GISCA. Gruppo Italiano per lo Studio Cardiologico dei pazienti affetti da AIDS. AIDS. 2000;14:827–38. doi: 10.1097/00002030-200005050-00009. [DOI] [PubMed] [Google Scholar]
- 24.Kan H, Xie Z, Finkel MS. HIV gp120 enhances NO production by cardiac myocytes through p38 MAP kinase-mediated NF-kappaB activation. Am J Physiol Heart Circ Physiol. 2000;279:H3138–43. doi: 10.1152/ajpheart.2000.279.6.H3138. [DOI] [PubMed] [Google Scholar]
- 25.Warpeha KM, Xu W, Liu L, et al. Genotyping and functional analysis of a polymorphic (CCTTT) (n) repeat of NOS2A in diabetic retinopathy. FASEB J. 1999;13:1825–32. doi: 10.1096/fasebj.13.13.1825. [DOI] [PubMed] [Google Scholar]
- 26.Xu W, Humphries S, Tomita M, et al. Survey of the allelic frequency of a NOS2A promoter microsatellite in human populations: assessment of the NOS2A gene and predisposition to infectious disease. Nitric Oxide. 2000;4:379–83. doi: 10.1006/niox.2000.0290. [DOI] [PubMed] [Google Scholar]
- 27.Hogan CM, Hammer SM. Host determinants in HIV infection and disease. Part 2. genetic factors and implications for antiretroviral therapeutics. Ann Intern Med. 2001;134:978–96. doi: 10.7326/0003-4819-134-10-200105150-00012. [DOI] [PubMed] [Google Scholar]
- 28.Burgner D, Xu W, Rockett K, et al. Inducible nitric oxide synthase polymorphism and fatal cerebral malaria. Lancet. 1998;352:1193–4. doi: 10.1016/S0140-6736(05)60531-4. [DOI] [PubMed] [Google Scholar]
- 29.Xu W, Liu L, Emson P, et al. The CCTTT polymorphism in the NOS2A gene is associated with dementia with Lewy bodies. Neuroreport. 2000;11:297–9. doi: 10.1097/00001756-200002070-00015. [DOI] [PubMed] [Google Scholar]
- 30.Yee LJ, Knapp S, Burgner D, et al. Inducible nitric oxide synthase gene (NOS2A) haplotypes and the outcome of hepatitis C virus infection. Genes Immun. 2004;5:183–7. doi: 10.1038/sj.gene.6364054. [DOI] [PubMed] [Google Scholar]