Abstract
The C1/RIPE3b1 (−118/−107 bp) binding factor regulates pancreatic-β-cell-specific and glucose-regulated transcription of the insulin gene. In the present study, the C1/RIPE3b1 activator from mouse βTC-3 cell nuclear extracts was purified by DNA affinity chromatography and two-dimensional gel electrophoresis. C1/RIPE3b1 binding activity was found in the roughly 46-kDa fraction at pH 7.0 and pH 4.5, and each contained N- and C-terminal peptides to mouse MafA as determined by peptide mass mapping and tandem spectrometry. MafA was detected in the C1/RIPE3b1 binding complex by using MafA peptide-specific antisera. In addition, MafA was shown to bind within the enhancer region (−340/−91 bp) of the endogenous insulin gene in βTC-3 cells in the chromatin immunoprecipitation assay. These results strongly suggested that MafA was the β-cell-enriched component of the RIPE3b1 activator. However, reverse transcription-PCR analysis demonstrated that mouse islets express not only MafA but also other members of the large Maf family, specifically c-Maf and MafB. Furthermore, immunohistochemical studies revealed that at least MafA and MafB were present within the nuclei of islet β cells and not within pancreas acinar cells. Because MafA, MafB, and c-Maf were each capable of specifically binding to and activating insulin C1 element-mediated expression, our results suggest that all of these factors play a role in islet β-cell function.
Insulin is an essential regulator of metabolism. This hormone, which is synthesized by the β cells of the islets of Langerhans, increases the storage of glucose, fatty acids, and amino acids through its actions in liver, adipose tissue, and muscle. Experiments performed in vivo with transgenic animals have established that the cis-acting elements controlling β-cell-selective expression are located within the insulin enhancer region, which is found between nucleotides −340 and −91 relative to the transcription start site. Several key control elements within the enhancer have been identified, including C2 (−317/−311 bp), A3 (−201/−196 bp), C1 (−118/−107 bp), and E1 (−100/−91 bp) (37, 60, 67). Mutations that decrease the binding affinity of the A3, C1, and E1 activators also reduce glucose-regulated transcription (37, 60, 67).
The activator of insulin C2-element stimulated transcription is Pax6 (61). Proteins in the Pax family all contain a paired box bipartite DNA-binding domain, although Pax6 also has a homeodomain. The Pdx-1 homeodomain protein (formerly known as IPF-1, STF-1, and IDX-1) is the regulator of A3 element-activated expression (46, 48, 49, 50), whereas the E1 activator is a heterodimer composed of proteins in the basic helix-loop-helix family that are enriched in islets (i.e., BETA2 [42]) and generally distributed (i.e., HEB [51] and E2A [2, 10, 17, 65]). In the adult pancreas, Pax6 (61) and BETA2 (42) are found in all islet cell types, whereas Pdx-1 appears to be found only in β cells, a subset of islet δ cells (48, 49), and exocrine acinar cells (71, 80). These transcription factors are necessary for maintaining physiologically functional islet cells, since Pax6, Pdx-1, and BETA2 contribute to insulin, β-glucokinase (39, 76, 77), islet amyloid polypeptide (6, 7, 61, 64, 75), glucose transporter type 2 (74), glucagon (12), and somatostatin (61) gene expression. Significantly, dysfunctional mutations in Pdx-1 (68), BETA2 (32), and Pax6 (81) impact on the development of diabetes in humans, presumably due to reduced expression of their target genes required in glucose sensing. In addition, each of these factors plays a critical role in islet cell development during embryogenesis (13, 16, 60). Collectively, the data strongly implicate the islet-enriched transcription factors of the insulin gene in processes important in both the formation and maintenance of physiologically functional islet cells.
In contrast to the Pdx-1, Pax-6, and BETA2 activators, little is known about the protein(s) involved in insulin C1-mediated stimulation. Two distinct factor-DNA complexes are formed with the C1 probe, termed RIPE3b1 and RIPE3b2 (65). The RIPE3b1 gel mobility complex is found exclusively in β-cell nuclear extracts (65, 82), whereas RIPE3b2-binding activity is present in a variety of cells types (65). The RIPE3b2 complex is composed of at least three subunits: p58, p62, and p110 (66). The p110 subunit of RIPE3b2 is Smbp-2, a protein with potential helicase motifs and a transcription activation domain (66). The RIPE3b2 activator is not believed to contribute to β-cell-specific expression of the insulin gene (65, 66). However, the enriched presence of RIPE3b1-binding activity in β cells and the ability of glucose to regulate this activity strongly suggest a significant role in controlling insulin C1-directed activation. The DNA-binding component of the RIPE3b1 activator was recently shown to be a 46-kDa protein, whose binding activity is likely induced by tyrosine phosphorylation (36, 82).
In the present study, the β-cell-enriched 46-kDa subunit of the RIPE3b1 activator was biochemically isolated from a mouse β-cell line nuclear extract and identified by mass spectrometry. Our results revealed that MafA, a member of the large Maf family of developmental regulators, was a DNA-binding component of this βTC-3 cell line activator. Interestingly, reverse transcription-PCR (RT-PCR) analysis detected not only MafA expression within islets but also the expression of two other large Mafs: c-Maf and MafB. In addition, immunohistochemical studies revealed the presence of MafA and MafB within the nuclei of islet β cells and not pancreatic acinar cells. Since MafA, MafB, and c-Maf had similar DNA-binding and activation characteristics, our data indicate that these factors may influence both the developmental and metabolic state of the β cell.
MATERIALS AND METHODS
Cell culture and nuclear extract preparation.
Monolayer cultures of pancreatic islet (β:βTC-3 [14], MIN-6 [38], and αTC-6 [20]) and non-islet (HeLa [78]) cell lines were grown under conditions described previously. Human islets were provided by the Juvenile Diabetes Research Foundation Distribution Program at the Washington University and were cultured in CMRL medium (Gibco-BRL, Gaithersburg, Md.) with 10% heat-inactivated fetal calf serum. Human islet and cell line nuclear extracts were prepared by the procedure described by Schreiber et al. (63), except that 1 mM phenylmethylsulfonyl fluoride (PMSF) was included in the high-salt nuclear resuspension buffer.
DNA affinity chromatography.
The oligonucleotide trapping method (15) was modified for use with double-stranded insulin C1 DNA (InsC1 [−126TGGAAACTGCAGCTTCAGCCCCTCTG−101]) containing a single GTGTGTGTGT tail, termed C1:(GT)5. βTC-3 nuclear extract (600 mg) dialyzed in gel shift binding buffer (10 mM Tris-Cl [pH 7.4], 1.0 mM EDTA, 10% glycerol, 150 mM NaCl, 1 mM dithiothreitol [DTT], 1 mM PMSF) was mixed with C1:(GT)5 (60 pmol/mg of protein), poly(dI-dC) (20 μg/mg of protein), and protease inhibitor cocktail (one tablet/50 ml of sample; Roche-Mannheim) and then rocked for 15 min at 4°C. The complexes were applied to a Sepharose 4B column containing covalently linked single-stranded (AC)5, and washed with binding buffer. The RIPE3b1 activator binding complex was eluted in a 0.15 to 1.5 M NaCl gradient, dialyzed in binding buffer, and reapplied to the (AC)5 column. C1:(GT)5 and poly(dI-dC) (10 μg/mg of protein) was added to the column flowthrough, and the DNA affinity step was repeated. RIPE3b1 activator purification was monitored in the InsC1 electrophoretic mobility shift assay.
Two-dimensional (2-D) gel electrophoresis and Southwestern blotting.
The DNA affinity-purified RIPE3b1 binding fractions and βTC-3 nuclear extract (300 μg) were dialyzed in no-salt buffer (20 mM Tris-Cl [pH 7.9], 1.0 mM EDTA, 1.0 mM EGTA, 10% glycerol, 1.0 mM DTT, 1.0 mM PMSF) and precipitated with 20% trichloroacetic acid. The pellet was resuspended in rehydration buffer (8.0 M urea, 4.0% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 2.0 mM tributyl phosphine, 0.2% [wt/vol] Biolytes 3/10) and run at 85,000 V · h on a pH 3 to 10 linear gradient IPG strip (17 cm; Bio-Rad Laboratories, Hercules, Calif.). The electrofocused proteins were resolved in the second dimension by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 10% polyacrylamide gel run at 200 V. For detection of InsC1-protein binding by Southwestern analysis, the 2-D resolved proteins were transferred onto a nitrocellulose membrane (Bio-Rad) at 200 mA in TG buffer (20 mM Tris-Cl [pH 8.0], 150 mM glycine). The proteins were then gently rocked first in renaturation buffer (10 mM Tris-Cl [pH 7.4], 1.0 mM EDTA, 100 mM NaCl, 10% glycerol, 2.0 mM DTT, 1 mM PMSF, 1.0% Triton, 1.0 mM sodium orthovanadate) for 30 min at room temperature, then in binding buffer plus β nuclear extract (10 mM Tris-Cl [pH 7.4], 1.0 mM EDTA, 100 mM NaCl, 10% glycerol, 2.0 mM DTT, 1 mM PMSF, 1.0% Triton, 10.0 mM sodium orthovanadate plus βTC-3 or MIN6 nuclear extract [500 μg/ml of binding buffer]) for 30 min at room temperature, and finally in hybridization buffer (10 mM Tris-Cl [pH 7.4], 1.0 mM EDTA, 100 mM NaCl, 10% glycerol, 2.0 mM DTT, 1 mM PMSF, 1.0 mM sodium orthovanadate) for 5 min at 4°C. The membrane was incubated with the InsC1 probe in hybridization buffer overnight at 4°C in the presence of poly(dI-dC) (5 μg/ml). The hybridized membrane was washed three times with hybridization buffer (for 10 min each time), dried, and subjected to autoradiography.
SDS-PAGE fractionation.
βTC-3 nuclear proteins resolved by 2-D gel electrophoresis were eluted from the gel after the SDS-PAGE step. The approximately 46-kDa range fraction was cut into slices spanning the pH 3 to 10 gradient. The proteins were eluted from the crushed gel in renaturation buffer (plus 100 μg of bovine serum albumin/ml) and analyzed for InsC1 binding activity in the electrophoretic mobility shift assay.
Protein identification by mass spectrometry.
Proteins were separated by 2-D gel electrophoresis and visualized by using the Colloidal Blue staining kit (Invitrogen, Carlsbad, Calif.). The 46-kDa protein spots at pH 7 and pH 4.5 were excised, equilibrated in 100 mM NH4HCO3, reduced with DTT (3 mM in 100 mM NH4HCO3 at 37°C for 15 min), and alkylated with iodoacetamide (6 mM in 100 mM NH4HCO3 for 15 min). The gel slice was then dehydrated with acetonitrile and rehydrated with 15 μl of 25 mM NH4HCO3 containing 0.01 μg of modified trypsin (Promega)/μl, and trypsin digestion was carried out for 2 h at 37°C. Peptides were extracted with 60% acetonitrile-0.1% trifluoroacetic acid, dried by vacuum centrifugation, and reconstituted in 10 μl of 0.1% trifluoroacetic acid. The peptides were then desalted and concentrated into 2 μl of 60% acetonitrile-0.1% trifluoroacetic acid by using ZipTipC18 pipette tips (Millipore), and then 0.4 μl was applied to a target plate and overlaid with 0.4 μl of α-cyano-4-hydroxycinnamic acid matrix. Matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry and TOF/TOF tandem mass spectrometry was carried out by using a Voyager 4700 mass spectrometer (Applied Biosystems, Foster City, Calif.) operated in reflectron mode. For intact peptide mass analysis, the mass spectrum was calibrated to within 20 ppm by using trypsin autolytic peptides present in the sample (m/z = 842.50, 1,045.56, and 2,211.09 Da). Ions ([M+H]) corresponding to peptide masses were entered into the mouse expressed sequence tag (EST) database, which was searched by using the MS-FIT algorithm (prospector.ucsf.edu). MafA-related peptides were confirmed by MALDI-TOF/TOF tandem mass spectrometry.
Electrophoretic mobility shift assay.
Binding reactions (20 μl of total volume) were conducted at 4°C for 30 min with nuclear extract protein, InsC1 probe (1 ng, 10−5 cpm) in binding buffer plus 1 μg of poly(dI-dC). The fractionated proteins were analyzed in the absence of poly(dI-dC). The conditions for the competition analyses were the same, except that excess of the specific competitor DNA was included in the mixture prior to addition of extract. MafA, MafB (P-20; Santa Cruz Biotechnology, Inc.), c-Maf (Maf#153; Santa Cruz Biotechnology), and c-Maf (N-15; Santa Cruz Biotechnology) antibodies were preincubated with extract protein for 15 min prior to the addition of the DNA probe. Each of the large Maf antisera recognizes a specific family member, except c-Maf M-153, which cross-reacts with both mammalian MafA and MafB (see Fig. 4C). The mouse MafA antiserum was raised to a C-terminal region peptide (332AGGAGFPREPS342) at Bethyl Laboratories (Montgomery, Tex.). The InsC1-protein complexes were resolved on a 6% nondenaturing polyacrylamide gel (acrylamide/bisacrylamide ratio of 29:1) and run in TGE buffer (50 mM Tris, 380 mM glycine, 2 mM EDTA [pH 8.5]). The gel was dried and subjected to autoradiography.
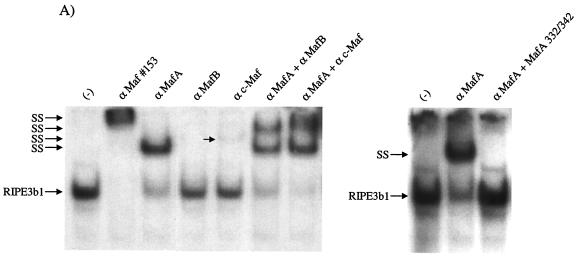
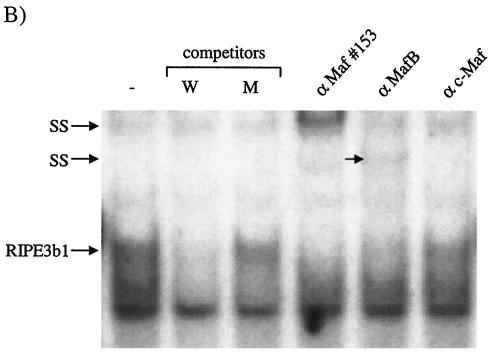
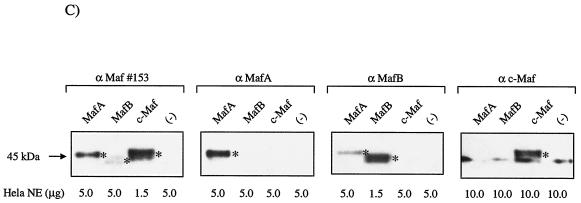
FIG. 4.
MafA is found in the βTC-3 and human islet InsC1/RIPE3b1 activator complex. Gel shift binding to the InsC1 probe was conducted with βTC-3 (A) or human islet (B) nuclear extract in the absence (−) or presence of αMafA, αlarge Maf (Maf#153), αMafB, and/or αc-Maf antibody. The arrows represent the supershifted (SS) complexes. The more broadly recognizing Maf#153 antisera (see panel C) completely altered RIPE3b1 mobility, whereas αMafA affected a portion of the (A) βTC-3 nuclear extract activity. The αMafA supershift in panel A was blocked by the addition of the antigenic MafA332/342 peptide. αc-Maf and αMafB also affected RIPE3b1 in βTC-3 (A) or human islet (B) nuclear extract, respectively. The RIPE3b1 complex in panel B was identified by competition with wild-type InsC1 (lane W) and the −111/−108 bp binding mutant (lane M). (C) Nuclear extracts from MafA-, MafB-, c-Maf-, and pcDNA3.1 [lanes (−)]-transfected HeLa cells were analyzed with αMafA, αMafB, αc-Maf, and/or αlarge Maf antisera by Western analysis. The asterisk denotes the location of the large Maf product.
Immunoblot analysis.
Large Maf expression was analyzed by Western blotting with antisera specific to MafA (1:10,000 dilution), c-Maf (1:2,000), and MafB (1:25,000), as well as a more broadly recognizing one, termed c-Maf M-153 (1:10,000). Nuclear proteins were fractionated by SDS-10% PAGE, transferred to nitrocellulose, and probed with antibody. Maf antibody binding was detected by using horseradish peroxidase coupled to goat anti-rabbit immunoglobulin G (IgG) or donkey anti-goat IgG antibody (1:10,000 dilution). The antibody complex was visualized by incubation with the Lumi-Light Western blotting substrate (Roche-Mannheim, Mannheim, Germany).
Preparation of expression plasmids and transient transfections.
The mouse MafA cDNA was isolated from βTC-3 total RNA by RT-PCR by using the One-Step RT-PCR kit (Clontech, Palo Alto, Calif.). The primers sets were designed based upon the sequence of the mouse MafA 5′ (forward [ATGGCCGCGGAGCTGGCGATGG]; accession no. BB646062) and 3′ (reverse [TCAGAAAGAAGTCGGGT]; accession no. BG798952) ESTs. MafA cDNA se-quences were subcloned into the polylinker of the cytomegalovirus (CMV) en-hancer-driven expression vector, pcDNA3.1/Zeo(+) (Invitrogen, San Diego, Calif.). Mouse MafB and c-Maf cDNA sequences were obtained by PCR from the SkmuMafB and RcRSVcMaf plasmids and subcloned into pcDNA3.1/Zeo(+). InsC1 mediated activation was assayed from rat insulin II-driven firefly luciferase expression constructs that contained either wild-type sequences from bp −238 to +2 (82) or the InsC1 −111/−108 mutant. The −111/−108 mutant (5′-TGGAAACTGCAGCTTACTACCCTCTG-3′; the mutated nucleotides are underlined) was constructed by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). Enzyme restriction and DNA sequencing analyses was used to determine the correctness of each construct.
The Lipofectamine reagent (Gibco-BRL) was used to introduce plasmids containing a large Maf (0.5 μg), −238 luciferase (0.5 μg), and TK-Renilla luciferase (phRL-TK; 10 ng) into HeLa cells. The luciferase activity from phRL-TK served as an internal transfection control. The preparation of cell extracts and luciferase measurements were carried out 40 to 48 h after transfection according to the manufacturer's protocol (Promega, Madison, Wis.). Each experiment was carried out more than three times with at least two independently isolated DNA preparations.
RT-PCR analysis.
Total cellular RNA was isolated from hand-picked mouse islets, βTC-3 cells, and αTC-6 cells by using the Trizol reagent (Gibco-BRL) and treated with the MessageClean kit to remove DNA (GenHunter Corp., Nashville, Tenn.). The One-Step RT-PCR kit was used to detect large Maf RNA expression. Islet (100 ng), βTC-3 (500 ng), and αTC-6 (500 ng) RNAs were reverse transcribed with 45 pmol of oligo(dT) primer at 50°C for 60 min and then PCR amplified for 32 cycles with 20 pmol of the 5′ and 3′ MafA primers (94°C for 45 s, 62°C for 45 s [MafA], 59°C for 45 s [MafB], 58°C for 45 s [c-Maf], and 72°C for 1 min). The mouse large Maf primer sets were as follows: MafA (numbering relative to ATG; forward, +485AGGCCTTCCGGGGTCAGAG; reverse, +887TGGAGCTGGCACTTCTCGCT; 403-bp product), MafB (forward, +541CAACAGCTACCCACTAGCCA; reverse, +906GGCGAGTTTCTCGCACTTGA; 366 bp), and c-Maf (forward, +919GTGCAGCAGAGACACGTCCT-3′; reverse, +1190CAACTAGCAAGCCCACTC; 272 bp). Quantitative competitive RT-PCR was conducted with islet RNA (200 ng) by adding various amounts of large Maf competitor DNA (1, 0.10, and 0.01 fmol) to RT-PCRs performed with 45 pmol of the primer sets described above and a competitor primer. Competitor DNA was synthesized by PCR from MafApcDNA3.1, SkmuMafB, and RcRSVcMaf expression plasmids. The sequences of the competitor primers were as follows: MafA (forward competitor, +575ATCATCACTCTGCCCACCAT; 312-bp competitor product), MafB (reverse competitor, +787TCTGCTGGACGCGTTTATAC; 246-bp competitor product), and c-Maf (reverse competitor, +1125CGCGTGTCACACTCACATGA; 206-bp competitor product). The prod-ucts were resolved on a 1.5% agarose gel run in TAE buffer (40 mM Tris-acetate, 1 mM EDTA) and visualized by ethidium bromide staining. The correctness of the amplified products was determined by diagnostic restriction-enzyme digestion and DNA sequencing.
Chromatin immunoprecipitation analysis.
βTC-3 cells (∼108) were formaldehyde cross-linked, and the sonicated chromatin-DNA complexes were isolated as described previously (19). αMafA (10 μg), normal rabbit IgG (10 μg; Santa Cruz Biotechnology), or no antibody was added to the sonicated chromatin. Antibody-protein-DNA complexes were isolated by incubation with A/G-agarose (Santa Cruz Biotechnology). PCR was performed on one-tenth of the purified immunoprecipitated DNA by using Ready-to-Go PCR beads (Amersham Pharmacia Biotech, Piscataway, N.J.) and 15 pmol of each primer. The primers used for amplifying mouse I and II insulin gene sequences were located at −378 (5′-GGAACTGTGAAACAGTCCAAGG-3′) and −46 (5′-CCCCCTGGACTTTGCTGTTTG), and mouse phosphoenolpyruvate carboxykinase (PEPCK) was located at −434 (5′-GAGTGACACCTCACAGCTGTGG-3′) and −96 (5′-GGCAGGCCTTTGGATCATAGCC-3′). The PCR cycling parameters were 1 cycle of 95°C for 2 min and 28 cycles of 95°C for 30 s, 61°C for 30 s, and 72°C for 30 s. Amplified products were electrophoresed through a 1.5% agarose gel in TAE buffer and then visualized by ethidium bromide staining.
Immunohistochemistry.
Pancreata from 6- to 8-week-old mice were fixed for 4 to 5 h in 4% paraformaldehyde 4°C, washed, dehydrated, and embedded in paraffin, and then 5-μm sections were mounted on glass slides. Double immunofluorescence was performed by using guinea pig anti-human insulin (Linco), guinea pig anti-glucagon (Linco), sheep α-somatostatin (American Research Products), guinea pig anti-pancreatic polypeptide (Linco), rabbit anti-MafA, and goat anti-human MafB (P-20) primary antibodies at dilutions of 1:2,000, 1:2,000, 1:2,000, 1:2,000, 1:1,500, and 1:8,000, respectively. Secondary antibodies were Cy3- or Cy5-labeled donkey anti-guinea pig, anti-rabbit, and anti-goat antibodies diluted 1:500 (Jackson ImmunoResearch Laboratories). Nuclear counterstaining was performed with the monomeric cyanine YoPro1 nucleic acid stain (Molecular Probes). Fluorescent images were captured on a Zeiss LSM510 confocal microscope at an optical depth of 1 μm.
RESULTS
RIPE3b1 binding activity is detected at pH 4.5 and pH 7.
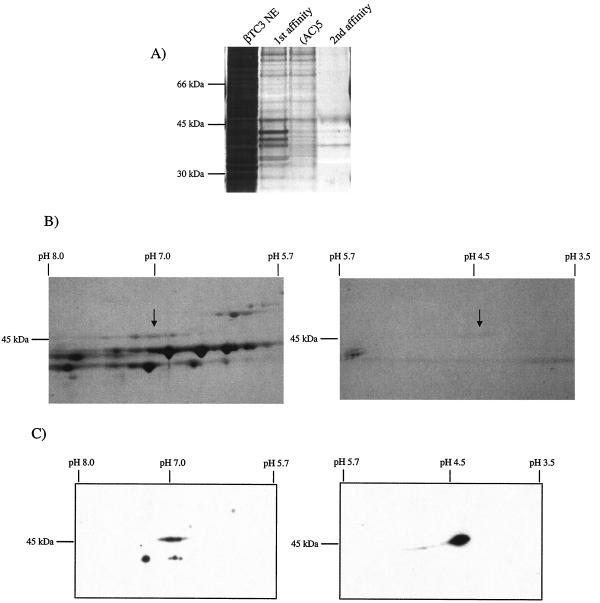
A Southwestern blotting strategy was developed to determine the isoelectric point of the 46-kDa RIPE3b1 binding protein. βTC-3 nuclear extracts were separated by using a linear pH 3 to 10 gradient in the first dimension and then a SDS-10% PAGE step. The 2-D resolved proteins were transferred to a nitrocellulose membrane and InsC1 binding to them analyzed in the presence β-cell nuclear extract. Binding was found at two distinct locations, centered at pH 7 and pH 4.5 (Fig. 1A). The pattern was also observed with 2-D resolved MIN6 β nuclear proteins but not with non-β cells (data not shown). In addition, binding was not detected at pH 7 or pH 4.5 with a binding defective InsC1 probe (data not shown).
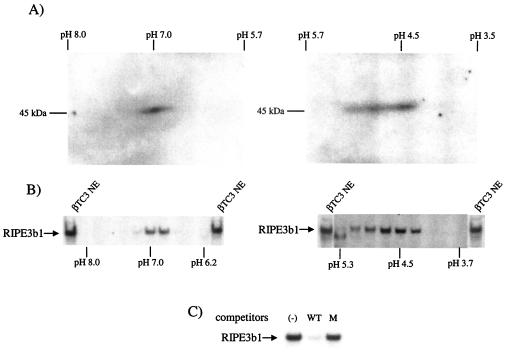
FIG. 1.
InsC1/RIPE3b1 binding activity localized to pH 7 and pH 4.5 after 2-D electrophoresis. (A) Southwestern analysis was performed by using 2-D electrophoresis resolved βTC-3 nuclear extract (NE). InsC1 binding of a 46-kDa protein(s) was found at roughly pH 7 and pH 4.5. (B) The proteins within the 46-kDa range were eluted from 2-D slices representing different pH ranges, renatured, and tested for InsC1 binding in the gel shift assay. The arrow marks the mobility of the RIPE3b1 complex in unfractionated, control βTC-3 nuclear extract (NE). (C) Gel shift reactions were conducted with the pH 7 fraction obtained from 2-D resolved βTC-3 nuclear extract. A 10-fold molar excess of unlabeled wild-type and RIPE3b1 binding-defective −111/−108 InsC1 competitor was used in gel shift reactions.
To further examine the specificity of the InsC1 binding activity observed after 2-D electrophoresis, the 46-kDa part of the gel was cut into slices to represent distinct pH ranges. The separated proteins were then eluted from the gel, renatured, and tested for InsC1 binding in the gel shift assay. The RIPE3b1 binding activity in crude βTC-3 nuclear extracts comigrated with the pH 7 and pH 4.5 activities detected by Southwestern analysis (Fig. 1B). These 2-D complexes were unable to bind to RIPE3b1/InsC1 mutant competitors (Fig. 1C; data not shown). These results suggested that the 46-kDa DNA-binding subunit of the RIPE3b1 activator was composed of a protein(s) with isoelectric points at pH 4.5 and 7.
Mass spectrometry identification of a RIPE3b1/InsC1 binding protein.
A combined DNA affinity chromatography and 2-D gel electrophoresis strategy was used to obtain the pH 4.5 and 7 InsC1 binding fractions for mass spectrometry analysis. The oligonucleotide trapping method was used in the DNA affinity chromatography step, which involved performing the RIPE3b1 activator gel shift reaction with an InsC1 element binding probe containing a (TG)5 tail. The activator binding complex was then applied to a Sepharose column containing the cross-linked and complementary single-stranded (AC)5 oligonucleotide and eluted in a linear NaCl gradient. RIPE3b1 activator binding activity eluted between 500 and 700 mM NaCl (data not shown). After the first DNA affinity purification step, the pooled RIPE3b1 activity fractions were applied directly to the (AC)5 column to remove additional contaminating proteins, and then a second oligonucleotide trapping enrichment step was performed. The specific binding activity and relative level of the roughly 46-kDa protein(s) band (Fig. 2A) was significantly enriched after each chromatography step, with an overall purification of ca. 2,000-fold. The pooled RIPE3b1 binding fractions were next subjected to 2-D gel electrophoresis, and the identity of the 46-kDa colloidal blue staining proteins at pH 7 and 4.5 (Fig. 2B) was determined by MALDI-TOF analysis. Eight tryptic peptides corresponding to the A member of the large Maf family of basic leucine zipper DNA-binding proteins (Fig. 3) were found in the pH 7 and pH 4.5 samples. In addition, the A+U-rich RNA-binding-1 protein (AUF1) was found in both. The MafA specific peptides were confirmed by MALDI-TOF tandem mass spectrometry (data not shown). (The mammalian form of MafA had not been isolated previously, although the ortholog in chicken [45] and quail [3] had been termed l-Maf and MafA, respectively. This protein will only be referred to as MafA.)
FIG. 2.
Biochemical isolation of the InsC1/RIPE3b1 DNA-binding subunit. (A) InsC1 affinity chromatography was performed on βTC-3 nuclear extract (NE). The protein composition of each step was determined by SDS-PAGE and silver staining. First affinity, InsC1-(AC)5 Sepharose chromatography; (AC)5, Sepharose chromatography alone; second affinity, InsC1-(AC)5 Sepharose chromatography. (B) The pooled InsC1/RIPE3b1 binding fractions from the second affinity step were subjected to 2-D electrophoresis. The Coomassie blue-stained spots at pH 7.0 and 4.5 (arrow) of approximately 46-kDa were subjected to MALDI-TOF mass spectrometry. (C) βTC-3 nuclear extract (NE) was resolved by 2-D electrophoresis and probed by Western analysis with αMafA.
FIG. 3.
Alignment of mouse MafA with other large Maf family members. The amino acid identity between mouse MafA, mouse MafB, mouse c-Maf, and mouse NRL is depicted by the gray shading. The underlined amino acids represent the eight tryptic peptides identified by mass spectrometry. All of these peptides were found in the pH 7 and pH 4.5 samples. Amino acids 232 to 324 of mouse MafA span the conserved basic leucine-zipper region involved in DNA binding and dimerization, whereas the less-characterized S/T-rich activation domain is likely found between amino acids 1 and 75 (4, 73).
MafA is a component of the InsC1/RIPE3b1 activator complex.
To determine whether MafA and/or AUF1 was a part of the RIPE3b1 activator complex in βTC-3 and human islet nuclear extracts, the effect of MafA- and AUF1-recognizing antisera were analyzed in the gel mobility shift assay. Two distinct large Maf antisera were used, one raised to a unique C-terminal epitope in MafA (termed αMafA) and the other raised to an N-terminal region of c-Maf antigenically shared with other large Maf proteins (antibody Maf#153). Upon comparing the Western immunoreactivity of these antisera to various large Mafs, αMafA only recognized MafA, whereas Maf#153 effectively detected both c-Maf and MafA, as well as MafB to a lesser extent (Fig. 4C). Incubation with either of these MafA recognizing antisera specifically “supershifted” the RIPE3b1 complex in βTC-3 (Fig. 4A) and human islet (Fig. 4B) extracts. (The human islet RIPE3b1 complex was identified in Fig. 4B by wild-type and binding defective InsC1 competition analysis.) Interestingly, Maf#153 completely changed the mobility of the RIPE3b1 complex, whereas αMafA did not. In addition, providing more αMafA to the gel shift reaction had no further influence on the level of shifted complex (data not shown). These antibody responses appear to be specific to the large Maf proteins, as judged by the ability of the antigenic MafA332/342 peptide to block αMafA activity and the selective effect on RIPE3b1 mobility, and not RIPE3b2, in islet extracts (Fig. 4). Furthermore, MafA immunoreactivity was detected at pH 7 and 4.5 in 2-D electrophoresis fractionated βTC-3 extracts with the αMafA antibody (Fig. 2C). In contrast, neither αAUF1 nor preimmune serum had an effect on RIPE3b1 formation (data not shown).
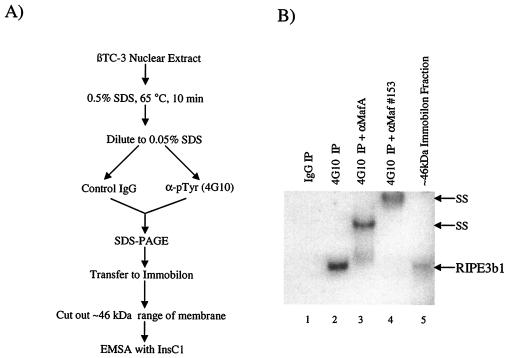
RIPE3b1 binding activity can be immunoprecipitated from β nuclear extracts with the anti-phosphotyrosine immunospecific monoclonal antibody 4G10 (36). This property is consistent with the observed inhibition of binding by the actions of a β cell or exogenously added tyrosine phosphatase (36). To test whether MafA was present in the 46-kDa fraction immunoprecipitated from βTC-3 nuclear extracts with 4G10, this molecular mass range sample was incubated with αMafA and Maf#153 antisera in a gel shift reaction. Both antisera supershifted the 46-kDa RIPE3b1 complex in an manner identical to that observed in crude nuclear extracts (compare the results with αMafA in Fig. 5B and 4A). Collectively, these results strongly suggest that MafA, and possibly another antigenically related large Maf protein, was present in the RIPE3b1 activator complex found in both human and mouse β cells.
FIG. 5.
MafA is present in the RIPE3b1 activator complex immunoprecipitated by anti-phosphotyrosine antisera. (A) Schematic of the antiphosphotyrosine antibody immunoprecipitation procedure. (B) βTC-3 nuclear extract was immunoprecipitated with normal IgG (lane 1) or antiphosphotyrosine 4G10 monoclonal antibody (lanes 2 to 4). The immunoprecipitated material was washed, and the released proteins were electrotransferred from an SDS-PAGE gel onto an Immobilon polyvinylidene difluoride membrane. The proteins eluted from the roughly 46-kDa membrane slice were assayed for InsC1 binding activity in the absence or presence of αMafA (lane 3) or Maf#153 (lane 4) antibodies. The RIPE3b1 complex eluted directly after SDS-PAGE and Immobilon fractionation is also shown (lane 5). These results are representative of an experiment repeated on several separate occasions.
MafA binds to the insulin enhancer region in vivo.
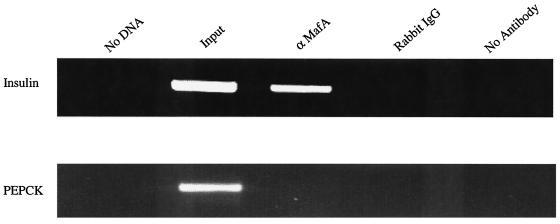
To directly determine whether MafA binds within the enhancer region of the endogenous insulin gene, a chromatin immunoprecipitation assay was performed with formaldehyde cross-linked chromatin from βTC-3 cells. Insulin 5′-flanking control sequences (nucleotides −378 to −46) were selectively amplified by PCR from chromatin precipitated by αMafA but not from chromatin treated with normal rabbit IgG or in the absence of antiserum (Fig. 6). In contrast, the MafA antisera did not immunoprecipitate the control sequences from the PEPCK gene (nucleotides −434 to −96), which is not expressed in β cells (Fig. 6). Importantly, these results demonstrate that MafA is physically associated with the region spanning InsC1 (bp −118 to −107) in the endogenous insulin gene in βTC-3 cells.
FIG. 6.
MafA binds within the enhancer region of the endogenous insulin gene. The cross-linked DNA immunoprecipitated from βTC-3 cells with αMafA antibody was analyzed by PCR with insulin (−378/−46) and PEPCK control region-specific primers (lane 3). As controls, PCRs were run with no DNA (lane 1), on input chromatin (1:100 dilution, lane 2), or with DNA obtained after precipitation with rabbit IgG (lane 4) or no antibody (lane 5). The same pattern was obtained with the Maf#153 antisera (data not shown).
Pancreatic islets express MafA and two other large Mafs, c-Maf and MafB.
Transcription factors of the large Maf family, which includes MafA, c-Maf, MafB, and NRL, contain a conserved basic region leucine-zipper domain that is responsible for their dimerization and DNA-binding properties, and an N-terminal transactivation domain that is rich in proline, serine, and threonine residues (Fig. 3) (5, 73). The large Maf proteins are emerging as regulators of cell differentiation in various tissues, as best illustrated in the eye (5). Thus, c-Maf, MafA, and NRL appear to be important in photoreceptor visual functions through their actions in activating transcription of key differentiation targets (3, 23, 26, 28, 45, 53, 55). In addition, MafB may be involved in rhombencephalon development (33) and monocytic differentiation (24).
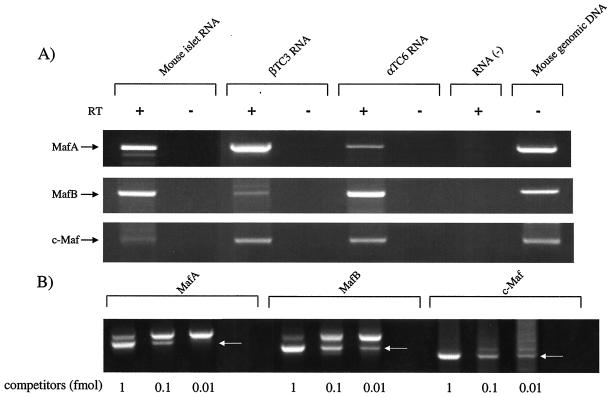
To examine large Maf expression in adult islet cells, RT-PCR analysis with MafA, c-Maf, MafB, and NRL specific primer sets was performed. Islets contained MafA, MafB, and c-Maf mRNA (Fig. 7) but not NRL (data not shown). Quantitative RT-PCR analysis showed that the relative ratios of MafA to MafB and Maf were approximately 1, 0.5, and 0.01, respectively (Fig. 7B). The authenticity of each large Maf RT-PCR product was confirmed by DNA sequence analysis. The MafA mRNA signal was higher than that of the other large Mafs in both islets and βTC-3 cells, while much less MafA RNA was detected in the islet α cell line, αTC-6 (Fig. 7A). This same pattern was observed in gel shift reactions performed with MafA-, MafB-, or c-Maf-recognizing antisera by using human islet and βTC-3 nuclear extracts. Thus, both MafA and MafB mRNA and gel shift binding activity were apparent in islet samples, whereas c-Maf mRNA expression was low and did not result in clear gel shift activity (compare the islet results in Fig. 7 to Fig. 4B). In contrast to islets, c-Maf was more clearly present in βTC-3 cells, a finding consistent with the finding that addition of both MafA and c-Maf antisera have a more significant effect on the RIPE3b1 complex than αMafA alone (Fig. 4A). Because these proteins can bind to InsC1 as either homo- or heterodimers (73; data not shown), these results strongly indicated that MafA, MafB, and/or c-Maf regulate RIPE3b1-mediated activation in the islet.
FIG. 7.
Islets express MafA, MafB, and c-Maf mRNA. (A) RT-PCR analysis was performed on RNA isolated from mouse islets, βTC3 cells, and αTC-6 cells with large Maf-specific primers. MafA, MafB, and c-Maf, but not NRL, were amplified to various degrees from each source. These RNAs were not detected without (− lanes) the addition of reverse transcriptase or in the absence of RNA. The correctness of the RT-PCR products was determined by DNA sequencing and by amplification of the same size PCR product from the intronless primer spanning sequences by using mouse genomic DNA. (B) Determination of MafA, MafB, and c-Maf levels in the adult islet by quantitative competitive RT-PCR. Increasing amounts of the large Maf DNA competitor were added to the RT-PCRs. The arrow denotes the location of the competitor band. The relative levels of MafA, MafB, and c-Maf were determined by densitometric quantification and correspond to approximately 1.0, 0.5, and 0.01, respectively.
Large Mafs activate InsC1-mediated expression.
To determine whether the β-cell-encoded MafA, c-Maf, and/or MafB transcription factors could activate insulin via the InsC1 binding site, we analyzed their effect upon the activity of −238-Insulin, a firefly luciferase reporter plasmid driven by rat insulin II gene sequences spanning bp −238 to +2. The large Maf cDNAs were cloned into the CMV enhancer-driven pcDNA3.1 plasmid, and their effect on wild-type −238-Insulin and a InsC1/RIPE3b1 binding defective mutant activity compared in HeLa cells. A cotransfected Renilla luciferase expression plasmid was used to normalize −238-Insulin activity.
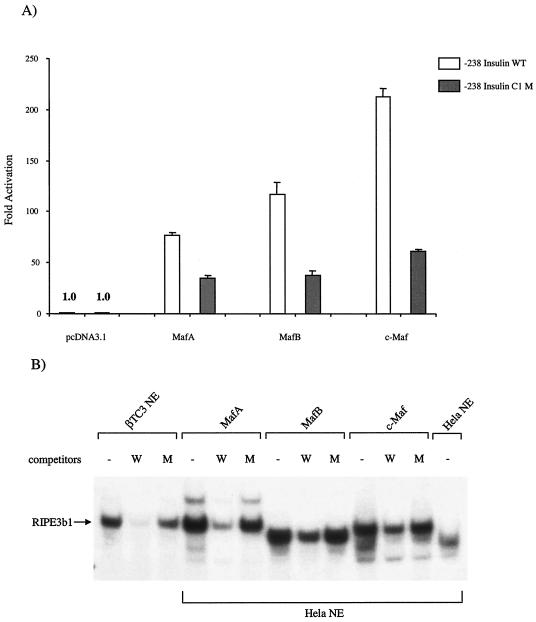
As expected, low-level activity was obtained in HeLa cells transfected with wild-type −238-Insulin in the absence of the large Mafs. The limited expression is due to the absence of β-cell-enriched transactivators. Cotransfection with plasmids expressing MafA, MafB, and c-Maf increased wild-type −238-Insulin activity by 76-, 117-, and 212-fold, respectively (Fig. 8A). Importantly, activation by these proteins was reduced in the InsC1/RIPE3b1 binding mutant. Since MafA, MafB, and c-Maf binding and dimerization properties are very similar (Fig. 8B; data not shown), the difference in stimulation observed between proteins may represent distinct activation domain potentials. Similar results were obtained upon transfection of this large Maf plasmid set in other non-insulin-producing cell types (data not shown). We conclude from these results that MafA, MafB, and c-Maf can specifically interact with and activate InsC1-mediated transcription.
FIG. 8.
MafA, MafB, and c-Maf stimulate InsC1-driven transcription. (A) HeLa cells were transfected with the wild-type (WT) or the RIPE3b1 binding defective −111/−108 InsC1 mutant (M) of the insulin-driven −238 LUC expression plasmid, the pRL-CMV internal transfection control, and MafA, MafB, and c-Maf subcloned into the CMV-driven pcDNA3.1 expression plasmid. The Maf activation signal is only partially reduced in the −238-Insulin C1 mutant due to the presence of additional sites in this region of the rat insulin II gene at −152/−140 bp and −94/−82 bp (data not shown). The data are presented as the large Maf transfected mean activity ± the standard error relative to the pcDNA3.1 alone from at least four separate experiments. The relative light unit activities of −238-Insulin and −238-Insulin C1 M alone were 13,361 ± 3,320 and 18,877 ± 4,258, respectively. (B) Competition analysis performed with nuclear extracts (NEs) prepared from MafA, MafB, and c-Maf transfected HeLa cells demonstrates RIPE3b1-like gel shift binding of the transfected proteins to the InsC1 wild type (lanes W) and −111/-108 bp mutant (lanes M). The c-Maf binding reactions contain three times more nuclear extract (6 μg) than did the MafA or MafB samples. No detectable l-Maf-binding activity was found in the untransfected HeLa nuclear extract.
MafA and MafB are nuclear proteins selectively expressed in pancreatic islet cells.
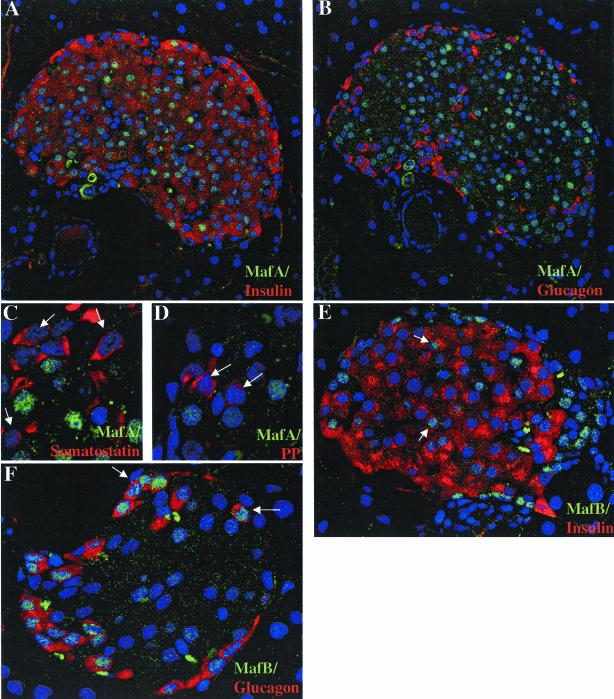
The presence and localization of MafA and MafB were examined immunohistochemically and their distribution compared to islet insulin-, glucagon (α)-, somatostatin (δ)-, and pancreatic polypeptide (PP)-producing cells. MafA staining was found in the nuclei of the majority of cells in the core of the islet (Fig. 9A and B), the principal location of β cells. Double-label immunofluorescence revealed that MafA was only expressed in β cells and not in α, δ, or PP cells (compare panel A to panels B, C, and D in Fig. 9). MafA expression was detected in most, but not all, β cells (Fig. 9A and B; 83% ± 5%). In contrast, MafB nuclear staining was detected in a smaller fraction of β cells (37% ± 4.5%) and essentially all α cells (91% ± 7%) (Fig. 9; data not shown). In contrast, no MafA or MafB staining was detected in the acinar cells surrounding the islet (Fig. 9).
FIG. 9.
MafA and MafB are localized in the nuclei of islet β cells. Immunohistochemical detection of MafA (A to D) and MafB (E and F) expression in adult mouse pancreas. Double immunofluorescence was used to analyze coexpression of these factors with hormones (red): insulin (A and E), glucagon (B and F), somatostatin (C), and/or PP (D). The nuclei are counterstained in blue. Note that MafA staining (green) was only detected in insulin-producing cells, whereas MafB was expressed in both insulin- and glucagon-producing cells. In addition, MafA and MafB immunoreactivity was not observed in surrounding exocrine cells. The arrows in panels C and D denote the absence of MafA staining in δ and PP cells and the presence of MafB expression in β (E) and α (F) cells. The c-Maf-specific antiserum (N-15) did not stain control tissues, preventing the determination of c-Maf expression in islets (data not shown).
The immunohistochemical data are consistent with the gel shift (Fig. 4) and RT-PCR (Fig. 7) results regarding the distribution of the large Maf proteins in the islet. These results demonstrate that MafA and MafB are nuclear proteins preferentially expressed in islet cells.
DISCUSSION
Diabetes results from a loss in the capacity of β cells to produce enough insulin to meet the body's needs, either as a result of autoimmune destruction of β cells (i.e., in type I diabetes mellitus [35]) or of reduced function (i.e., in type II [72]). Here, we have isolated and shown that the MafA transcription factor represents a β-cell-enriched DNA-binding component of the C1/RIPE3b1 activator complex that is involved in cell type-specific and glucose-regulated expression of the insulin gene. MafA is a member of the large Maf family of basic leucine-zipper containing DNA-binding proteins. In addition to MafA, there are three other closely related members of this family—c-Maf, MafB, and NRL—each of which have very similar DNA-binding (basic leucine zipper) and N-terminal transactivation domains (5, 73).
Phosphorylation of conserved serine and threonine residues in the large Maf proteins is important in activation domain function (4). In addition, phosphorylation of a tyrosine(s) appears to potentiate MafA binding, as suggested by our ability to both immunoprecipitate this protein with the α-phosphotyrosine 4G10 monoclonal antibody (Fig. 5) and prevent the loss in C1/RIPE3b1 binding in β-cell extracts with tyrosine phosphatase inhibitors (36). Because the MafA protein coding region is found on a single exon (data not shown), phosphorylation and/or another type of posttranslational modification(s) most likely caused this 46-kDa protein to localize to both pH 7 and 4.5 after 2-D electrophoresis (Fig. 2). However, it is also possible that the effect of the 4G10 antibody and phosphatases (82) on MafA binding does not result from direct tyrosine phosphorylation but instead occurs through association with a tyrosine-phosphorylated coregulatory factor.
Islet β cells exposed to stimulating glucose concentrations induce a series of signaling events that increase insulin secretion and insulin gene transcription. Each of these processes is potentiated by insulin in an autocrine manner (30), and mice lacking either the insulin receptor (27) or insulin receptor substrate-2 (79) in their β cells are diabetic. Autocrine stimulation of insulin gene expression appears to be mediated by the Pdx-1 and BETA2:E47 activators, although C1/RIPE3b1-targeted activation was not examined directly (31). Since Pdx-1, BETA2:E47 and C1/RIPE3b1 mediate the insulin gene transcriptional response to glucose (37, 60, 67), MafA activation may also be controlled by insulin. As a consequence, we are investigating whether MafA is involved in reduced β-cell function in insulin receptor (27)- or insulin receptor substrate-2 (79)-null mice. The recent observations showing that transgenic Pdx-1 expression rescues the β-cell deficiency in insulin receptor substrate-2-null mice (29) and that RIPE3b1/Maf directly activates pdx-1 transcription (59) provide additional support for this possibility.
MafA was purified from βTC-3 cell line nuclear extract by InsC1 affinity chromatography and 2-D electrophoresis and identified by mass spectroscopy. Several distinct observations allowed us to link MafA (versus the copurifying AUF1 RNA binding factor) to the RIPE3b1 activator, including the demonstration that: (i) MafA recognizing antibodies detected a protein in the RIPE3b1 activator gel shift complex (Fig. 4); (ii) MafA activated InsC1 element-driven reporter expression (Fig. 8); (iii) the α-phosphotyrosine 4G10 monoclonal antibody immunoprecipitated a MafA containing RIPE3b1 binding complex (Fig. 5); and (iv) MafA bound within the enhancer region of the endogenous insulin gene (Fig. 6). Based on these criteria, we concluded that MafA was the principal β-cell-enriched DNA-binding subunit of the RIPE3b1 activator. Primarily based on the criteria described above in points i and ii, a similar conclusion was recently independently drawn by two other groups while the present study was in progress (22, 47). However, the RT-PCR (Fig. 7) and large Maf antibody (Fig. 4 and 9) analyses presented here also indicated that the closely related c-Maf and MafB proteins are normally expressed in islet cells. The relatively higher levels of MafA in β-cell lines and the difficulty in obtaining large amounts of purified material probably explain why this was the only family member identified from the biochemical purification (47) (Fig. 2).
Since the large Mafs have similar activation properties (Fig. 8) and were capable of codimerizing through their conserved homoamphipathic C-terminal leucine zipper region (73; data not shown), RIPE3b1 activator function during pancreatic development and in the β cell is presumably mediated by both homo- and heterodimers of MafA, c-Maf, and MafB. The data presented here and elsewhere demonstrated that the insulin and pdx-1 (59) genes are bona fide transcriptional targets for RIPE3b1/Maf control in β cells. Interestingly, our immunohistochemical data suggest that MafA and/or MafB are presented in the entire islet β-cell population, with most cells containing MafA. In contrast, only MafB appears to be found in α cells (Fig. 9). The presence of a consensus large Maf-binding motif, TGC(N)6-7GCA (25), in the transcription control region of other islet cell-enriched genes suggests a more general and significant role, e.g., the −183/−165-bp islet-specific glucose-6-phosphatase catalytic subunit-related protein gene (34), the −540/−528-bp islet amyloid polypeptide gene (11, 18, 40), and the −63/−58-bp glucagon gene (52). Furthermore, the functional cooperativity observed between the large Mafs and Pax6 in activating both glucagon expression in α cells and lens crystallin gene expression (8, 41, 54, 58) implies that MafA and MafB act in concert with more widely distributed islet-enriched factors (e.g., Pax6) to mediate selective expression.
Strikingly, each of the characterized islet-enriched transcriptional activators of the insulin gene also have a profound effect on islet cell development. Thus, Pdx-1 expression in a common precursor cell population is essential for the development of the endocrine and exocrine compartments of the pancreas, permitting proliferation, branching, and differentiation of the pancreatic epithelium (21, 44). Humans (69) or mice (21, 44) with an inactivating mutation in Pdx-1 are born without a pancreas. In contrast, Pax6 and BETA2 act downstream of Pdx-1 in the generation of specific islet cell types (43, 70). All of these factors are also likely to be essential in the maintenance of functional islet cell types, since selective elimination of Pdx-1 in mouse β cells in vivo caused glucose intolerance and diabetes (1). Moreover, mutations in Pdx-1 (68), BETA2 (32), and Pax6 (81) caused diabetes in humans. Interestingly, and in contrast to each of these factors, MafA appears to be expressed uniquely in β cells in the adult islet (Fig. 9). This selective distribution pattern has only been observed for one other islet-enriched transcription factor, Nkx6.1, which is essential for β-cell development (62).
The role of MafA, MafB, and c-Maf in pancreas development has not been evaluated. The isolation of MafA represented the first description of the mammalian gene (22, 47). The chicken l-Maf (for Lens-specific) ortholog is expressed in the lens placode as early as the onset of lens formation (45). In addition to islets, MafB and c-Maf are expressed in the developing eye, spinal cord, and chondrocytes in mammals (57). Chicken MafA has been proposed to be a master regulator of lens differentiation due to its ability upon ectopic expression in chick embryos to induce lens-specific α-, β-, and γ-crystallin gene expression and promote transdifferentiation of neuroretinal cells into lens fibers (45). The analysis of MafB and c-Maf mutant mice has also revealed their importance in eye, kidney, and brain development (9, 26, 55, 56). Collectively, the data indicate that the large Maf proteins are regulators of processes important in cell differentiation. Our results suggest a specific and central role in islet β-cell development and function.
Acknowledgments
We thank Eva Henderson, Yasuyuki Asai, and Kevin C. Ray for invaluable technical support and members of the Stein lab for stimulating discussions. The mouse MafB (SkmuMafB) amd c-Maf (RcRSVcMaf) expression vectors were generously provided by Linda Shapiro and Paul Overbeek, respectively. In addition, we thank Gary Brewer for providing the αAUF-1 antiserum.
This work was supported by grants from the National Institutes of Health (P01 DK42502 to R.S. and GM43609 to H.W.J.) and the Juvenile Diabetes Research Foundation (JDRF 32001678 to T.-A.M.). Partial support was also provided to the Molecular Biology Core Laboratory by the Vanderbilt University Diabetes Research and Training Center (Public Health Service grant P60 DK20593).
REFERENCES
- 1.Ahlgren, U., J. Jonsson, L. Jonsson, K. Simu, H. Edlund. 1998. Beta-cell-specific inactivation of the mouse Ipf1/Pdx1 gene results in loss of the beta-cell phenotype and maturity onset diabetes. Genes Dev. 12:1763-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aronheim, A., H. Ohlsson, C. W. Park, T. Edlund, and M. D. Walker. 1991. Distribution and characterization of helix-loop-helix enhancer-binding proteins from pancreatic beta cells and lymphocytes. Nucleic Acids Res. 19:3893-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benkhelifa, S., S. Provot, O. Lecoq, C. Pouponnot, G. Calothy, and M. P. Felder-Schmittbuhl. 1998. mafA, a novel member of the maf proto-oncogene family, displays developmental regulation and mitogenic capacity in avian neuroretina cells. Oncogene 17:247-254. [DOI] [PubMed] [Google Scholar]
- 4.Benkhelifa, S., S. Provot, E. Nabais, A. Eychene, G. Calothy, and M. P. Felder-Schmittbuhl. 2001. Phosphorylation of MafA is essential for its transcriptional and biological properties. Mol. Cell. Biol. 14:4441-4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blank, V., and N. C. Andrews. 1997. The Maf transcription factors: regulators of differentiation. Trends Biochem. Sci. 22:437-441. [DOI] [PubMed] [Google Scholar]
- 6.Bretherton-Watt, D., N. Gore, and D. S. W. Boam. 1996. Insulin upstream factor 1 and a novel ubiquitous factor bind to the human islet amyloid polypeptide/amylin gene promoter. Biochem. J. 313:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carty, M. D., J. S. Lillquist, M. Peshavaria, R. Stein, and W. C. Soeller. 1997. Identification of cis- and trans-active factors regulating islet amyloid polypeptide expression in pancreatic β cells. J. Biol. Chem. 272:11986-11993. [DOI] [PubMed] [Google Scholar]
- 8.Civil, A., S. T. van Genesen, and N. H. Lubsen. 2002. c-Maf, the γD-crystallin Maf-responsive element and growth factor regulation. Nucleic Acids Res. 30:975-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cordes, S. P., and G. S. Barsh. 1994. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell 79:1025-1034. [DOI] [PubMed] [Google Scholar]
- 10.Cordle, S. R., E. Henderson, H. Masuoka, P. A. Weil, and R. Stein. 1991. Pancreatic β-cell-type-specific transcription of the insulin gene is mediated by basic helix-loop-helix DNA-binding proteins. Mol. Cell. Biol. 11:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Wit, L., A. D. van Mansfeld, H. A. van Teeffelen, C. J. Lips, and J. W. Hoppener. 1993. Strong promoter activity of human and rat islet amyloid polypeptide/amylin gene constructs in mouse beta cells (βTC3). Biochem. Biophys. Res. Commun. 192:840-848. [DOI] [PubMed] [Google Scholar]
- 12.Dumonteil, E., B. Laser, I. Constant, and J. Philippe. 1998. Differential regulation of the glucagon and insulin I gene promoters by the basic helix-loop-helix transcription factors E47 and BETA2. J. Biol. Chem. 273:19945-19954. [DOI] [PubMed] [Google Scholar]
- 13.Edlund, H. 2002. Pancreatic organogenesis-developmental mechanisms and implications for therapy. Nat. Rev. Genet. 3:524-532. [DOI] [PubMed] [Google Scholar]
- 14.Efrat, S., S. Linde, H. Kofod, D. Spector, M. Delannoy, S. Grant, D. Hanahan, and S. Baekkeskov. 1988. Beta-cell lines derived from transgenic mice expressing a hybrid insulin gene-oncogene. Proc. Natl. Acad. Sci. USA 85:9037-9041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gadgil, H., and H. W. Jarrett HW. 2002. Oligonucleotide trapping method for purification of transcription factors. J. Chromatogr. A 966(1-2):99-110. [DOI] [PubMed]
- 16.Gannon, M., and C. V. E. Wright. 1999. Endoderm patterning and organogenesis, p. 583-615. In Sally Moody (ed.), Cell lineage and fate determination. Academic Press, Inc., San Diego, Calif.
- 17.German, M. S., M. A. Blanar, C. Nelson, L. G. Moss, and W. J. Rutter. 1991. Two related helix-loop-helix proteins participate in separate cell-specific complexes that bind the insulin enhancer. Mol. Endocrinol. 5:292-299. [DOI] [PubMed] [Google Scholar]
- 18.German, M. S., L. G. Moss, J. Wang, and W. J. Rutter. 1992. The insulin and islet amyloid polypeptide genes contain similar cell-specific promoter elements that bind identical beta-cell nuclear complexes. Mol. Cell. Biol. 12:1777-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerrish, K., M. A. Cissell, and R. Stein. 2001. The role of hepatic nuclear factor 1α and PDX-1 in transcriptional regulation of the pdx-1 gene. J. Biol. Chem. 276:47775-47784. [DOI] [PubMed] [Google Scholar]
- 20.Hamaguchi, K., and E. H. Leiter. 1990. Comparison of cytokine effects on mouse pancreatic alpha-cell and beta-cell lines: viability, secretory function, and MHC antigen expression. Diabetes 39:415-425. [DOI] [PubMed] [Google Scholar]
- 21.Jonsson, J., L. Carlsson, T. Edlund, and H. Edlund. 1994. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371:606-609. [DOI] [PubMed] [Google Scholar]
- 22.Kataoka, K., S. I. Han, S. Shioda, M. Hirai, M. Nishizawa, and H. Handa. 2002. MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J. Biol. Chem. 277:49903-49910. [DOI] [PubMed] [Google Scholar]
- 23.Kawauchi, S., S. Takahashi, O. Nakajima, H. Ogino, M. Morita, M. Nishizawa, K. Yasuda, and M. Yamamoto M. 1999. Regulation of lens fiber cell differentiation by transcription factor c-Maf. J. Biol. Chem. 274:19254-19260. [DOI] [PubMed] [Google Scholar]
- 24.Kelly, L. M., U. Englmeier, I. Lafon, M. H. Sieweke, and T. Graf. 2000. MafB is an inducer of monocytic differentiation. EMBO J. 19:1987-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kerppola, T. K., and T. Curran. 1994. A conserved region adjacent to the basic domain is required for recognition of an extended DNA binding site by Maf/Nrl family proteins. Oncogene 9:3149-3158. [PubMed] [Google Scholar]
- 26.Kim, J. I., T. Li, I. C. Ho, M. J. Grusby, and L. H. Glimcher. 1999. Requirement for the c-Maf transcription factor in crystallin gene regulation and lens development. Proc. Natl. Acad. Sci. USA 96:3781-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kulkarni, R. N., J. C. Bruning, J. N. Winnay, C. Postic, M. A. Magnuson, and C. R. Kahn. 1998. Tissue-specific knockout of the insulin receptor in the pancreatic β cells creates an insulin secretory defect similar to that in type 2 diabetes. Cell 96:329-339. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, R. S. Chen, D. Scheurer, Q. L. Wang, E. Duh, C. H. Sung, A. Rehemtulla, A. Swaroop, R. Adler, and D. J. Zack. 1996. The bZIP transcription factor Nrl stimulates rhodopsin promoter activity in primary retinal cell cultures. J. Biol. Chem. 271:29612-29618. [DOI] [PubMed] [Google Scholar]
- 29.Kushner, J. A., J. Ye, M. Schubert, D. J. Burks, M. A. Dow, C. L. Flint, S. Dutta, C. V. Wright, M. R. Montminy, and M. F. White. 2002. Pdx1 restores beta cell function in Irs2 knockout mice. J. Clin. Investig. 109:1193-1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leibiger, I. B., B. Leibiger, and P. O. Berggren. 2002. Insulin feedback action on pancreatic beta-cell function. FEBS Lett. 532:1-6. [DOI] [PubMed] [Google Scholar]
- 31.Leibiger, I. B., B. Leibiger, T. Moede, and P. O. Berggren. 1998. Exocytosis of insulin promotes insulin gene transcription via the insulin receptor/PI-3 kinase/p70 s6 kinase and CaM kinase pathways. Mol. Cell 1:933-938. [DOI] [PubMed] [Google Scholar]
- 32.Malecki, M. T., U. S. Jhala, A. Antonellis, L. Fields, T. Orban, M. Saad, A. Doria, J. H. Warram, M. Montminy, and A. S. Krolewski. 1999. Mutations in NeuroD1/BETA2 are associated with the development of type 2 diabetes mellitus. Nat. Genet. 23:323-328. [DOI] [PubMed] [Google Scholar]
- 33.Manzanares, M., S. Cordes, L. Ariza-McNaughton, V. Sadl, K. Maruthainar, G. Barsh, and R. Krumlauf. 1999. Conserved and distinct roles of kreisler in regulation of the paralogous Hoxa3 and Hoxb3 genes. Development 126:759-769. [DOI] [PubMed] [Google Scholar]
- 34.Martin, C. C., L. J. Bischof, B. Bergman, L. A. Hornbuckle, C. Hilliker, C. Frigeri, D. Wahl, C. A. Svitek, R. Wong, J. K. Goldman, J. K. Oeser, F. Lepretre, P. Froguel, R. M. O'Brien, and J. C. Hutton. 2001. Cloning and characterization of the human and rat islet-specific glucose-6-phosphatase catalytic subunit-related protein (IGRP) genes. J. Biol. Chem. 276:25197-25207. [DOI] [PubMed] [Google Scholar]
- 35.Mathis, D., L. Vence, and C. Benoist. 2001. Beta-cell death during progression to diabetes. Nature 414:792-798. [DOI] [PubMed] [Google Scholar]
- 36.Matsuoka, T., L. Zhao, and R. Stein. 2001. The DNA binding activity of the RIPE3b1 transcription factor of the insulin gene appears to be influenced by tyrosine phosphorylation. J. Biol. Chem. 276:22071-22076. [DOI] [PubMed] [Google Scholar]
- 37.Melloul, D., S. Marshak, and E. Cerasi. 2002. Regulation of insulin gene transcription. Diabetologia 45:309-326. [DOI] [PubMed] [Google Scholar]
- 38.Miyazaki, J., K. Araki, E. Yamato, H. Ikegami, T. Asano, Y. Shibasaki, Y. Oka, and K. Yamamura. 1990. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 127:126-132. [DOI] [PubMed] [Google Scholar]
- 39.Moates, J. M., S. Nanda, M. A. Cissell, M. J. Tsai, and R. Stein. 2003. BETA2 activates transcription from the upstream glucokinase gene promoter in islet beta-cells and gut endocrine cells. Diabetes 52:403-408. [DOI] [PubMed] [Google Scholar]
- 40.Mosselman, S., J. W. Hoppener, L. de Wit, W. Soeller, C. J. Lips, and H. S. Jansz. 1990. IAPP/amylin gene transcriptional control region: evidence for negative regulation. FEBS Lett. 271:33-36. [DOI] [PubMed] [Google Scholar]
- 41.Muta, M., Y. Kamachi, A. Yoshimoto, Y. Higashi, and H. Kondoh. 2002. Distinct roles of SOX2, Pax6, and Maf transcription factors in the regulation of lens-specific δ1-crystallin enhancer. Genes Cells 7:791-805. [DOI] [PubMed] [Google Scholar]
- 42.Naya, F. J., C. M. M. Stellrecht, and M.-J. Tsai. 1995. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 9:1009-1019. [DOI] [PubMed] [Google Scholar]
- 43.Naya, F. J., H.-P. Huang, Y. Qiu, H. Mouth, F. J. DeMayo, A. B. Leiter, and M.-J. Tsai. 1997. Diabetes, defective pancreatic morphogenesis, and abnormal enteroendocrine differentiation in BETA2/NeuroD-deficient mice. Genes Dev. 11:2323-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Offield, M. F., T. L. Jetton, R. Stein, T. Labosky, M. Ray, M. Magnuson, B. Hogan, and C. V. E. Wright. 1996. PDX-1 is required for development of the pancreas and differentiation of the rostral duodenum. Development 122:983-995. [DOI] [PubMed] [Google Scholar]
- 45.Ogino, H., and K. Yasuda. Induction of lens differentiation by activation of a bZIP transcription factor, l-Maf. Science 280:115-158. [DOI] [PubMed]
- 46.Ohlsson, H., K. Karlsson, and T. Edlund. 1993. IPF-1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 12:4251-4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olbrot, M., J. Rud, L. G. Moss, and A. Sharma. 2002. Identification of β-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc. Natl. Acad. Sci. USA 99:6737-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peers, B., J. Leonard, S. Sharma, G. Teitelman, and M. R. Montminy. 1994. Insulin expression in pancreatic islet cells relies on cooperative interactions between the helix loop helix factor E47 and the homeobox factor STF-1. Mol. Endocrinol. 8:1798-1806. [DOI] [PubMed] [Google Scholar]
- 49.Peshavaria, M., L. Gamer, E. Henderson, G. Teitelman, C. V. E. Wright, and R. Stein. 1994. XlHbox 8, an endoderm-specific Xenopus homeodomain protein, is closely related to a mammalian insulin gene transcription factor. Mol. Endocrinol. 8:806-816. [DOI] [PubMed] [Google Scholar]
- 50.Petersen, H. V., P. Serup, J. Leonard, B. K. Michelsen, and O. D. Madsen. 1994. Transcriptional regulation of the human insulin gene is dependent of the homeodomain proteins STF1/IPF1 acting through the CT boxes. Proc. Natl. Acad. Sci. USA 91:10465-10469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peyton, M., L. Moss, and M.-J. Tsai. 1994. Two distinct class A helix-loop-helix transcription factors, E2A and BETA1, form separate DNA-binding complexes on the insulin E-box. J. Biol. Chem. 269:25936-25941. [PubMed] [Google Scholar]
- 52.Planque, N., L. Leconte, F. M. Coquelle, S. Benkhelifa, P. Martin, M. P. Felder-Schmittbuhl, and S. Saule. 2001. Interaction of Maf transcription factors with Pax-6 results in synergistic activation of the glucagon promoter. J. Biol. Chem. 276:35751-35760. [DOI] [PubMed] [Google Scholar]
- 53.Rehemtulla, A., R. Warwar, R. Kumar, X. Ji, D. J. Zack, and A. Swaroop. 1996. The basic motif-leucine zipper transcription factor Nrl can positively regulate rhodopsin gene expression. Proc. Natl. Acad. Sci. USA 93:191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reza, H. M., H. Ogino, and K. Yasuda. 2002. l-Maf, a downstream target of Pax6, is essential for chick lens development. Mech. Dev. 116:61-73. [DOI] [PubMed] [Google Scholar]
- 55.Ring, B. Z., S. P. Cordes, P. A. Overbeek, and G. S. Barsh. 2000. Regulation of mouse lens fiber cell development and differentiation by the Maf gene. Development 127:307-317. [DOI] [PubMed] [Google Scholar]
- 56.Sadl, V., F. Jin, J. Yu, S. Cui, D. Holmyard, S. Quaggin, G. Barsh, and S. Cordes. 2002. The mouse Kreisler (Krml1/MafB) segmentation gene is required for differentiation of glomerular visceral epithelial cells. Dev. Biol. 249:16-29. [DOI] [PubMed] [Google Scholar]
- 57.Sakai, M., J. Imaki, K. Yoshida, A. Ogata, Y. Matsushima-Hibaya, Y. Kuboki, M. Nishizawa, and S. Nishi. 1997. Rat maf related genes: specific expression in chondrocytes, lens, and spinal cord. Oncogene 14:745-750. [DOI] [PubMed] [Google Scholar]
- 58.Sakai, M., M. S. Serria, H. Ikeda, K. Yoshida, J. Imaki, and S. Nishi. 2001. Regulation of c-maf gene expression by Pax6 in cultured cells. Nucleic Acids Res. 29:1228-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Samaras, S. E., L. Zhao, A. Means, E. Henderson, T. A. Matsuoka, and R. Stein. 2003. The islet β-cell-enriched RIPE3b1/Maf transcription factor regulates pdx-1 expression. J. Biol. Chem. 278:12263-12270. [DOI] [PubMed]
- 60.Sander, M., and M. S. German. 1997. The beta cell transcription factors and development of the pancreas. J. Mol. Med. 75:327-340. [DOI] [PubMed] [Google Scholar]
- 61.Sander, M., A. Neubuser, J. Kalamaras, H. C. Ee, G. R. Martin, and M. S. German. 1997. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Gene Dev. 11:1662-1673. [DOI] [PubMed] [Google Scholar]
- 62.Sander, M., L. Sussel, J. Conners, D. Scheel, J. Kalamaras, F. Dela Cruz, V. Schwitzgebel, A. Hayes-Jordan, and M. German. 2000. Homeobox gene Nkx6.1 lies downstream of Nkx2.2 in the major pathway of beta-cell formation in the pancreas. Development 127:5533-5540. [DOI] [PubMed] [Google Scholar]
- 63.Schreiber, E., P. Matthias, M. M. Muller, and W. Schaffner. 1989. Eukaryotic expression vectors for the analysis of mutant proteins. Nucleic Acids Res. 17:6418.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Serup, P., J. Jensen, F. G. Andersen, M. C. Jorgensen, N. Blume, J. J. Holst, and O. D. Madsen. 1996. Induction of insulin and islet amyloid polypeptide production in pancreatic islet glucagonoma cells by insulin promoter factor 1. Proc. Natl. Acad. Sci. USA 93:9015-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shieh, S.-Y., and M.-J. Tsai. 1991. Cell-specific and ubiquitous factors are responsible for the enhancer activity of the rat insulin II gene. J. Biol. Chem. 266:16708-16714. [PubMed] [Google Scholar]
- 66.Shieh, S.-Y., M. M. Stellrecht, and M.-J. Tsai. 1995. Molecular characterization of the rat insulin enhancer-binding complex 3b2. J. Biol. Chem. 270:21503-21508. [DOI] [PubMed] [Google Scholar]
- 67.Stein, R. 2001. Insulin gene transcription: the factors involved in cell-type-specific and glucose-regulated expression in islet β cells are also essential during pancreatic development, p. 25-78. In A. Cherrington and J. Jefferson (ed.), Handbook of physiology, vol. II. American Physiology Society, Washington, D.C.
- 68.Stoffers, D. A., J. Ferrer, W. L. Clarke, and J. F. Habener. 1997. Early-onset type-II diabetes mellitus (MODY4) linked to IPF1. Nat. Genet. 17:138-139. [DOI] [PubMed] [Google Scholar]
- 69.Stoffers, D. A., N. T. Zinkin, V. Stanojevic, W. L. Clarke, and J. F. Habener. 1997. Pancreatic agenesis attributable to a single nucleotide deletion in the human IPF1 gene coding sequence. Nat. Genet. 15:106-110. [DOI] [PubMed] [Google Scholar]
- 70.St-Onge, L., B. Sosa-Pineda, K. Chowdhury, A. Mansouri, and P. Gruss. 1997. Pax6 is required for differentiation of glucagon-producing α-cells in mouse pancreas. Nature 387:406-409. [DOI] [PubMed] [Google Scholar]
- 71.Swift, G. H., Y. Liu, S. D. Rose, L. J. Bischof, S. Steelman, A. M. Buchberg, C. V. Wright, and R. J. MacDonald. 1998. An endocrine-exocrine switch in the activity of the pancreatic homeodomain protein PDX1 through formation of a trimeric complex with PBX1b and MRG1 (MEIS2). Mol. Cell. Biol. 18:5109-5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Taylor, S. I. 1999. Deconstructing type 2 diabetes. Cell 97:9-12. [DOI] [PubMed] [Google Scholar]
- 73.Vinson, C., M. Myakishev, A. Acharya, A. A. Mir, J. R. Moll, and M. Bonovich. 2002. Classification of human B-ZIP proteins based on dimerization properties. Mol. Cell. Biol. 22:6321-6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waeber, G., N. Thompson, P. Nicod, and C. Bonny. 1996. Transcriptional activation of the GLUT2 gene by the IPF-1/STF-1/IDX-1 homeobox factor. Mol. Endocrinol. 10:1327-1334. [DOI] [PubMed] [Google Scholar]
- 75.Watada, H., Y. Kajimoto, H. Kaneto, T. Matsuoka, Y. Fujitani, J. Miyazaki, and Y. Yamasaki. 1996. Involvement of the homeodomain-containing transcription factor PDX-1 in islet amyloid polypeptide gene transcription. Biochem. Biophys. Res. Commun. 229:746-751. [DOI] [PubMed] [Google Scholar]
- 76.Watada, H., Y. Kajimoto, J. Miyagwa, T. Hanafusa, K. Hamaguchi, T. Matsuoka, K. Yamamoto, Y. Matsuzawa, R. Kawamori, and Y. Yamasaki. 1996. PDX-1 induces insulin and glucokinase gene expression in αTC1 clone 6 cells in the presence of betacellulin. Diabetes 45:1826-1831. [DOI] [PubMed] [Google Scholar]
- 77.Watada, H., Y. Umayahara, T. Matsuoka, H. Kaneto, Y. Fujitani, T. Kamada, R. Kawamori, and Y. Yamasaki. 1996. The human glucokinase gene β-cell-type promoter: an essential role of insulin promoter factor 1/PDX-1 in its activation in HIT-T15 cells. Diabetes 45:1478-1488. [DOI] [PubMed] [Google Scholar]
- 78.Whelan, J., S. R. Cordle, E. Henderson, P. A. Weil, and R. Stein. 1990. Identification of a pancreatic β-cell insulin gene transcription factor that binds to and appears to activate cell-type-specific expression: its possible relationship to other cellular factors that bind to a common insulin gene sequence. Mol. Cell. Biol. 10:1564-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Withers, D. J., D. J. Burks, D. J., H. H. Towery, S. L. Altamuro, C. L. Flint, and M. F. White. 1999. Irs-2 coordinates Igf-1 receptor-mediated β-cell development and peripheral insulin signalling. Nat. Genet. 23:32-40. [DOI] [PubMed] [Google Scholar]
- 80.Wu, K.-L., M. Peshavaria, M. Gannon, E. Henderson, M. F. Offield, E. Henderson, M. Ray, A. Marks, L. W. Gamer, C. V. E. Wright, and R. Stein. 1997. HNF3β is involved in pancreatic β-cell-specific transcription of the pdx-1 gene. Mol. Cell. Biol. 17:6002-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yasuda, T., Y. Kajimoto, Y. Fujitani, H. Watada, S. Yamamoto, T. Watarai, Y. Umayahara, M. Matsuhisa, S. Gorogawa, Y. Kuwayama, Y. Tano, Y. Yamasaki, and M. Hori. 2002. PAX6 mutation as a genetic factor common to aniridia and glucose intolerance. Diabetes 51:224-230. [DOI] [PubMed] [Google Scholar]
- 82.Zhao, L., M. A. Cissell, E. Henderson, R. Colbran, and R. Stein. 2000. The RIPE3b1 activator of the insulin gene is composed of a protein(s) of approximately 43 kDa, whose DNA binding activity is inhibited by protein phosphatase treatment. J. Biol. Chem. 275:10532-10537. [DOI] [PubMed] [Google Scholar]