Abstract
Budding yeast Rad53 is an essential protein kinase that is phosphorylated and activated in a MEC1- and TEL1-dependent manner in response to DNA damage. We studied the role of Rad53 phosphorylation through mutation of consensus phosphorylation sites for upstream kinases Mec1 and Tel1. Alanine substitution of the Rad53 amino-terminal TQ cluster region reduced viability and impaired checkpoint functions. These substitution mutations spared the basal interaction with Asf1 and the DNA damage-induced interactions with Rad9. However, they caused a decrease in DNA damage-induced Rad53 kinase activity and an impaired interaction with the protein kinase Dun1. The Dun1 FHA (Forkhead-associated) domain recognized the amino-terminal TQ cluster of Rad53 after DNA damage or replication blockade. Thus, the phosphorylation of Rad53 by upstream kinases is important not only for Rad53 activation but also for creation of an interface between Rad53 and Dun1.
Failure in maintenance of genomic integrity allows the accumulation of mutations that may promote carcinogenesis. Cells employ many mechanisms to ensure complete and accurate transmission of genomic DNA to daughter cells. For example, DNA replication checkpoint pathways prevent cells with incompletely replicated DNA from entering mitosis, and DNA damage checkpoint pathways slow or arrest the cell cycle upon DNA damage. Activation of either pathway results in induction of transcription of genes required for DNA repair (43). Inherited mutations in DNA checkpoint pathways are found in multiple cancer-predisposing disorders, with examples including mutation of ATM in ataxia telangiectasia and of TP53 and CHK2 in Li-Fraumeni syndrome (8, 35, 51).
DNA checkpoint pathways are well conserved from yeasts to humans. The conserved mechanisms include protein kinase cascades in which large phosphoinositide 3′-kinase-like kinases (PIKKs), Mec1 and Tel1 in Saccharomyces cerevisiae, Rad3 in Schizosaccharomyces pombe, and ATM and ATR in mammals, phosphorylate effector kinases that act in parallel, ScChk1 and ScRad53, SpChk1 and SpCds1, and mammalian Chk1 and Chk2, respectively (40).
In budding yeast, the signal initiated by DNA damage is conveyed to the effector kinases Rad53 and Chk1 through Rad9 (note that ScRad9 is unrelated to SpRad9 and human Rad9) (47, 59). Upon DNA damage, Rad9 undergoes a MEC1- and TEL1-dependent phosphorylation that enables binding of Rad53 through the Rad53 FHA (Forkhead-associated) domains, which are modular phosphopeptide binding domains, and occurs concomitantly with binding to Chk1 (16, 53, 59, 62). Replicative stress activates Rad53 through an intermediary distinct from Rad9, possibly Mrc1 (4). DNA damage in S phase may signal through Rad9-dependent and -independent pathways (4, 42). In mammals, there is no single Rad9 homolog, but candidate orthologs that have similarly situated carboxyl-terminal BRCT domains and are phosphorylated with checkpoint activation include 53BP1 (6, 12, 64), MDC1 (21, 33, 46, 55, 56, 68), and BRCA1 (70).
Activated Rad53 is involved in cell cycle arrest, transcriptional induction of repair genes, inhibition of late replication origin firing, and stabilization of stalled replication forks (10, 32, 49, 50, 61). Under conditions of replicative stress, Rad53 phosphorylates and negatively regulates the Dbf4/Cdc7 kinase, which is required for DNA replication origin firing (13, 14, 27, 67). Rad53 phosphorylates and activates Dun1, another FHA domain-containing kinase, upon DNA checkpoint activation (7, 10, 76), resulting in increased expression and activity of ribonucleotide reductase (26, 71-74).
While the activation of Rad53 (or its orthologs SpCds1 and Chk2) is one of the key steps in DNA checkpoint pathways, the detailed mechanism of Rad53 activation remains unclear. Activation of Rad53-like protein kinases occurs concomitantly with phosphorylation (11, 45, 48, 58). Mutation of genes thought to act upstream in the DNA checkpoint pathways, including MEC1 and RAD9, prevents the phosphorylation of Rad53 (11, 57, 65). Phosphorylated Rad53 isolated from cells with activated DNA replication or damage checkpoint pathways is associated with elevated kinase activity, even after protein denaturation, separation from other proteins in sodium dodecyl sulfate (SDS)-polyacrylamide gels, and renaturation (45). Since kinase-defective Rad53 is still partially phosphorylated after DNA damage or replicative stress (45, 48, 58), some of the Rad53 phosphorylations occur in trans. The similar phenotypes associated with mutation of the PIKKs Mec1 and, to a lesser extent, Tel1 and the dependency of Rad53 phosphorylation on these kinases have suggested that the PIKKs directly phosphorylate Rad53. Consistent with this idea, a carboxyl-terminal fragment of Rad53 is phosphorylated in vitro by Mec1 (63). However, Rad53, like other protein kinases, can also autophosphorylate, so that checkpoint activation of Rad53 likely involves both PIKK-dependent transphosphorylation and intra- or intermolecular autophosphorylation.
Critical phosphorylation sites have been identified in the Rad53 orthologs SpCds1 and mammalian Chk2, respectively. Threonine 11 in SpCds1 is phosphorylated by the PIKK SpRad3 and is required for SpCds1 activation (60). ATM and ATR phosphorylate Chk2 at several partially overlapping sites located within a cluster of PIKK consensus sites amino terminal to the FHA domain. Mutation of some of these sites, notably threonine 68, impedes Chk2 activation (3, 38, 39). Thus, phosphorylation of Rad53 by Mec1 at a similar cluster may be crucial for Rad53 activation and function. However, efforts to activate Rad53 and its orthologs solely through transphosphorylation by PIKKs have been unsuccessful (38).
On the other hand, Rad53 may also autoactivate upon DNA damage. Rad53 expressed at high levels in bacteria is heavily phosphorylated in the absence of other yeast proteins, presumably by autophosphorylation (20). It was proposed previously that phosphorylated Rad9 serves as an adaptor that delivers Rad53 to Mec1 (59), since Mec1-dependent phosphorylation of Rad9 enables phosphorylated Rad9 to bind Rad53. However, Rad9 may play an alternative or additional role in concentrating bound Rad53 (20). Binding of Rad53 to phosphorylated Rad9 may enable Rad53 cross-phosphorylation and activation. Work on Chk2 suggests that, once activated, Rad53 may activate other naive Rad53 molecules through a PIKK- and RAD9-independent mechanism. DNA damage-activated phosphorylated Chk2 can form homo-oligomers mediated by an interaction between phosphorylation sites in the [S/T]Q cluster domain on one molecule and the FHA domain on the other (2, 69). This can be reconstructed in vitro using bacterially expressed Chk2, which autophosphorylates on sites including threonine 68 (2, 69). Once Chk2 is activated by PIKKs, activated Chk2 may bind to inactive Chk2 through the FHA domain of the latter, phosphorylate, and activate this molecule. This may result in a transition from PIKK-dependent to PIKK-independent regulation of Chk2.
These observations suggest at least three models for the mechanism of Rad53 activation. First, phosphorylation by Mec1-Tel1 on critical phosphorylation sites in Rad53 may activate Rad53 directly. In this case, Rad9 may work as an adaptor to deliver Rad53 to Mec1. In the second model, Rad53 is activated through intermolecular autophosphorylation upon DNA damage. The main role of Mec1-Tel1 would be to phosphorylate Rad9, but the PIKKs would not be required for further activation of Rad53. Rad9 would participate as a scaffold that concentrates Rad53. The third model is that phosphorylation of Rad53 by Mec1-Tel1 is required for Rad53 activation but that Rad9 is required as a coactivator.
Despite the overall conservation of DNA checkpoint proteins, Rad53 is unique among FHA-containing protein kinases, including SpCds1 and Chk2, in having two FHA domains that flank the kinase catalytic domain, rather than a single amino-terminal FHA domain. A cluster of potential PIKK phosphorylation sites is located amino terminal to each of the Rad53 FHA domains. We investigated the role of phosphorylation of Rad53 in its activation and function in vivo. The two clusters of PIKK sites within Rad53 are required for the full checkpoint-dependent phosphorylation of Rad53 and associated biological functions. While mutation in the second cluster of sites had mild effects, mutation of the first cluster of sites significantly impaired Rad53 function and activation. In addition, we found that phosphorylation of the Rad53 amino-terminal TQ cluster by upstream kinases mediates direct interactions between Rad53 and the FHA domain of Dun1. We propose that Mec1 and Tel1 not only play an important role in catalytic activation of Rad53 but also create a binding interface between Rad53 and Dun1 through phosphorylation in the amino-terminal TQ cluster.
MATERIALS AND METHODS
Plasmids and strains.
Wild-type and kinase-defective RAD53 (rad53kd; rad53K227A,D339A) were expressed under the control of the endogenous promoter in pRS316 or pRS315, a low-copy-number CEN plasmid, as described previously (18, 58). Expression and regulation (amount and phosphorylation) of plasmid-encoded Rad53 were comparable to those of genomic RAD53. The plasmids encoding alanine replacements of Rad53 were produced by PCR-based mutagenesis (four-primer PCR with internal mutagenic primers). RAD53 open reading frames (ORFs) (wild type or with nucleotide substitutions) were amplified by PCR with primers that introduced EcoRI and SmaI sites at 5′ and 3′ ends of the ORF. These sequences were inserted at the corresponding restriction sites in pBluescript SKII (+) (pSK) 3xFLAG, which contains sequences encoding a carboxyl-terminal three-FLAG (3xFLAG) tag, yielding pSK RAD53 3xFLAG alleles. pET28b RAD53 3xFLAG plasmids were constructed by inserting an EcoRI-NotI fragment containing the RAD53 3xFLAG ORF into the corresponding sites in pET28b. To make pRS316 RAD53 3xFLAG, a HpaI-SpeI fragment from pSK RAD53 3xFLAG was inserted at the same sites in pRS316 RAD53 13xMYC, which contains a SpeI site between the RAD53 13xMYC ORF and its 3′ untranslated region (UTR). Sequences encoding two-hemagglutinin (2xHA) tags were introduced similarly at the 3′ end of RAD53 to produce pRS316 RAD53 2xHA.
pRS314 DUN1 with a 13-MYC (13xMYC) tag (pRS314 DUN1 13xMYC) under the control of the endogenous DUN1 promoter is described elsewhere (54). Mutations in the FHA domain of DUN1 (Dun1R60A/N103A; Dun1RN) were introduced by replacing NotI-StuI PCR fragments with PCR products having directed mutations at the conserved amino acids in the FHA domain. pGEX 4T3 GST-DUN1 FHA and dun1 FHA domains with mutations (residues 1 to 199; Dun1 FHAR60A/N103A; Dun1 FHARN) were constructed as follows. Mutagenic primers were used to introduce EcoRI sites 5′ to the initiation codon of DUN1 in a PCR with pRS314 DUN1 13xMYC (wild type or with FHA mutations) as templates. The EcoRI-StuI PCR fragments were cloned into EcoRI-SmaI sites in pGEX 4T3. Plasmids encoding the glutathione S-transferase (GST) CHK2 FHA domain were described previously (69). Plasmids for GST RAD53 FHA1 and GST RAD53 FHA2 (BamHI-HpaI) are described elsewhere (53, 54).
Plasmids carrying 18xMYC MEC1 in pRS315 were constructed by replacing SpeI-AatII fragments in pRS315 MEC1 with SpeI-AatII fragments amplified by PCR with genomic DNA from DMP3084/3C as a template. The DMP3084/3C strain, in which genomic Mec1 was tagged with 18xMYC, was described previously (44). pRS315 MEC1 containing ∼800 bp of 5′ UTR and ∼500 bp of 3′ UTR was constructed by recombination repair of a gapped plasmid. All plasmids constructed from PCRs were sequenced and tested for expression and function.
The strains used herein are in the W303 background: U960-5C (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sml1-1 rad53-XB::HIS3), yJKD 103 (MATa cdc13-1 cdc15-2 ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sml1Δ::TRP1 rad53Δ::HIS3), yJKD 201 (MATa cdc15-2 ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura1-3 sml1Δ::TRP1 rad53Δ::HIS3 dun1Δ::Kanr), yJKD 403 (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sml1-1 rad53-XB::HIS3 ASF1-3xHA::Kanr), and yJKD 415 (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 sml1-1 rad53-XB::HIS3 dun1Δ::Kanr). DMP3084/3C (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 MEC1-MYC18::LEU2::med1 DDC2-HA3::URA3) and U960-5C were generously provided by M. Longhese and R. Rothstein, respectively. Construction of yeast strains is described elsewhere (54). Strains containing the rad53-XB::HIS3 deletion express a residual carboxyl-terminal fragment of Rad53. yJKD 103 and yJKD 201 have a complete deletion of RAD53.
yEF569HA-212 (MATa leu2 trp1 ura3 his3 mec1::TRP1 sml1-1 RAD9-3xHA pipo [pRS315 18xMYC-MEC1]) is in the A364a background. yEF569, the parental strain for yEF569HA-212, was generously provided by E. Foss. DZ6-1 (MATa/MATα ura3-53/ura3-53 leu2-3,112/leu2-3,112 his3-Δ200/his3-Δ200 lys2-801/+ ade4/+ rad53ΔXB::HIS/+ [pRS316-RAD53]) and DZ71 (MATa rad53ΔXB::HIS [pRS316-RAD53]; segregant from sporulation of DZ6-1) were previously described (75). Most experiments were performed with at least two different strains, and consistent results were observed. Where appropriate, these strains were transformed with the plasmids described above. Yeast strains with cdc13-1 and/or cdc15-2 were grown at 23°C in selectable medium or YPAD (1% yeast extract, 2% Bacto Peptone, 2% dextrose, and 0.05% adenine). Other strains were grown at 30°C.
Cell lysis, immunoprecipitation (IP), and Western blot analysis.
Cells treated with or without 0.1% methyl methanesulfonate (MMS) for 1 h or 200 mM hydroxyurea (HU) for 2 h were washed with washing buffer (phosphate-buffered saline [PBS], 10% glycerol, and 1% Triton X-100). Cell pellets were resuspended in 700 μl of lysis buffer (PBS, 10% glycerol, 1% Triton X-100, 1 mM EDTA, 1% aprotinin, 1 mM phenylmethylsulfonyl fluoride, 10 mM NaF, 20 mM β-glycerophosphate, 5 mM sodium vanadate, and protease inhibitor cocktail [Roche]). Cells were mechanically disrupted in the presence of zirconium beads (Biospec) in a mini-Bead Beater-8 (Biospec). The extract was clarified in a microcentrifuge at 4°C for 10 min. Two to three milligrams of protein was used per IP with 2.5 μg of purified antibody and 40 μl of protein G- plus protein A-agarose beads (Oncogene). IP mixtures were rotated for 2 to 4 h at 4°C, centrifuged, and washed three times in washing buffer. SDS-polyacrylamide gel electrophoresis loading buffer was added to each sample, and the samples were boiled for 5 min. Proteins were separated in 6 or 5 to 15% acrylamide gradient gels and transferred to a polyvinylidene difluoride membrane (Millipore). After 1 h of blocking with 5% milk in TBST (20 mM Tris [pH 7.5], 150 mM NaCl, 0.1% Tween 20), membranes were incubated with primary antibodies overnight and secondary antibodies for 1 h to detect endogenous Rad53 or Rad9. Antibodies to Rad53 or Rad9 were affinity-purified polyclonal rabbit antibodies, as previously described (53, 54, 75). Phosphorylated forms of Rad53 were detected using a rabbit polyclonal antibody that recognizes phosphorylated [S/T]Q motifs (Cell Signaling Technology). HA-, MYC-, or FLAG-tagged proteins were detected by incubation for 1 h with horseradish peroxidase (HRP)-conjugated antibodies (anti-HA-HRP from Roche, anti-MYC-HRP from Santa Cruz, and anti-FLAG-HRP from Sigma).
Kinase assays.
The in situ autophosphorylation assay (ISA) was performed as described previously (45). For Rad53 kinase assays in immune complexes (IP kinase assays) (58), cell lysates from strains expressing FLAG-tagged Rad53 were prepared using HEPES lysis buffer (25 mM HEPES [pH 7.5], 10% glycerol, 0.1% Triton X-100, 1 mM EDTA, 1% aprotinin, 1 mM phenylmethylsulfonyl fluoride, 10 mM NaF, 20 mM β-glycerophosphate, 5 mM sodium vanadate, and protease inhibitor cocktail [Roche]). Two milligrams of protein was incubated for 2 to 3 h with mouse anti-FLAG antibody (Sigma) and protein G- plus protein A-agarose beads. Beads were washed three times with kinase washing buffer (25 mM HEPES [pH 7.5], 10% glycerol, and 0.1% Triton X-100). IP kinase assays were carried out on ice for 1 h in kinase reaction buffer (25 mM HEPES [pH 7.5], 1 mM MnCl2, 1 mM MgCl2, 10 μCi of [γ-32P]ATP, 1 μM nonradioactive ATP) with 4 μg of histone H1 as substrate per reaction. Kinase reactions were stopped by adding SDS-PAGE loading buffer. Samples were resolved in SDS-5 to 15% polyacrylamide gels. The portion of the gel below the 50-kDa protein marker was cut, stained with Coomassie blue to visualize histone H1, dried, and exposed to film. Proteins over 50 kDa were transferred to a polyvinylidene difluoride membrane (Millipore), exposed to film for autoradiography, and immunoblotted for Rad53 and Rad9.
Mec1 IP kinase assays were performed similarly except that 18xMyc Mec1 was immunoprecipitated using mouse anti-Myc antibody (Sigma) and that recombinant Rad53kd 3xFLAG or Rad53kd1-4/9-12AQ 3xFLAG was used as substrate.
Plasmid shuffle assay.
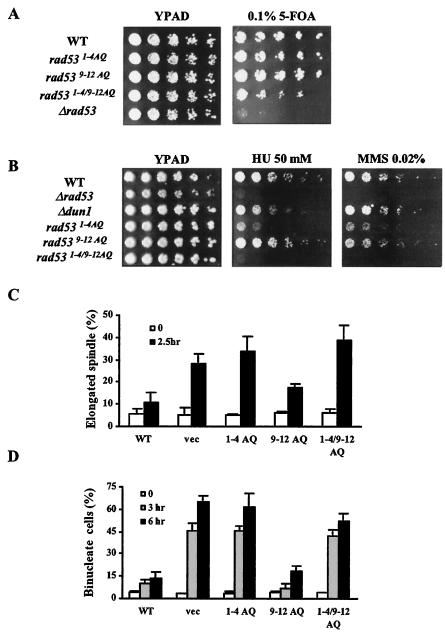
pRS315 RAD53, wild type or with alanine mutations, was introduced into strain DZ71, which carries RAD53 on a URA3 plasmid (pRS316) in a genomic rad53 deletion background. Cells were diluted fivefold serially, spotted onto plates containing 0.1% 5-fluoroorotic acid (5-FOA), and incubated at 30°C for 2 to 4 days.
Viability and checkpoint assays.
For the genotoxin sensitivity assays, cells were diluted fivefold serially and spotted onto YPAD plates containing various concentrations of HU or MMS. Cells were incubated at 30°C for 2 days.
The DNA replication checkpoint was monitored by measuring the percentage of elongated spindles after visualization by indirect immunofluorescence, as previously described (5). Cells were arrested in G1 phase by using α-factor (Sigma) for 2 h and 30 min. Cells were washed three times with water and released into YPAD containing 200 mM HU. Elongated spindles were counted after visualization by indirect immunofluorescence, by using rat antitubulin monoclonal antibody YOL1/34 and fluorescein isothiocyanate-conjugated donkey anti-rat secondary antibody (Jackson Immunoresearch).
G2/M checkpoint function was examined by scoring cells for arrest before or after anaphase in response to DNA damage as described previously (19, 34, 53). Briefly, cdc13-1 cdc15-2 strains were grown at 23°C and arrested in G1 phase for 3 h with α-factor in YPAD. Cells were washed extensively with YPAD and shifted to 37°C. Samples were collected at 0, 3, and 6 h after temperature shift; fixed; stained with DAPI (4′,6′-diamidino-2-phenylindole); and counted for single or double nuclei.
Pull-down experiments.
Expression of six-histidine (6xHis) Rad53 3xFLAG (wild type or kinase defective) in BL21(DE3)pLysS cells was induced by adding 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After 4 h of induction, the cells were pelleted and lysed by sonication in Tris lysis buffer (25 mM Tris [pH 7.5], 300 mM NaCl, 10 mM imidazole, 10% glycerol, and 1% Triton X-100). After centrifugation at 4°C, the supernatant was rotated for 4 to 8 h at 4°C with Ni-nitrilotriacetic acid beads (Qiagen) for 6xHis Rad53 3xFLAG. The beads were washed three times with Tris washing buffer (25 mM Tris [pH 7.5], 300 mM NaCl, 20 mM imidazole, 10% glycerol, and 1% Triton X-100) and eluted with imidazole elution buffer (25 mM Tris [pH 7.5], 300 mM NaCl, 250 mM imidazole, 10% glycerol, and 1% Triton X-100). Rad53 wild type was dephosphorylated with lambda phosphatase for 1 h at 30°C and repurified with the same procedure described above. Glycerol was added to 50%, and the final purified proteins were stored at either −20 or −80°C.
Expression of GST FHA fusion proteins in BL21 cells was induced with 1 mM IPTG. The cells were pelleted after 4 h and lysed by sonication in bacterial PBS lysis buffer (PBS, 10% glycerol, and 1% Triton X-100 with protease inhibitor cocktail [Roche]). Cell lysates were clarified by centrifugation at 4°C and rotated overnight at 4°C with glutathione Sepharose beads (Amersham) to purify GST fusion proteins. The beads were washed three times and resuspended in bacterial PBS lysis buffer. Glycerol was added to 50%, and the purified proteins with the beads were stored at −20°C.
For each pull-down assay, 10 μg of 6xHis Rad53 3xFLAG or 20 μg of GST FHA fusion protein was incubated for 4 h with 2 mg of yeast lysate. The beads were then washed three times with IP washing buffer, and SDS-PAGE loading buffer was added. Approximately half of each sample was used for Western blot analysis.
RESULTS
Rad53 has two major phosphorylation site clusters in vivo.
Rad53 has 16 potential phosphorylation sites with the [S/T]Q sequence favored for phosphorylation by PIKKs (numbered 1 to 16, Fig. 1A). These include a TQ cluster amino terminal to FHA1 situated similarly as regulatory sites in Chk2 and Cds1 (1-4 TQ sites; Fig. 1A), and an SQ cluster amino terminal to FHA2 (9-12 SQ sites; Fig. 1A). Phosphorylation of Rad53 correlates with elevated kinase activity (55), so we evaluated candidate regulatory site mutants for alterations in Rad53 phosphorylation.
FIG.1.
Phosphorylation of Rad53 with [S/T]Q mutations. (A) Schematic diagram of Rad53 showing each [S/T]Q site. SCD, [S/T]Q cluster domain. (B) Western blot analysis of Rad53, Rad9, and Dun1 in cells expressing kinase-active Rad53 combined with alanine substitutions. Strains analyzed are yJKD 415 background plus pRS314 DUN1 13xMYC and pRS316 RAD53, wild type or with mutations. Rad53 (top) and Rad9 (middle) were detected using polyclonal rabbit antibodies for each protein, and Dun1 13xMYC (bottom) was detected with anti-Myc-HRP. Cells were mock treated (lanes C) or treated with 200 mM HU for 2 h (lanes H) or 0.1% MMS for 1 h (lanes M) prior to analysis, and trichloroacetic acid lysates were prepared for Western blotting. (C) Western blot analysis of phosphorylated forms of Rad53 in RAD53, rad53kd, or rad531-4/9-12AQ cells. pRS314 DUN1 13xMYC and pRS316 RAD53 3xFLAG, wild type or with alanine substitution mutations, were introduced into strain yJKD 415. Cells were treated in the same way as described for panel B, and Rad53 was immunoprecipitated with mouse anti-FLAG antibody. Phosphorylated forms of Rad53 were detected by immunoblotting with polyclonal rabbit anti-phosphorylated [S/T]Q antibody. (D) In vitro phosphorylation of Rad53 by Mec1. Mec1 was immunoprecipitated from MMS-treated yEF569HA-212 cell extracts with mouse anti-Myc antibody and incubated at 30°C with recombinant Rad53kd 3xFLAG or Rad53kd1-4/9-12AQ 3xFLAG in the presence of [γ-32P]ATP. The gel was exposed to film overnight for autoradiography (middle). Mec1 or recombinant Rad53 proteins were detected by Western blotting (top and bottom, respectively). (E) Western blot analysis of Rad53 in cells expressing kinase-defective Rad53 with alanine substitutions. Shown is strain U960-5C with pRS316 RAD53 alleles as marked. The experiment was performed as described for panel B. WT, wild type.
Cells with wild-type or mutated Rad53 were exposed to HU (DNA replication-blocking agent) or MMS (alkylating agent). Consistent with previous reports (58), Rad53 was moderately phosphorylated (moderate mobility shift) in response to HU and hyperphosphorylated upon MMS treatment in wild-type RAD53 strains (Fig. 1B, lanes 2 and 3 in top panel), while strains expressing kinase-defective Rad53 (rad53kd) showed enhanced phosphorylation with HU (Fig. 1B, lanes 17 and 18 in top panel). Rad53 hyperphosphorylation upon HU treatment in the rad53kd strain may have resulted from amplification of DNA damage signals in the presence of replicative stress due to the loss of RAD53 function in this strain. The extent of the MMS-dependent mobility shift of Rad53 was significantly diminished in rad531-4AQ (the strain expressing Rad53 with alanine substitutions at T5, T8, T12, and T15; Fig. 1B, lane 9 in top panel) and in rad539-12AQ (the strain expressing Rad53 with alanine substitutions at S473, S480, S485, and S489; Fig. 1B, lane 12 in top panel). However, the Rad53 HU-dependent shift was enhanced in rad531-4AQ cells (Fig. 1B, compare lanes 2 and 8; note that the basal form of Rad53 was decreased and that the majority of Rad53 was hyperphosphorylated), similarly to that in rad53kd strains (Fig. 1B, lane 17 in top panel). There was not a significant change in rad539-12AQ strains compared to RAD53 strains (Fig. 1B, lane 11 in top panel). The MMS- and HU-dependent mobility shifts of Rad53 were largely eliminated in rad531-4/9-12AQ cells (Fig. 1B, lanes 14 and 15 in top panel), which expressed Rad53 with combined substitutions of 1-4 TQ sites and 9-12 SQ sites. In strains expressing Rad53 with single or double alanine substitutions at the remaining [S/T]Q sites (sites 5, 6, 7, 8, 13, 14, and 15-16, Fig. 1A), there were no significant effects on migration of Rad53 in comparison to wild-type Rad53 upon HU or MMS treatment (data not shown).
HU- and MMS-induced phosphorylation of Rad53 was detected using an antibody that recognizes phosphorylated [S/T]Q sequences (PIKKs' consensus phosphorylation sites) in strains with RAD53 or rad53kd (Fig. 1C, lanes 5, 6, 8, and 9) but was not detected in rad531-4/9-12AQ strains (Fig. 1C, lanes 11 and 12).
The simplest explanation for substitutions that reduce mobility shift and phosphorylation at [S/T]Q sites is that they eliminate in vivo phosphorylation by upstream PIKKs. Thus, we examined in vitro phosphorylation of recombinant Rad53kd 3xFLAG or Rad53kd1-4/9-12AQ 3xFLAG by Mec1 immune complexes. Phosphorylation of Rad53kd1-4/9-12AQ 3xFLAG by Mec1 was significantly reduced compared to that of recombinant Rad53kd 3xFLAG (Fig. 1D, compare lanes 3 and 6 in middle panel), despite the presence of many other phosphorylatable amino acids, including eight other consensus PIKK phosphorylation sites.
Alternative explanations would include the possibility that the 1-4 TQ sites and 9-12 SQ sites are regulatory sites that affect Rad53 autophosphorylation or that these mutations may affect the localization of the protein such that it cannot be targeted for phosphorylation by the upstream kinases. To determine how Rad53 autophosphorylation activity contributes to the altered phosphorylation profiles of Rad53 [S/T]Q site mutants, we examined Rad53 phosphorylation in cells expressing kinase-defective Rad53 (rad53kd), with 1-4 TQ and/or 9-12 SQ alanine substitution (rad53kd1-4AQ, rad53kd9-12AQ, and rad53kd1-4/9-12AQ). Cluster mutations with kinase-defective Rad53 caused similar changes in Rad53 phosphorylation regardless of Rad53 kinase activity. Combined TQ and SQ cluster site mutations abolished the slowly migrating forms of Rad53 produced upon HU or MMS treatment (Fig. 1E, lanes 11 and 12). rad53kd or rad53kd1-4AQ strains showed enhanced Rad53 phosphorylation upon HU treatment in a similar manner (note that basal forms of Rad53 are not apparent in Fig. 1E, lanes 2 and 5). In rad53kd9-12AQ cells, Rad53 displayed reduced phosphorylation upon HU treatment, implying that phosphorylation in 9-12 SQ sites is responsible for slow-migrating forms of Rad53 in rad53kd or rad53kd1-4AQ strains. However, the overall phosphorylation was reduced in these strains (Fig. 1B and E). These observations suggested, at minimum, that trans-phosphorylation by upstream kinases occurs on two major clusters of [S/T]Q sites in Rad53 in vivo as well as in vitro and that Rad53 autophosphorylation, possibly on the same 1-4 TQ and 9-12 SQ sites, is required for the full DNA checkpoint-induced phosphorylation of Rad53 in vivo.
The amino-terminal TQ cluster is important for Rad53 functions.
We next investigated whether Rad53 function is compromised by substitutions at the cluster phosphorylation sites. Since Rad53 is an essential gene, we examined the effect of alanine substitutions on the Rad53 essential function with a plasmid shuffle assay. Strains with wild-type Rad53 or alanine substitutions were able to grow on 0.1% 5-FOA plates, while strains with vector only barely grew (Fig. 2A). However, when cells recovered from 0.1% 5-FOA plates were replated on YPAD plates, rad531-4/9-12AQ strains showed slightly slower growth (data not shown). We concluded that alanine substitutions of Rad53 spare the essential function.
FIG. 2.
1-4 TQ sites are important for Rad53 checkpoint function. (A) Viability of cells expressing Rad53 with alanine substitutions. DZ71 strains with rad53 alleles derived from pRS315 were spotted on 0.1% 5-FOA plates. Plates were scored after 3 days at 30°C. (B) Cells with Rad53 mutated in 1-4 TQ sites are sensitive to HU and MMS. yJKD 415 strains with pRS314 DUN1 13xMYC and pRS316 RAD53 or rad53 alleles were used for these experiments (B and C). Cells were diluted serially and spotted on YPAD with or without HU (50 mM) or MMS (0.02%). Plates were photographed after 2 days at 30°C. (C) S/M checkpoint assay. G1-arrested cells were released into HU-containing YPAD medium. Cells were collected and fixed after 2.5 h, and elongated spindles were scored after visualization by indirect immunofluorescence. Experiments were performed three times independently. Averages and standard deviations in the graph were from a representative experiment performed three times. (D) G2/M checkpoint assay. cdc13-1 cdc15-2 cells (yJKD 103) were exposed to DNA damage by being shifted to a restrictive temperature for 3 or 6 h after release from G1 arrest. At each time point, cells were collected, fixed, and stained with DAPI to visualize nuclear morphology. WT, wild type.
Rad53 is phosphorylated and activated upon DNA damage or replicative stress, and Rad53 activation is essential for DNA damage and intra-S-phase checkpoint pathways. One indication of the loss of Rad53 function with genotoxin treatment in alanine substitution mutants was the enhanced Rad53 phosphorylation in rad531-4AQ cells upon HU treatment. Loss of Rad53 function (e.g., with mutations that eliminate kinase activity) similarly enhances Rad53 phosphorylation (Fig. 1B, compare lanes 2 and 17 in top panel), probably by activation of DNA damage checkpoint pathways in S phase as a result of secondary DNA damage caused by replication fork collapse or improper late origin firing (32, 49, 50, 61). Consistent with this conclusion, mutation of the Rad53 1-4 TQ cluster caused Rad9 to become hyperphosphorylated and associated with Rad53 in response to HU (Fig. 1B, compare lanes 2 and 8 in middle panel), a hallmark of DNA damage checkpoint activation. These data indicate that the rad531-4AQ strain is deficient for the response to replicative stress.
We examined the effect of alanine substitutions of Rad53 on viability when cells were spotted on MMS- or HU-containing plates. The assays were conducted with strains with the sml1-1 allele, which bypasses the essential requirement for RAD53 to transcriptionally activate production of ribonucleotide reductase at S phase (71). rad531-4AQ strains were highly sensitive to HU and moderately sensitive to MMS treatment, whereas rad539-12AQ strains were as resistant as strains with wild-type RAD53 (Fig. 2B). rad531-4/9-12AQ cells were more sensitive to HU or MMS than were rad531-4AQ cells. Hence, Rad53 1-4 TQ sites are important for resistance to HU and MMS, and the 9-12 SQ sites are largely redundant in these assays unless 1-4 TQ sites are eliminated.
Since RAD53-dependent resistance to DNA damage and replication is mediated in part through the activation of DNA checkpoint pathways, we determined whether these rad53 strains have DNA checkpoint defects. For the replication checkpoint assay, cells were synchronized in G1 phase with α-factor and released into YPAD containing 200 mM HU. After 150 min, most (90%) of the cells with wild-type Rad53 were arrested as large budded cells with short spindles, while approximately 33% of rad531-4AQ cells had elongated spindles, similar to rad53 deletion strains (28%) (Fig. 2C). rad539-12AQ cells were only slightly impaired in DNA replication checkpoint function.
G2/M DNA damage checkpoint function was assayed by monitoring nuclear division after DNA damage caused by the temperature-sensitive cdc13-1 allele. Cells were synchronized with α-factor and released at a restrictive temperature (34). Cells escaping G2/M checkpoint arrest will arrest instead at telophase due to the cdc15-2 mutation. RAD53 strains exhibited delayed nuclear division and accumulated cells with one nucleus in large budded cells, while rad531-4AQ cells proceeded into mitosis at the same rate as rad53 deletion cells (Fig. 2D). rad539-12AQ cells had normal G2/M checkpoint function. These results indicate that the Rad53 1-4 TQ sites are important for replication and DNA damage checkpoint functions. Rad53 9-12 SQ sites have a minor role in the replication checkpoint arrest and are not required for the G2/M checkpoint.
1-4 TQ sites work redundantly for Rad53 function in genotoxin sensitivity.
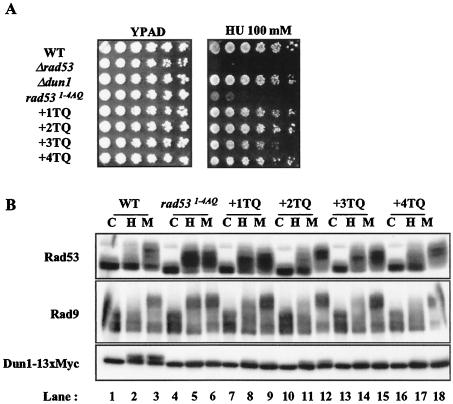
The simultaneous alanine substitution of Rad53 1-4 TQ sites resulted in HU sensitivity. To determine whether individual sites within this cluster are more important for Rad53 function, “add-back” alleles were produced, wherein an individual threonine within the cluster was restored. The individual reintroduction of each site restored viability in HU (Fig. 3A), with a slight defect in the third TQ site add-back allele. Concomitant with restoration of partial or complete HU resistance, the add-back Rad53 proteins displayed HU- and MMS-dependent phosphorylation that was more similar to wild-type Rad53, i.e., with a smaller HU-dependent mobility shift than was seen in rad531-4AQ cells and a greater MMS-dependent mobility shift (Fig. 3B). These observations suggest that individual phosphorylation sites in the Rad53 1-4 TQ cluster are functionally redundant.
FIG. 3.
1-4 TQ sites work redundantly. yJKD 415 strains with pRS314 DUN1 13xMYC and pRS316 RAD53 or rad53 alleles were used for these experiments. (A) Viability test on HU plates. The experiment was performed as described for Fig. 2B. (B) Western blot analysis for the indicated proteins. Cells were mock treated (lanes C) or treated with HU (lanes H) or MMS (lanes M) prior to analysis. WT, wild type.
Effects of phosphorylation site mutations on Rad53 kinase activity.
Alteration of the 1-4 TQ phosphorylation sites caused defects in Rad53 phosphorylation, DNA checkpoint functions, and viability in response to HU or MMS. Loss of the [S/T]Q clusters could alter cellular responses by impairing Rad53 activation or by interference with coupling of Rad53 to downstream responses.
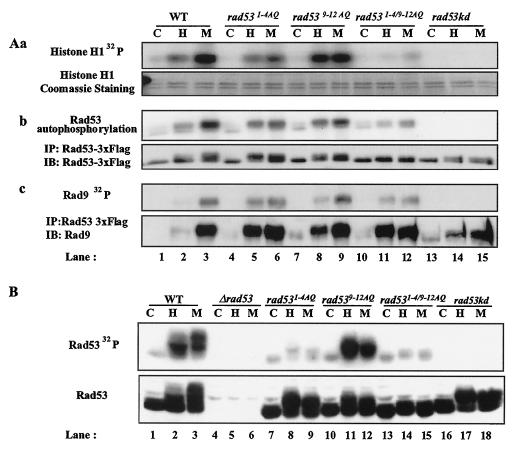
First, we determined the contribution of the Rad53 [S/T]Q cluster sites to Rad53 kinase activity and regulated activation. We used immune complex kinase assays to measure autophosphorylation and transphosphorylation of Rad53 isolated from strains with Rad53 mutations (Fig. 4Aa and Ab). In wild-type RAD53 cells, MMS, and to a lesser extent HU, enhanced total Rad53 autophosphorylation and transphosphorylation activity (Fig. 4Aa and Ab, lanes 2 and 3). Upon HU or MMS treatment, rad531-4AQ cells retained kinase activity and showed regulated activation measured by autophosphorylation or histone H1 phosphorylation in trans (Fig. 4Aa and Ab, lanes 5 and 6). However, in five out of six experiments, the observed extent of Rad53 autophosphorylation and transphosphorylation of histone H1 was partially decreased by mutation of the TQ cluster relative to that of wild-type Rad53. The reduction of kinase activity in rad531-4AQ cells was more significant in response to MMS (Fig. 4Aa and Ab, compare lanes 3 and 6). Since rad531-4AQ cells displayed hypershifted forms of Rad53 upon HU treatment, possibly owing to strong phosphorylation of 9-12 SQ sites, it is possible that phosphorylation in 9-12 SQ sites can partially compensate for the absence of phosphorylation in 1-4 TQ sites in Rad53 activation.
FIG. 4.
Kinase activity of Rad53 mutants. (A) Rad53 immune complex kinase assays. yJKD 415 strains with pRS314 DUN1 13xMYC and pRS316 RAD53 3xFLAG alleles were used for these experiments. (Aa) trans-phosphorylation activity of Rad53 with histone substrate. Rad53 was immunoprecipitated with antibody against FLAG from cells treated with or without HU or MMS and incubated on ice with histone H1 in the presence of [γ-32P]ATP. Gels were exposed to film overnight (top). Histone H1 was visualized by Coomassie blue staining after SDS-5 to 15% PAGE (bottom). (Ab) Autophosphorylation activity of Rad53. (Top) Autoradiogram of Rad53 autophosphorylation activity; (bottom) Western blot of Rad53. (Ac) Rad9, coimmunoprecipitated with Rad53 and phosphorylated in Rad53 immune complexes. (Top) Autoradiogram of Rad9 phosphorylated by Rad53; (bottom) Rad9 Western blot after IP with anti-Rad53 antibodies. Cells were mock treated (lanes C) or treated with HU (lanes H) or MMS (lanes M) prior to analysis. (B) Rad53 ISA. Experiments were performed as described for Fig. 1B with the same strains. Rad53 was detected by Western blotting following ISA. WT, wild type.
rad539-12AQ cells retained comparable levels of Rad53 kinase activation or showed increased kinase activity with HU treatment (Fig. 4Aa and Ab, lanes 8 and 9). rad531-4/9-12AQ cells displayed significant decreases in kinase activation, although some residual regulation by HU and MMS was retained (Fig. 4Aa and Ab, lanes 11 and 12).
Similar results were obtained using ISA to measure Rad53 activation (Fig. 4B). These assays measure Rad53 phosphorylation after denaturing gel electrophoresis and renaturation in situ. With this assay Rad53 activation was severely impaired with 1-4 alanine substitutions (Fig. 4B, compare lanes 2 and 3 and lanes 8 and 9), which is slightly different from the results of IP kinase assays. It is possible that the 1-4 TQ sites are important phosphorylation sites in the ISA, that 1-4 substitutions impair refolding in situ, or that other proteins present in immune complexes account for this difference. Together, these results indicate that combined phosphorylation in 1-4 TQ and 9-12 SQ sites is required for Rad53 activation, and between them, mutation of 1-4 TQ sites has a greater effect on Rad53 activation.
Rad53 1-4 TQ mutants interact with Asf1 and Rad9.
We next investigated whether the Rad53 [S/T]Q cluster mutations interfere with binding of other proteins to Rad53. DNA damage induces interaction of Rad53 with Rad9, which is required to couple Rad53 to the damage response (53, 59, 62). The MMS-induced Rad9 phosphorylation and coimmunoprecipitation with Rad53 were largely normal in rad531-4AQ, rad539-12AQ, and rad531-4/9-12AQ cells (Fig. 4Ac). With HU treatment, Rad53 interacted with Rad9 in rad531-4AQ and rad531-4/9-12AQ cells, and to a lesser extent in rad539-12AQ cells, as Rad53 with [S/T]Q mutations caused secondary DNA damage due to defective checkpoint functions. In addition, Rad9 coimmunoprecipitated with Rad53 was phosphorylated in Rad53 immune complex kinase assays in rad531-4AQ and rad539-12AQ strains, with reduced phosphorylation in rad531-4/9-12AQ cells (Fig. 4Ac), consistent with results in other assays (Fig. 4Aa and Ab).
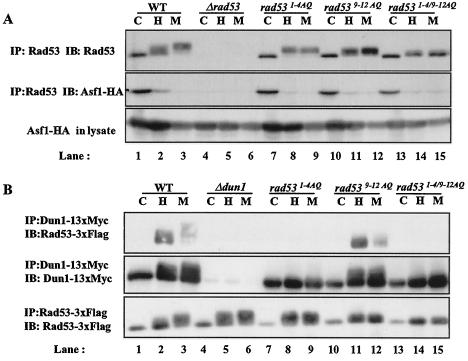
Rad53 interacts with the chromatin assembly factor Asf1 in the absence of DNA damage (17, 25). The basal interaction and the regulated dissociation of Rad53 and Asf1 were not greatly impaired in cells with Rad53 alanine substitutions (Fig. 5A). These results indicate that the Rad53 [S/T]Q clusters are not required for the interaction of Rad53 with Rad9 or Asf1.
FIG. 5.
1-4 TQ sites are required for interaction between Rad53 and Dun1. Alanine substitutions do not disrupt basal interaction between Rad53 and Asf1. (A) Asf1 coimmunoprecipitated with Rad53. yJKD 403 strains were transformed with rad53 alleles derived from pRS316. Rad53 was immunoprecipitated with polyclonal rabbit antibody against Rad53, and copurified Asf1 was detected using mouse anti-HA antibody conjugated to HRP. (B) Rad53 coimmunoprecipitated with Dun1. pRS314 DUN1 13xMYC and pRS316 RAD53 3xFLAG with mutations were introduced into yJKD 415. Dun1 was immunoprecipitated with mouse anti-Myc antibody, and Rad53 was detected using mouse anti-FLAG antibody conjugated with HRP. Cells were mock treated (lanes C) or treated with HU (lanes H) or MMS (lanes M) prior to analysis. WT, wild type.
1-4 TQ site mutations in Rad53 disrupt interaction between Rad53 and Dun1.
Dun1 is a protein serine/threonine kinase with a single FHA domain (7, 76). Dun1 is phosphorylated in a RAD53-dependent manner upon DNA damage and replication blockade and forms a complex with phosphorylated Rad53 (24). Mutation of the FHA domain in Dun1 reduces DNA damage-dependent Dun1 phosphorylation and impairs checkpoint functions and survival in the presence of HU or MMS (7). Dun1 allows activation of transcriptional targets of the DNA damage response pathway by phosphorylation and inhibition of the transcriptional repressor Crt1 (26). Dun1 also phosphorylates Sml1, the inhibitor of the large subunit of ribonucleotide reductase, targeting it for degradation (71, 74).
Since Dun1 is a likely in vivo substrate of Rad53, we measured Dun1 phosphorylation as an indicator of in vivo Rad53 kinase activity. HU- and MMS-induced phosphorylation of Dun1 was normal in rad539-12AQ strains (Fig. 1B, lanes 11 and 12). However, Dun1 phosphorylation was inhibited in rad531-4AQ (Fig. 1B, lanes 8 and 9). Deletion of DUN1 slightly decreased viability upon HU treatment in a sml1-1 background, which suppresses the DNA damage sensitivity due to deletion of DUN1, though it was not as severe as in rad531-4AQ cells (Fig. 2B). Restoration of single TQ sites in the Rad53 1-4 TQ cluster only partially restored the phosphorylation of a Dun1, suggesting that single-site phosphorylation in this portion of Rad53 is not sufficient to induce full Dun1 phosphorylation (Fig. 3B). These results can be interpreted to mean either that Rad53 with 1-4 TQ mutations does not have in vivo kinase activity or that these mutations cause disruption in DNA damage-induced interaction between Dun1 and Rad53.
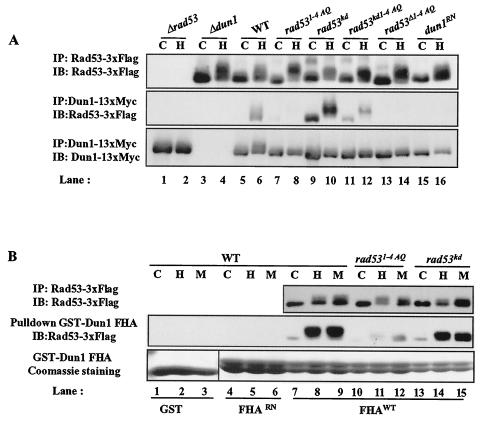
Since Rad53 kinase was partially activated in rad531-4AQ cells (Fig. 4A), we examined interactions between Dun1 and Rad53. This interaction was largely eliminated in rad531-4AQ cells (Fig. 5B, lanes 8 and 9) but was unaffected in rad539-12AQ cells (Fig. 5B, lanes 11 and 12). In rad531-4/9-12AQ strains, HU- and MMS-induced interactions between Rad53 and Dun1 were attenuated (Fig. 5B, lanes 14 and 15). These results suggest that the phosphorylation of the Rad53 1-4 TQ sites is required for coupling Dun1 to Rad53.
Dun1 FHA domain interacts with phosphorylated 1-4 TQ cluster of Rad53 in a Rad53 kinase-independent manner.
Because Dun1 interacts with Rad53 in a phosphorylation-dependent manner (24), and FHA domains generally recognize phosphorylated proteins, we determined if the FHA domain of Dun1 interacts with the phosphorylated 1-4 TQ cluster region of Rad53.
Either mutation of Rad53 1-4 TQ sites or an inactivating amino acid substitution in the Dun1 FHA domain disrupted HU-dependent interaction between Rad53 and Dun1 (Fig. 6A, lanes 8 and 16). Furthermore, GST-Dun1 FHA domain fusion proteins expressed in bacteria were sufficient to pull down Rad53 from HU- or MMS-treated cell lysates but not from control cell lysates (Fig. 6B, lanes 7 to 9). A GST-Dun1 FHA fusion protein with the inactivating RN mutation (Dun1 FHARN; Dun1 FHAR60A/N103A) did not react with phosphorylated Rad53 (Fig. 6B, lanes 4 to 6), confirming that the FHA domain of Dun1 is responsible for interaction between Dun1 and phosphorylated Rad53. The precipitation of Rad53 by the GST-Dun1 FHA domain fusion protein was reduced by substitution of the Rad53 1-4 TQ cluster (Fig. 6B, lanes 10 to 12). Since it is formally possible that the 1-4 TQ region harbors a basal inhibitory domain that keeps Rad53 from interacting with Dun1 prior to checkpoint activation, we determined whether deletion of the 1-4 TQ sites would restore interaction between Rad53 and Dun1 upon HU treatment. This Rad53 deletion mutant did not interact efficiently with Dun1 in vivo (Fig. 6A, lane 14), nor with GST-Dun1 FHA proteins (data not shown), suggesting that 1-4 TQ sites are directly involved in the interaction of Dun1 with Rad53.
FIG. 6.
Binding of Rad53 mutants to Dun1 FHA domains. yJKD 415 (A) and yJKD 201 (B) with pRS314 DUN1 13xMYC and pRS316 RAD53 3xFLAG were used. (A) Rad53 coimmunoprecipitated with Dun1. (B) GST-Dun1 FHA and Dun1 FHA with mutation were purified from bacteria. Twenty micrograms of GST fusion proteins was used to pull down Rad53 from 2 mg of yeast lysate. GST fusion proteins were visualized by Coomassie blue staining. Cells were mock treated (lanes C) or treated with HU (lanes H) or MMS (lanes M) prior to analysis. WT, wild type.
Interestingly, the interaction between Dun1 and Rad53 does not require Rad53 kinase activity, implying that phosphorylation by other kinases, possibly Mec1 and Tel1, is sufficient to induce this interaction (Fig. 6A, lanes 9 and 10, and 6B, lanes 13 to 15). Thus, the PIKKs are required not only for Rad53 activation but also for creation of an interface joining Rad53 and Dun1. The increased association of kinase-defective Rad53 with Dun1 may reflect the secondary activation of DNA damage checkpoint pathways as a result of the loss of Rad53 function.
Specificity and sequence context dependence of FHA domains.
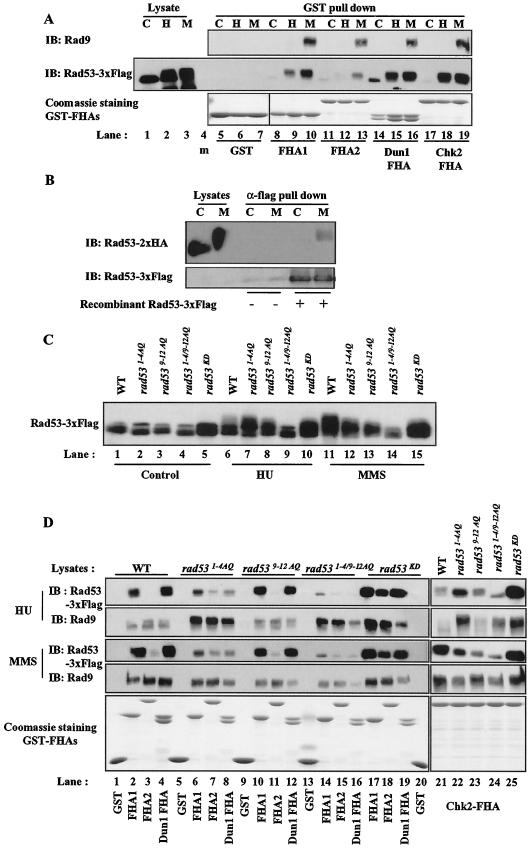
Chk2, the mammalian homolog of Rad53, oligomerizes upon DNA damage in a Chk2 phosphorylation-dependent manner that involves the FHA domain on one Chk2 molecule interacting with the phosphorylated [S/T]Q cluster on another Chk2 molecule (2, 69). Similarly, we isolated a carboxyl-terminal fragment of Rad53 (beginning with residue 432) in a two-hybrid screen using Rad53 as bait (Z. Sun and D. Stern, unpublished data) and observed that Rad53 FHA1 preferentially interacts with phosphorylated Rad53 compared to Rad53 FHA2 (Fig. 7A) (54). We also observed that endogenous Rad53 from yeast interacted with dephosphorylated recombinant Rad53 from bacteria upon MMS treatment (Fig. 7B).
FIG.7.
Specific binding of FHA domains to Rad53 mutants. yJKD 415 with pRS314 DUN1 13xMYC and pRS316 RAD53 3xFLAG was used. (A, C, and D) Rad53 and Rad9 pulled down by GST-Rad53 FHA1 (FHA1), GST-Rad53 FHA2 (FHA2), GST-Dun1 FHA (Dun1 FHA), and GST-Chk2 FHA (Chk2 FHA). Twenty micrograms of GST fusion protein was used to pull down Rad53 and Rad9 from 2 mg of yeast lysate from cells expressing wild-type RAD53 (A) or rad53 mutations (C and D) with or without treatment. Rad53 and/or Rad9 was detected by Western blotting with polyclonal rabbit antibody against each protein (A, C, and D). GST fusion proteins were visualized by Coomassie blue staining (A and D). Rad53 in the lysates from cells with wild-type or mutated Rad53 is shown in panel C. Rad53 and Rad9 Western blotting from GST pull-down assays from each lysate (top four panels) and Coomassie blue staining of GST proteins (bottom) are shown in panel D. (B) Interaction between endogenous Rad53 in yeast and dephosphorylated recombinant Rad53 from bacteria upon DNA damage. U960-5C strains were transformed with pRS316 RAD53 2xHA. Ten micrograms of recombinant Rad53 3xFLAG was used to pull down endogenous Rad53 2xHA in 2 mg of yeast lysate. Cells were mock treated (lanes C) or treated with HU (lanes H) or MMS (lanes M) prior to analysis (m, marker). WT, wild type.
To determine if Rad53 FHA1, like the Chk2 FHA domain, interacts with a PIKK phosphorylation site cluster, we performed pull-down assays with the GST-Rad53 FHA1 domain (Fig. 7A). The Rad53 FHA1 domain bound to Rad53 from HU-treated and MMS-treated cells but not from control cells (Fig. 7A, lanes 8 to 10). The 1-4 TQ substitution in Rad53 slightly impaired the isolation of Rad53 from HU- and MMS-treated cells by the FHA1 domain, without affecting the interaction with phosphorylated Rad9 from the same cell extracts (Fig. 7D, lanes 2 and 6). Mutation of Rad53 9-12 SQ sites had a minor effect, or in HU treatment, increased on binding of Rad53 FHA1 to Rad53, despite the substantial contribution of these sites to MMS-dependent Rad53 mobility shift (Fig. 7C, compare lanes 11 and 13; Fig. 7D, lane 10). Double mutation of 1-4 TQ and 9-12 SQ sites inhibited the interaction between Rad53 and the Rad53 FHA1 domain (Fig. 7D, lane 14). Taken together, these data suggest that the Rad53 1-4 TQ sites are important for regulated Rad53-Rad53 interaction, probably through direct binding of the FHA domain to this phosphopeptide, with a minor contribution from the 9-12 SQ sites.
The binding specificity of FHA domains was studied in vitro using phosphopeptide libraries (9, 15, 23, 66), but there are few in vivo binding sites known for FHA domain proteins (53, 69). Both Rad53 FHA1 and Dun1 FHA apparently recognize phosphorylated threonine residues in the 1-4 TQ cluster. In contrast, FHA2 had a similar ability to recognize phosphorylated Rad9 but weakly detected hyperphosphorylated Rad53 from MMS-treated wild-type cell lysates (Fig. 7A, lane 13, and 7D, lane 3) (54). The relatively weak binding of Rad53 FHA2 to Rad53 was barely affected by 9-12 SQ substitution or the double substitution of the Rad53 [S/T]Q clusters but was slightly increased by 1-4 TQ cluster substitution in HU treatment (Fig. 7D, lanes 3, 7, 11, 15, and 18). These results demonstrate site-specific binding preferences of FHA domains in vivo and show that, unlike Rad53 FHA1 and Dun1 FHA, Rad53 FHA2 discriminates in vivo between phosphorylated Rad9 and phosphorylated Rad53.
To further compare interactions between phosphorylated proteins and FHA domains from different proteins, we performed pull-down experiments with the human Chk2 FHA domain. The human Chk2 FHA domain displayed mixed characteristics in these assays (Fig. 7D, lanes 21 to 25). Like the homologous Rad53 FHA1, the Chk2 FHA bound Rad53 from HU- and MMS-treated cells. However, the Chk2 FHA domain interacted better with Rad53 from HU-treated rad531-4AQ cells than with that from HU-treated wild-type cells (Fig. 7D, HU panel, lanes 21 and 22), under conditions where FHA1 and Dun1 FHA binding were diminished. The simplest interpretation is that the Chk2 FHA domain preferentially recognizes the 9-12 SQ sites rather than the 1-4 TQ sites and that 9-12 SQ sites are hyperphosphorylated with HU treatment in rad531-4AQ cells (Fig. 7D, lanes 21 and 22). Consistent with this interpretation, in rad539-12AQ cells, interaction between Chk2 FHA and Rad53 was reduced in MMS- and HU-treated cells, whereas this mutation had little impact on interaction between Rad53 and either Rad53 FHA1 or Dun1 FHA. However, in vitro studies with peptide libraries have shown that FHA domains prefer phosphorylated threonine to serine. An alternative explanation is that rad539-12AQ may cause phosphorylation in other remaining TQ sites, creating binding sites for FHA2 and Chk2 FHA domains.
The interactions between GST fusion FHA domains and Rad53 were Rad53 phosphorylation dependent, since the interactions were greatly diminished in rad531-4/9-12AQ cells (Fig. 7D, lanes 14 to 16 and 24). Interactions with each FHA domain still occurred with kinase-defective Rad53 (Fig. 7D, lanes 17 to 19 and 25), so phosphorylation by other kinases is sufficient to induce these interactions. Overall, these observations show that FHA domains recognize phosphorylated threonines, and possibly serines, in sequence context in vivo and that FHA domains can discriminate among DNA checkpoint proteins at sites phosphorylated in vivo.
DISCUSSION
We conducted a mutational analysis to determine the function of consensus PIKK phosphorylation sites within Rad53. We found that the Rad53 amino-terminal 1-4 TQ cluster region is important for Rad53 function. Alanine substitution mutations in these sites caused decreased viability after HU treatment and impaired replication blockade and DNA damage checkpoint function of Rad53. These mutations caused a reduction in DNA damage-induced Rad53 kinase activation and impaired interactions between Rad53 and Dun1 upon DNA damage or replication blockade. Phosphorylated Rad53 1-4 TQ sites are directly recognized by the FHA domain of Dun1, regardless of Rad53 kinase activity. Thus, Mec1 and Tel1 are not only important for Rad53 activation but also create a Rad53-Dun1 interaction interface.
Different FHA domains show different specificities toward in vivo-phosphorylated proteins.
Several proteins involved in DNA checkpoint signaling pathways contain FHA domains. Although binding of FHA domains to artificial phosphopeptides has been extensively characterized in vitro, only a few FHA-binding sites have been identified in vivo. These include the sites of interaction of Chk2 with its own SCD ([S/T]Q cluster domain) (2, 69), of Rad53 with phosphorylated Rad9 (53, 59), and now of Rad53 FHA1 and Dun1 with the Rad53 1-4 TQ cluster.
Earlier work with in vitro peptide binding assays suggested that different FHA domains have different binding specificities (9, 15, 23, 66). We now provide in vivo evidence for the importance of FHA domain specificity, which is suggested by the fact that different FHA domains recognize different substrates in vivo. The differences in binding nature and specificity were predicted previously (15): the interaction between Rad53 FHA2 and Rad9 is not decreased in the presence of phosphorylated peptides which can compete the interaction of Rad53 FHA1 with Rad9. Based on biological evidence and the results of GST pull-down experiments, it appears that Rad53 FHA2 has in vivo binding preferences distinct from those of Rad53 FHA1 and the Dun1 FHA domain. The well-conserved Rad53 FHA1 and Dun1 FHA evidently prefer the phosphorylated Rad53 1-4 TQ cluster. The more divergent Rad53 FHA2 domain prefers a highly phosphorylated form of Rad53, which presumably includes phosphorylation at the 9-12 SQ cluster and other sites. In further support of in vivo FHA domain specificity, it has recently been found that FHA1, but not FHA2, mediates interactions between Rad53 and Asf1 and also Ptc2/3 (30, 54). The Chk2 FHA domain showed mixed characteristics of those seen with Rad53 FHA1 and FHA2 domains (Fig. 7D). In vitro, the FHA domains tested have a strong preference for phosphorylated TQ over phosphorylated SQ sites, consistent with the importance of Rad9 phosphorylated TQ sites for binding of Rad53 FHA1 and FHA2 and of the Rad53 1-4 TQ cluster for binding of Rad53 FHA1 and Dun1 FHA. Hence, it will be of interest to determine whether the phosphorylated Rad53 9-12 SQ cluster is recognized in vitro by Rad53 FHA2 and Chk2 FHA domains.
Phosphorylation of Rad53 [S/T]Q clusters is important for Rad53 activation and function.
Alanine substitution of the Chk2 amino-terminal [S/T]Q cluster including threonine 68 (3, 38, 39) and SpCds1 threonine 11 (60), which lie in [S/T]Q clusters similarly situated as the 1-4 TQ cluster in Rad53, reduces phosphorylation by upstream PIKKs ATM and Rad3, phosphorylation of downstream targets, and functional activity in vivo. Our results indicate that phosphorylation of the [S/T]Q clusters within Rad53 has a similar function.
Mutation of both sets of Rad53 [S/T]Q cluster sites greatly affected HU- and MMS-dependent Rad53 phosphorylation. Based on precedent with Chk2 (2, 3, 38, 39, 69), [S/T]Q cluster site phosphorylation is likely to be mediated by a combination of trans-phosphorylation by PIKKs and homotypic intra- or intermolecular autophosphorylation. The greater effect of Rad53 9-12 SQ site substitutions on MMS-dependent mobility shift of kinase-defective Rad53 may indicate that these sites are preferential targets for PIKKs. Consistent with this idea, a carboxyl-terminal fragment of Rad53 is phosphorylated in vitro by Mec1 (63). However, an alternative interpretation is that phosphorylation of the 9-12 SQ sites may have a greater impact on electrophoretic mobility of Rad53 than phosphorylation of the 1-4 TQ sites does.
Substitution of both phosphorylation site clusters eliminated HU- and MMS-dependent Rad53 mobility shift and all of the biological activities measured. These substitutions did not compromise the basal binding of Rad53 to Asf1 or the DNA damage-dependent binding of Rad53 to phosphorylated Rad9. The latter finding indicates that the phosphorylation site mutations do not directly affect modulation by this upstream regulator of the DNA damage checkpoint.
Phosphorylation directly regulates Rad53 activity (45) and affects the formation of phosphorylation-dependent homo- and heteromeric complexes. DNA damage enables Chk2 oligomerization through FHA-phosphopeptide interactions. This may enable a transition from upstream PIKK-dependent activation of Chk2 to PIKK-independent activation mediated by other active Chk2 molecules (2, 69). Our results now suggest a similar interaction of Rad53 FHA1, and perhaps FHA2, with phosphorylated Rad53 (Fig. 7) (54). At least one of the [S/T]Q cluster sites is required for this interaction. Dephosporylated recombinant Rad53 produced in bacteria can bind to phosphorylated Rad53 from HU- or MMS-treated yeast cells (Fig. 7B). Finally, Rad53 can phosphorylate other Rad53 molecules efficiently. However, Rad53-dependent activation of Rad53 may also be fostered through binding to Rad9 oligomers (20; S.-J. Lee and D. F. Stern, unpublished data). Phosphorylated Rad9-Rad53 interactions are intact in Rad53 1-4 TQ site mutants, which have severe phenotypes. Perhaps the 1-4 TQ sites are involved exclusively in Rad53-dependent, Rad9-, and PIKK-independent activation of Rad53 molecules that may occur after Rad53 has been released from sites of DNA damage.
Mutation of the amino-terminal Rad53 TQ cluster impairs Rad53 function and activation, whereas mutation of the 9-12 SQ sites has only minor effects. The effects of Rad53 9-12 SQ substitutions were most evident in the absence of Rad53 1-4 TQ sites, suggesting that they play partially redundant roles. Thus, there are important functional differences between these two Rad53 phosphorylation site clusters independent of the overall extent of Rad53 phosphorylation. Curiously, rad539-12AQ cells consistently showed increased Rad53 kinase activity after HU treatment (Fig. 4). This may be due to an enhanced phosphorylation at the functionally superior Rad53 1-4 TQ sites in the absence of 9-12 SQ sites or an overall signal amplification resulting from indirect DNA damage due to loss of the 9-12 SQ sites.
Overall, mutation of Rad53 1-4 TQ sites had greater effects than did mutation of 9-12 SQ sites, including a moderate effect on catalytic activation of Rad53. While amino-terminal TQ sites are important for activation and function of SpCds1 (60), mammalian Chk2 (3, 38, 39), and ScRad53 (this study), the underlying mechanism is not clear. It was suggested that intramolecular interaction basally inhibits Chk2 and that this intramolecular interaction is relieved upon phosphorylation at threonine 68. However, deletion of the Rad53 1-4 TQ domain did not rescue viability on HU and led to a more severe phenotype than did deletion of DUN1, just as mutation of the 1-4 TQ sites did (Fig. 2) (Lee and Stern, unpublished). Chk2 threonine 68 phosphorylation is required for initial activation of Chk2 that leads to autophosphorylation in the catalytic activation loop (29), but once Chk2 is activated, phosphorylation at this site is not required for maintenance of elevated Chk2 kinase activity (1). In a similar way, Rad53 1-4 TQ sites may be involved in initiation or maintenance of Rad53 kinase activity.
Substitution of Rad53 1-4 TQ sites severely affected resistance to HU and MMS and DNA checkpoint functions. A component of these effects is contributed by the failure to activate Dun1. However, the sml1-1 mutation, which was used to maintain the viability of rad53 strains, suppresses genotoxin sensitivity caused by the loss of DUN1 (73). Hence, it is not yet clear what causes the greatly decreased viability in rad531-4AQ cells on HU and MMS treatment. Rad53 activation in response to HU treatment in rad531-4AQ cells was only moderately decreased compared to that of the wild type (Fig. 4A). Was the activation of Rad53 insufficient to carry out checkpoint functions of Rad53 in rad531-4AQ cells? Or is phosphorylation of Rad53 1-4 TQ sites involved in other binding interactions or dissociation of Rad53 from its activators or downstream targets? It will be of interest to determine whether mutation of the Rad53 1-4 TQ sites affects phosphorylation of other Rad53 downstream targets or impairs their dynamic interactions with Rad53.
Mec1 and/or Tel1 creates a binding interface between Rad53 and Dun1.
PIKKs have at least four roles in DNA damage and replicative stress checkpoint pathways. They may be involved in direct recognition of at least some types of DNA lesions, since Mec1 localizes to sites of DNA damage in the absence of other checkpoint proteins (28, 41). They phosphorylate coupling-activator proteins like ScRad9 (16, 59, 62). PIKK-dependent phosphorylation is important for catalytic activation of Rad53, Chk2 (3, 38, 39; this study), and Chk1 (22, 31, 36) and finally Rad53 and Chk2 oligomerization (2, 69). Here, we found that upstream kinases create a binding interface between Rad53 and its downstream Dun1 by phosphorylating Rad53 1-4 TQ sites. This novel function of Rad53 extends the known functionality of PIKKs and demonstrates that Rad53-like kinases can serve as stable platforms for recruitment of substrate proteins.
DNA damage signaling cascades have been hypothesized to resemble signaling pathways regulated by receptor kinases. The analogy is apt, since it is now known that DNA damage induces formation of dynamic signaling complexes stabilized in part by protein-phosphopeptide interactions. Oligomerization-induced autophosphorylation of receptor kinases creates binding sites for adaptors and other signaling proteins containing SH2 and PTB domains, which recognize phosphorylated tyrosine residues (52). In transforming growth factor β signaling pathways, ligand-activated transforming growth factor β receptor type II activates receptor type I by phosphorylating the GS domain, which creates a binding interface for R-Smads (37). In a similar way, phosphorylation by PIKKs enables activating interactions between Rad9 and Rad53 and plays a direct role in Rad53 activation. Moreover, it creates an interface for binding the effector kinase Dun1 by phosphorylating the Rad53 1-4 TQ sites. Signal amplification by these cascades is accelerated by the polyvalency of these interactions and by the fact that these interactions, by scaffolding kinase-kinase complexes, create additional active kinase molecules.
Acknowledgments
We thank R. Rothstein, T. Weinert, E. Foss, M. Longhese, and M. Snyder for providing strains and reagents. We thank Jia Li, Jonathan McMenamin-Balano, Lyuben Tsvetkov, and Xingzhi Xu for critical reading and JoAnn Falato for assistance.
This work was supported by R01CA USPHS R01CA82257 (D.F.S.). S.-J.L. was supported by USAMRMC Predoctoral Training Program in Breast Cancer Research DAMD17-99-1-946 and by USAMRMC DAMD 17-03-1-0355. M.F.S. was supported by NIH NRSA T32GM07223 from the NIGMS and by USAMRMC DAMD 17-99-1-9460.
REFERENCES
- 1.Ahn, J., and C. Prives. 2002. Checkpoint kinase 2 (Chk2) monomers or dimers phosphorylate Cdc25C after DNA damage regardless of threonine 68 phosphorylation. J. Biol. Chem. 277:48418-48426. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, J. Y., X. Li, H. L. Davis, and C. E. Canman. 2002. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the Forkhead-associated domain. J. Biol. Chem. 277:19389-19395. [DOI] [PubMed] [Google Scholar]
- 3.Ahn, J. Y., J. K. Schwarz, H. Piwnica-Worms, and C. E. Canman. 2000. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 60:5934-5936. [PubMed] [Google Scholar]
- 4.Alcasabas, A. A., A. J. Osborn, J. Bachant, F. Hu, P. J. Werler, K. Bousset, K. Furuya, J. F. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3:958-965. [DOI] [PubMed] [Google Scholar]
- 5.Allen, J. B., Z. Zhou, W. Siede, E. C. Friedberg, and S. J. Elledge. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2401-2415. [DOI] [PubMed] [Google Scholar]
- 6.Anderson, L., C. Henderson, and Y. Adachi. 2001. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol. Cell. Biol. 21:1719-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bashkirov, V. I., E. V. Bashkirova, E. Haghnazari, and W. D. Heyer. 2003. Direct kinase-to-kinase signaling mediated by the FHA phosphoprotein recognition domain of the Dun1 DNA damage checkpoint kinase. Mol. Cell. Biol. 23:1441-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell, D. W., J. M. Varley, T. E. Szydlo, D. H. Kang, D. C. Wahrer, K. E. Shannon, M. Lubratovich, S. J. Verselis, K. J. Isselbacher, J. F. Fraumeni, J. M. Birch, F. P. Li, J. E. Garber, and D. A. Haber. 1999. Heterozygous germ line hCHK2 mutations in Li-Fraumeni syndrome. Science 286:2528-2531. [DOI] [PubMed] [Google Scholar]
- 9.Byeon, I. J., S. Yongkiettrakul, and M. D. Tsai. 2001. Solution structure of the yeast Rad53 FHA2 complexed with a phosphothreonine peptide pTXXL: comparison with the structures of FHA2-pYXL and FHA1-pTXXD complexes. J. Mol. Biol. 314:577-588. [DOI] [PubMed] [Google Scholar]
- 10.de la Torre Ruiz, M. A., and N. F. Lowndes. 2000. DUN1 defines one branch downstream of RAD53 for transcription and DNA damage repair in Saccharomyces cerevisiae. FEBS Lett. 485:205-206. [DOI] [PubMed] [Google Scholar]
- 11.de la Torre-Ruiz, M. A., C. M. Green, and N. F. Lowndes. 1998. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 17:2687-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DiTullio, R. A., Jr., T. A. Mochan, M. Venere, J. Bartkova, M. Sehested, J. Bartek, and T. D. Halazonetis. 2002. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat. Cell Biol. 4:998-1002. [DOI] [PubMed] [Google Scholar]
- 13.Dohrmann, P. R., G. Oshiro, M. Tecklenburg, and R. A. Sclafani. 1999. RAD53 regulates DBF4 independently of checkpoint function in Saccharomyces cerevisiae. Genetics 151:965-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duncker, B. P., K. Shimada, M. Tsai-Pflugfelder, P. Pasero, and S. M. Gasser. 2002. An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing. Proc. Natl. Acad. Sci. USA 99:16087-16092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durocher, D., I. A. Taylor, D. Sarbassova, L. F. Haire, S. L. Westcott, S. P. Jackson, S. J. Smerdon, and M. B. Yaffe. 2000. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol. Cell 6:1169-1182. [DOI] [PubMed] [Google Scholar]
- 16.Emili, A. 1998. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell 2:183-189. [DOI] [PubMed] [Google Scholar]
- 17.Emili, A., D. M. Schieltz, J. R. Yates III, and L. H. Hartwell. 2001. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol. Cell 7:13-20. [DOI] [PubMed] [Google Scholar]
- 18.Fay, D. S., Z. Sun, and D. F. Stern. 1997. Mutations in SPK1/RAD53 that specifically abolish checkpoint but not growth-related functions. Curr. Genet. 31:97-105. [DOI] [PubMed] [Google Scholar]
- 19.Gardner, R., C. W. Putnam, and T. Weinert. 1999. RAD53, DUN1 and PDS1 define two parallel G2/M checkpoint pathways in budding yeast. EMBO J. 18:3173-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert, C. S., C. M. Green, and N. F. Lowndes. 2001. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell 8:129-136. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg, M., M. Stucki, J. Falck, D. D'Amours, D. Rahman, D. Pappin, J. Bartek, and S. P. Jackson. 2003. MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421:952-956. [DOI] [PubMed] [Google Scholar]
- 22.Guo, Z., A. Kumagai, S. X. Wang, and W. G. Dunphy. 2000. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 14:2745-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hammet, A., B. L. Pike, K. I. Mitchelhill, T. Teh, B. Kobe, C. M. House, B. E. Kemp, and J. Heierhorst. 2000. FHA domain boundaries of the dun1p and rad53p cell cycle checkpoint kinases. FEBS Lett. 471:141-146. [DOI] [PubMed] [Google Scholar]
- 24.Ho, Y., A. Gruhler, A. Heilbut, G. D. Bader, L. Moore, S. L. Adams, A. Millar, P. Taylor, K. Bennett, K. Boutilier, L. Yang, C. Wolting, I. Donaldson, S. Schandorff, J. Shewnarane, M. Vo, J. Taggart, M. Goudreault, B. Muskat, C. Alfarano, D. Dewar, Z. Lin, K. Michalickova, A. R. Willems, H. Sassi, P. A. Nielsen, K. J. Rasmussen, J. R. Andersen, L. E. Johansen, L. H. Hansen, H. Jespersen, A. Podtelejnikov, E. Nielsen, J. Crawford, V. Poulsen, B. D. Sorensen, J. Matthiesen, R. C. Hendrickson, F. Gleeson, T. Pawson, M. F. Moran, D. Durocher, M. Mann, C. W. Hogue, D. Figeys, and M. Tyers. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180-183. [DOI] [PubMed] [Google Scholar]
- 25.Hu, F., A. A. Alcasabas, and S. J. Elledge. 2001. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 15:1061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang, M., Z. Zhou, and S. J. Elledge. 1998. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94:595-605. [DOI] [PubMed] [Google Scholar]
- 27.Kihara, M., W. Nakai, S. Asano, A. Suzuki, K. Kitada, Y. Kawasaki, L. H. Johnston, and A. Sugino. 2000. Characterization of the yeast Cdc7p/Dbf4p complex purified from insect cells. Its protein kinase activity is regulated by Rad53p. J. Biol. Chem. 275:35051-35062. [DOI] [PubMed] [Google Scholar]
- 28.Kondo, T., T. Wakayama, T. Naiki, K. Matsumoto, and K. Sugimoto. 2001. Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science 294:867-870. [DOI] [PubMed] [Google Scholar]
- 29.Lee, C. H., and J. H. Chung. 2001. The hCds1 (Chk2)-FHA domain is essential for a chain of phosphorylation events on hCds1 that is induced by ionizing radiation. J. Biol. Chem. 276:30537-30541. [DOI] [PubMed] [Google Scholar]
- 30.Leroy, C., S. E. Lee, M. B. Vaze, F. Ochsenbien, R. Guerois, J. E. Haber, and M. C. Marsolier-Kergoat. 2003. PP2C phosphatases Ptc2 and Ptc3 are required for DNA checkpoint inactivation after a double-strand break. Mol. Cell 11:827-835. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Q., S. Guntuku, X. S. Cui, S. Matsuoka, D. Cortez, K. Tamai, G. Luo, S. Carattini-Rivera, F. DeMayo, A. Bradley, L. A. Donehower, and S. J. Elledge. 2000. Chk1 is an essential kinase that is regulated by Atr and required for the G2/M DNA damage checkpoint. Genes Dev. 14:1448-1459. [PMC free article] [PubMed] [Google Scholar]
- 32.Lopes, M., C. Cotta-Ramusino, A. Pellicioli, G. Liberi, P. Plevani, M. Muzi-Falconi, C. S. Newlon, and M. Foiani. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557-561. [DOI] [PubMed] [Google Scholar]
- 33.Lou, Z., K. Minter-Dykhouse, X. Wu, and J. Chen. 2003. MDC1 is coupled to activated CHK2 in mammalian DNA damage response pathways. Nature 421:957-961. [DOI] [PubMed] [Google Scholar]
- 34.Lydall, D., and T. Weinert. 1997. Use of cdc13-1-induced DNA damage to study effects of checkpoint genes on DNA damage processing. Methods Enzymol. 283:410-424. [DOI] [PubMed] [Google Scholar]
- 35.Malkin, D., F. P. Li, L. C. Strong, J. F. Fraumeni, Jr., C. E. Nelson, D. H. Kim, J. Kassel, M. A. Gryka, F. Z. Bischoff, M. A. Tainsky, et al. 1990. Germ line p53 mutations in a familial syndrome of breast cancer, sarcomas, and other neoplasms. Science 250:1233-1238. [DOI] [PubMed] [Google Scholar]
- 36.Martinho, R. G., H. D. Lindsay, G. Flaggs, A. J. DeMaggio, M. F. Hoekstra, A. M. Carr, and N. J. Bentley. 1998. Analysis of Rad3 and Chk1 protein kinases defines different checkpoint responses. EMBO J. 17:7239-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Massague, J. 2000. How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 1:169-178. [DOI] [PubMed] [Google Scholar]
- 38.Matsuoka, S., G. Rotman, A. Ogawa, Y. Shiloh, K. Tamai, and S. J. Elledge. 2000. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. Proc. Natl. Acad. Sci. USA 97:10389-10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melchionna, R., X. B. Chen, A. Blasina, and C. H. McGowan. 2000. Threonine 68 is required for radiation-induced phosphorylation and activation of Cds1. Nat. Cell Biol. 2:762-765. [DOI] [PubMed] [Google Scholar]
- 40.Melo, J., and D. Toczyski. 2002. A unified view of the DNA-damage checkpoint. Curr. Opin. Cell Biol. 14:237-245. [DOI] [PubMed] [Google Scholar]
- 41.Melo, J. A., J. Cohen, and D. P. Toczyski. 2001. Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev. 15:2809-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Navas, T. A., Y. Sanchez, and S. J. Elledge. 1996. RAD9 and DNA polymerase epsilon form parallel sensory branches for transducing the DNA damage checkpoint signal in Saccharomyces cerevisiae. Genes Dev. 10:2632-2643. [DOI] [PubMed] [Google Scholar]
- 43.Nyberg, K. A., R. J. Michelson, C. W. Putnam, and T. A. Weinert. 2002. Toward maintaining the genome: DNA damage and replication checkpoints. Annu. Rev. Genet. 36:617-656. [DOI] [PubMed] [Google Scholar]
- 44.Paciotti, V., M. Clerici, G. Lucchini, and M. P. Longhese. 2000. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 14:2046-2059. [PMC free article] [PubMed] [Google Scholar]
- 45.Pellicioli, A., C. Lucca, G. Liberi, F. Marini, M. Lopes, P. Plevani, A. Romano, P. P. Di Fiore, and M. Foiani. 1999. Activation of Rad53 kinase in response to DNA damage and its effect in modulating phosphorylation of the lagging strand DNA polymerase. EMBO J. 18:6561-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peng, A., and P. L. Chen. 2003. NFBD1, like 53BP1, is an early and redundant transducer mediating Chk2 phosphorylation in response to DNA damage. J. Biol. Chem. 278:8873-8876. [DOI] [PubMed] [Google Scholar]
- 47.Sanchez, Y., J. Bachant, H. Wang, F. Hu, D. Liu, M. Tetzlaff, and S. J. Elledge. 1999. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science 286:1166-1171. [DOI] [PubMed] [Google Scholar]
- 48.Sanchez, Y., B. A. Desany, W. J. Jones, Q. Liu, B. Wang, and S. J. Elledge. 1996. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271:357-360. [DOI] [PubMed] [Google Scholar]
- 49.Santocanale, C., and J. F. Diffley. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395:615-618. [DOI] [PubMed] [Google Scholar]
- 50.Santocanale, C., K. Sharma, and J. F. Diffley. 1999. Activation of dormant origins of DNA replication in budding yeast. Genes Dev. 13:2360-2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Savitsky, K., A. Bar-Shira, S. Gilad, G. Rotman, Y. Ziv, L. Vanagaite, D. A. Tagle, S. Smith, T. Uziel, S. Sfez, et al. 1995. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science 268:1749-1753. [DOI] [PubMed] [Google Scholar]
- 52.Schlessinger, J. 2000. Cell signaling by receptor tyrosine kinases. Cell 103:211-225. [DOI] [PubMed] [Google Scholar]
- 53.Schwartz, M. F., J. K. Duong, Z. Sun, J. S. Morrow, D. Pradhan, and D. F. Stern. 2002. Rad9 phosphorylation sites couple Rad53 to the Saccharomyces cerevisiae DNA damage checkpoint. Mol. Cell 9:1055-1065. [DOI] [PubMed] [Google Scholar]
- 54.Schwartz, M. F., S.-J. Lee, J. K. Duong, and D. F. Stern. 2003. FHA domain-mediated DNA checkpoint regulation of Rad53. Cell Cycle 2:384-396. [PubMed]
- 55.Shang, Y. L., A. J. Bodero, and P. L. Chen. 2003. NFBD1, a novel nuclear protein with signature motifs of FHA and BRCT, and an internal 41 amino acid repeat sequence, is an early participant in DNA damage response. J. Biol. Chem. 278:6323-6329. [DOI] [PubMed] [Google Scholar]
- 56.Stewart, G. S., B. Wang, C. R. Bignell, A. M. Taylor, and S. J. Elledge. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421:961-966. [DOI] [PubMed] [Google Scholar]
- 57.Sugimoto, K., S. Ando, T. Shimomura, and K. Matsumoto. 1997. Rfc5, a replication factor C component, is required for regulation of Rad53 protein kinase in the yeast checkpoint pathway. Mol. Cell. Biol. 17:5905-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun, Z., D. S. Fay, F. Marini, M. Foiani, and D. F. Stern. 1996. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 10:395-406. [DOI] [PubMed] [Google Scholar]
- 59.Sun, Z., J. Hsiao, D. S. Fay, and D. F. Stern. 1998. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science 281:272-274. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka, K., M. N. Boddy, X. B. Chen, C. H. McGowan, and P. Russell. 2001. Threonine-11, phosphorylated by Rad3 and ATM in vitro, is required for activation of fission yeast checkpoint kinase Cds1. Mol. Cell. Biol. 21:3398-3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tercero, J. A., and J. F. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412:553-557. [DOI] [PubMed] [Google Scholar]
- 62.Vialard, J. E., C. S. Gilbert, C. M. Green, and N. F. Lowndes. 1998. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 17:5679-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wakayama, T., T. Kondo, S. Ando, K. Matsumoto, and K. Sugimoto. 2001. Pie1, a protein interacting with Mec1, controls cell growth and checkpoint responses in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:755-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, B., S. Matsuoka, P. B. Carpenter, and S. J. Elledge. 2002. 53BP1, a mediator of the DNA damage checkpoint. Science 298:1435-1438. [DOI] [PubMed] [Google Scholar]
- 65.Wang, H., and S. J. Elledge. 1999. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 96:3824-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, P., I. J. Byeon, H. Liao, K. D. Beebe, S. Yongkiettrakul, D. Pei, and M. D. Tsai. 2000. II. Structure and specificity of the interaction between the FHA2 domain of Rad53 and phosphotyrosyl peptides. J. Mol. Biol. 302:927-940. [DOI] [PubMed] [Google Scholar]
- 67.Weinreich, M., and B. Stillman. 1999. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 18:5334-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu, X., and D. F. Stern. 2003. NFBD1/KIAA0170 is a chromatin-associated protein involved in DNA damage signaling pathways. J. Biol. Chem. 278:8795-8803. [DOI] [PubMed] [Google Scholar]
- 69.Xu, X., L. M. Tsvetkov, and D. F. Stern. 2002. Chk2 activation and phosphorylation-dependent oligomerization. Mol. Cell. Biol. 22:4419-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yarden, R. I., S. Pardo-Reoyo, M. Sgagias, K. H. Cowan, and L. C. Brody. 2002. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat. Genet. 30:285-289. [DOI] [PubMed] [Google Scholar]
- 71.Zhao, X., A. Chabes, V. Domkin, L. Thelander, and R. Rothstein. 2001. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 20:3544-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao, X., B. Georgieva, A. Chabes, V. Domkin, J. H. Ippel, J. Schleucher, S. Wijmenga, L. Thelander, and R. Rothstein. 2000. Mutational and structural analyses of the ribonucleotide reductase inhibitor Sml1 define its Rnr1 interaction domain whose inactivation allows suppression of mec1 and rad53 lethality. Mol. Cell. Biol. 20:9076-9083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao, X., E. G. Muller, and R. Rothstein. 1998. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2:329-340. [DOI] [PubMed] [Google Scholar]
- 74.Zhao, X., and R. Rothstein. 2002. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. USA 99:3746-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng, P., D. S. Fay, J. Burton, H. Xiao, J. L. Pinkham, and D. F. Stern. 1993. SPK1 is an essential S-phase-specific gene of Saccharomyces cerevisiae that encodes a nuclear serine/threonine/tyrosine kinase. Mol. Cell. Biol. 13:5829-5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou, Z., and S. J. Elledge. 1993. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell 75:1119-1127. [DOI] [PubMed] [Google Scholar]