Abstract
Autoimmune diseases are either tissue-specific like multiple sclerosis (MS) or multisystemic like systemic lupus erythematosus (SLE), although clinically both exhibit common features. To gain insight into the properties of the genes involved in each disease we have investigated the gene expression signature of peripheral blood mononuclear cells (PBMC) in MS and SLE in comparison to healthy subjects. Total RNA was purified, hybridized to Genechip array and analysed in 36 subjects (13 relapsing-remitting MS patients, five SLE patients and 18 age-matched healthy subjects that served as controls). Additional blood samples from 15 relapsing-remitting MS patients, 8 SLE patients and 10 healthy subjects were used for confirmation of microarray gene expression findings by ELISA and RT-PCR. MS and SLE patients demonstrated a common gene expression autoimmune signature of 541 genes which differentiated them from healthy subjects. The autoimmune signature included genes that encode proteins involved in apoptosis, cell cycle, inflammation and regulation of matrix metalloproteinase pathways. Specifically, decreased TIMP1 gene expression in the autoimmunity signature suggests increased MMP activity in target tissues as a result of the lack of feedback mechanism. An additional different disease specific signature identified the gene expression pattern for MS (1031 genes), mainly associated with over-expression of adhesion molecules and down-expression of heat shock proteins; the SLE specific signature (1146 genes) mainly involved DNA damage/repair pathways that result in production of nuclear autoantibodies.
These results provide insights into the genetic pathways underlying autoimmune diseases, and identify specific disease-associated signatures that may enable targetted disease-related specific therapies to be developed.
Keywords: Autoimmunity, multiple sclerosis, systemic lupus erythematosus, gene expression
INTRODUCTION
Autoimmune diseases occur in 3–5% of the population [1], as a result from a myriad of genetic and environmental factors that lead to altered immune reactivity [2,3]. The alterations in the immune system initiated by a loss of immunological tolerance to self-antigens, lead to the development of autoreactive phenomena that can be detected in the peripheral blood. Defining specific pathogenic mediators that may trigger the development or progression of an autoimmune disease remains a focus of increasing investigative efforts. Factors initiating or promoting an autoimmune disease may not be identical to factors that influence the severity or progression of the same disease. Identifying the specific mRNA transcript profile that is common to autoimmune diseases may suggest a group of genes universally involved with clinically distinct forms of autoimmune disease, while the specific disease related signature might provide clues about the disease pathogenesis and define disease severity or progression.
In here we focus on two autoimmune diseases, multiple sclerosis (MS) a chronic autoimmune inflammatory disease limited to the brain and spinal cord and systemic lupus erythematosus (SLE) a chronic autoimmune inflammatory disease that typically affects multiple systems including the skin, joints, kidney, lung, and central nervous system (CNS). In MS, autoreactive activated T-cells invade the CNS and initiate an inflammatory response that leads to myelin destruction and significant neurological disability [4]. The typical immune abnormalities in SLE result in the ability to produce pathogenic autoantibodies, impaired T- and B-lymphocyte regulation, and defective clearance of autoantigens and immune complexes. The majority of autoantibodies found in SLE are targeted at intracellular nucleoprotein particles, 98% of patients possess antinuclear antibodies and antidouble-stranded DNA antibodies are found in 50–80% of patients [5]. As SLE and MS share several common clinical features such as an appearance in young adults, a high female/male ratio, an unpredictable waxing and waning course and beneficial response to steroid treatment, they could share a common gene expression signature. Recently, using microarray analysis in several autoimmune diseases a common expression signature was described [6]. These authors emphasized that autoimmune expression pattern was unrelated to the normal immune response profile after immunization with influenza vaccine. In the present study we applied gene microarray to PBMC of untreated patients with MS and SLE in order to identify universal common autoimmune signature restricted from normal population and to distinct gene expression signatures that characterized each disease.
MATERIALS AND METHODS
Study participants
After written informed consent, blood was obtained from 13 relapsing-remitting MS patients (9 females, 4 males, mean age 40·7 ± 3·4 years, mean disease duration 6·6 ± 2·3 years), five SLE patients (4 females, 1 male, mean age 42·8 ± 12·6 years, mean disease duration 8·4 ± 5·6 years) and 18 age-matched healthy subjects (16 females, 2 males). All patients were free of cytotoxic agents or immunomodulatory drugs for at least 30 days before blood was withdrawn. All participants had peripheral blood counts within the normal range. The 13 MS patients and the 18 healthy controls were a part of our previous study related to gene transcriptional signatures of MS related disease activity [7].
RNA isolation and microarrays hybridization
Peripheral blood mononuclear cells (PBMC) were separated from heparinized blood by centrifugation on Ficol-Hypaque gradient. Total RNA was purified, labelled, and 10 µg were hybridized to Genechip array (U95Av2) and scanned (Hewlett Packard, GeneArray-TM scanner G2500A) according to the manufacturer's protocol (Affymetrix Inc, Santa Clara, CA, USA). MAS5 software (Affymetrix Inc.) was used to analyse the scanned arrays. All data was normalized by dChip software [8].
Data processing and statistical analysis
Data were analysed using Scoregene statistical package (http://compbio.cs.huji.ac.il/scoregenes). Genes were scored by the classic parametric t-test, and the nonparametric tests: (i) The threshold number of misclassifications (TNoM) method and (ii) the Info score [9,10]. TNoM score counts the number of classification errors that occur between compared groups for each gene of the dataset. The best threshold (TNoM = 0) implies that no errors have been counted and the distinction between the analysed groups in relation to the expression level of a specific gene is maximal. Info score is a defined version of TNoM that measures the misclassifications made by a simple threshold in terms of the information lost (or entropy) of the labels of samples in each side of the threshold. Analysis was performed between MS patients and healthy subjects, between SLE patients and healthy subjects and between MS patients and SLE patients, for each gene of the dataset. Fold ratios were calculated for each gene in the samples against the geometric mean of controls and log (base 2) transformed. We filtered probes that did not have an expression value of 100 in at least one of the arrays. This resulted in a file with 9225 probes, the raw transcript data is demonstrated in http://www.sheba.co.il/sclerosis
The hybridization of the arrays was done in eight batches. To control for artifacts of batches, we fitted a multiple effect model for each gene, where we model the log-ratio measurement as a sum of contributions of (a) batch (b) subject state (control, MS, SLE), and (c) array specific noise. We fitted the model to minimize the least sum of squares of the errors. We created a cleaned log-ratio file by removing from each log-ratio the associated batch effect parameter. The most informative differentially expressed genes were defined as those that pass 95% confidence interval on all three statistical tests (t-test, TNoM and Info), while top score genes were defined as genes that in addition have at least 1·5 fold change in comparison to the controls in either direction. We included in the common signature the most informative genes from both diseases with a fold change greater than 1·5 versus healthy subjects in at least one disease.
Consistency of the data
In order to determine consistency of the data we performed the leave-one-out-cross-validation (LOOCV) test [10]. The LOOCV statistical test simulates removal of a single sample from the dataset every trial and trains on the rest. The procedure is repeated until each sample is left out once and the number of correct and incorrect predictions is counted. According to LOOCV test, each dataset was re-arrayed and tested in an independent manner.
Microarray results verification
PBMC from additional 15 relapsing-remitting MS patients, 8 SLE patients and 10 healthy subjects were used for confirmation of microarray gene expression findings. Verification was performed for the top score genes by quantitative RT-PCR using commercial Applied Biosystems Tagman® Assay-on Demand™ Gene Expression products. GAPDH was used as an internal control. The reaction was carried out in ABI PRISM 7900 Sequence Detector (Applied Biosystems, Forster City, USA). Protein levels were measured in supernatant of PBMC short-term (24 h) cultures by commercial ELISA kits (Quantikine, R & D Systems, Inc, Minneapolis, USA).
RESULTS
We identified gene expression distinct signatures that distinguish MS and SLE patients from healthy subjects. Strikingly, both MS and SLE patients showed a common autoimmune gene expression signature that differentiated them from healthy subjects in addition to a specific MS or SLE related gene profile.
Autoimmunity specific signature
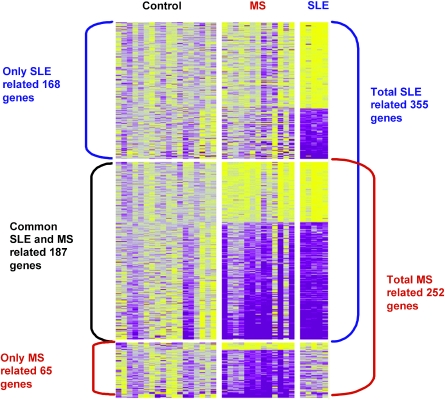
The universal common autoimmunity signature, which intersects between the SLE and MS signatures and differs from the healthy subjects’ gene expression, contains 541 genes (represented by 589 probes). There were 322 genes (339 probes) over-expressed and 219 genes (250 probes) down-expressed. All transcripts passed 95% confidence level on t-test, info and TNoM statistical scores. Further analysis identified 187 (207 probes) top score genes, 67 (69 probes) over-expressed and 120 (138 probes) down-expressed, all with at least 1·5-fold change difference in expression in each direction, Fig. 1. The autoimmunity top-score signature revealed transcripts mainly associated with genes involved in immune surveillance, including genes related to apoptosis pathways (TRAF5, CASP8, BCL2A, IER3 and IL1B), genes that encode proteins involved in stimulation of inflammation, proliferation and immune response (CTBP1, IL11RA, VEGF, BTG1 and 2, AREG and CD19), Table 1.
Fig. 1.
Top score gene expression profile of PBMC from 13 MS patients, 5 SLE patients and 18 healthy controls. All shown gene transcripts passed 95% confidence level on t-test, info and TNoM statistical scores and 1·5 fold change in two directions. Increased genes are shown in progressively brighter shades of yellow, and decreased genes are shown in progressively darker shades of purple.
Table 1.
Functional groups of genes contributing to the common autoimmunity, MS and SLE signatures
| Function | Expression | Common | MS | SLE |
|---|---|---|---|---|
| Apoptosis | Up | TRAF5, CASP8 | CTLA1 | OA48-18, AHR, TIAL1 |
| Down | BCL2A, CIAS1, IL1B, MYBL2, IER3 TNFAIP3, SERPINB2 | NFKB1,NFKBIA, BIRC2, BIRC3 | TNFSF9 | |
| Proliferation | Up | CTBP1 | IL15 | CDC25B, v-MYC |
| Down | KLF4, VEGF, OSM, BTG1, BTG2, AREG, INSIG1, FOSL2, FOSB | TOB1, ZFP36L2 | ||
| Inflammatory response | Up | DEFA3, NFATC3, PTGS2 | BAT1, IFI16 | |
| Down | NR4A1, PTX3, SCYA20 | IL1RN, CEBPB | CEBPD, CLECSF2 | |
| Immune response | Up | CD24 | ||
| Down | CD83 | AQP9 | ||
| Adhesion | Up | PKP4, SCYE1, PNN | ITGAL, ITGA6, LY75 | NELL2, EED, CCR2,TSC1 |
| Down | OLR1,SCYA4, CXCR4 | TNFAIP6, SCYA2, SCYA3 | ||
| Cell cycle | Up | BTG3, NXP2, TOPBP1 | ||
| Down | CCNH, CDKN1A, SCYA8 | CCND2, MAPK6, MCM6 | ||
| Signalling | Up | ADM, EREG, TIP3, NR4A2, MADH7, GNA13, PTGER4, RGS1, PBEF | IL8, GRO3, GNA15 | SMAP, FNTA, TNFS10, IL7, GPR65 |
| Down | ||||
| Heat-Shock | Up | SAFB | ||
| Down | HSPA1A, HSPA1B, HSPA5 | DNAJB1 | ||
| Immune cell receptors | Up | IL11RA, CD19 | CD1D | EGFL5 |
| Down | IL1R1, PLAUR, TIMP1 | CD69, PTGER4, TRD | ||
| DNA damage/repair | Up | ATR, PMS1 | POLS, MBD4, ERCC2, MSH3 | |
| Down | GADD45B, TARBP1 | FANCC, GADD45A, DDIT3 | ||
| Cell growth | Up | DDX17 | ||
| Down | NR4A3, MAP3K, DTR | G0S2 | DBY, GDF11, PTPRE, YES1 |
The most prominent cluster in the autoimmunity signature contained several genes associated with the MMP regulation pathways (TIMP1, JUN, FOS and Oncostatin M), Table 2.
Table 2.
MMPs associated genes in the common autoimmunity expression signature
| Identifier | Symbol | Name |
|---|---|---|
| D11139 | TIMP1 | Tissue inhibitor of metalloproteinase 1 |
| X89750 | TGIF | TGFB-induced factor |
| X04500 | IL1B | Interleukin 1, beta |
| M63978 | VEGF | VEGF |
| U27467 | BCL2A1 | BCL2-related protein A1 |
| X56681 | JUND | Jun D proto-oncogene |
| X51345 | JUNB | Jun B proto-oncogene |
| J04111 | JUN | V-jun avian sarcoma virus 17 oncogene homolg |
| L49169 | FOSB | FBJ murine osteosarcoma viral oncogene homolog B |
| X16706 | FOSL2 | FOS-like antigen 2 |
| U35113 | MTA1 | Metastasis associated 1 |
| M27288 | OSM | Oncostatin M |
| AB000734 | SSI-1 | JAK binding protein |
MS specific signature
The specific MS gene expression signature included 1031 unique genes (1105 probes); 594 genes (653 transcripts) down-expressed and 437 genes (442 probes) over-expressed, all passed 95% confidence level on all three statistical scores used. The top score genes (with at least 1·5 fold change, Fig. 1) included 14 genes (14 probes) over-expressed and 51 genes (55 probes) down-expressed. This signature revealed down regulation of cell death related genes (NFKB1, NFKBIA, BIRC2 and 3), heat shock proteins (HSPA1A, HSPA5, HSPA1B), and signal transduction (IL8, GRO3, CNA15). On the other hand over-expressed genes within the signature were associated with inflammation (CD24, IL15, DEFA3, NFATC3, and PTGS2), antigen recognition (CD1D), and adhesion (ITGA6, ITGAL, and LY75).
SLE specific signature
The SLE signature that differs from healthy subjects contains 1146 genes (1185 probes); 723 genes (750 probes) over-expressed and 423 gene (435 probes) down-expressed. All these genes passed 95% confidence level on all three statistical scores used. The top score genes (with at least 1·5 fold change, Fig. 1) included 106 over-expressed genes (112 probes) and 62 down-expressed genes (63 probes). SLE expression patterns included mainly up-regulated genes associated with inflammation (IFI16, BAT1), DNA damage or repair inducible genes (POLS, MBD4, ERCC2, MSH3), adhesion (NELL2, EED, CCR2, TSC1), genes responsible for negative regulation of proliferation (DDX17) and antiapoptosis pathways (TIAL1), Table 1.
Specifically, we identified SLE related genes previously reported to be involved in autoantigen production relevant to SLE pathogenesis. These include over-expression of genes known to encode antigens that serve as target for autoantibody: NXP2 (encodes for an antigen that leads to antinuclear matrix protein) and TOPBP1 (encodes for antigen for DNA topoisomerase II autoantibodies) and IFI16 (interferon inducible gene that is localized to the nucleus and is able to bind DNA).
Consistency of the data
The results of LOOCV analysis of our data demonstrated an impressive 0 classification error between MS and SLE groups, between MS and control groups, and between SLE and control groups, using minimal set of 163, 19, 343 genes, respectively, with P-value < 0·001 for INFO statistical test. This low error rate suggests that the gene expression signatures found are reliable and ensure consistency of data.
Verification of microarray results
Gene expression patterns measured by microarray analysis were verified by RT-PCR for the following top score selected genes: CCR2, CD1D, NR4A1, TOPBP1, IFI16, and NXP2. Protein secretion levels were performed for IL1B, and IL8 genes. Both methods correlated with microarray results and demonstrated significant difference in expression level between MS or SLE patients and healthy subjects, Table 3.
Table 3.
Verification of gene expression microarray results
| Microarray analysis | Verification | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Symbol | Log fold change | TNOM P-value | Info P-value | t-test P-value | Log fold change | t-test P-value | Method |
| 1520_s_at | IL1B* | −2·04522 | 3·01E-07 | 1·36E-07 | 3·59E-09 | −3·473931188 | 0·03 | ELISA |
| 35372_r_at | IL8* | −1·93409 | 4·36E-05 | 6·01E-05 | 9·86E-06 | −0·785875195 | 0·03 | ELISA |
| 280_g_at | NR4A1* | −1·34086 | 4·51E-06 | 2·80E-06 | 4·22E-07 | −1·514573173 | 0·04 | RT-PCR |
| 38567_at | CD1D* | 0·922984 | 0·0016476 | 0·00047093 | 6·45E-05 | 0·584962501 | 0·03 | RT-PCR |
| 36845_at | NXP2** | 0·655707 | 0·00136705 | 0·00035662 | 2·86E-05 | 3·232660757 | 0·01 | RT-PCR |
| 1456_s_at | IFI16** | 0·598806 | 5·94E-05 | 5·94E-05 | 3·50E-05 | 4·87774425 | 0·02 | RT-PCR |
| 38834_at | TOPBP1** | 0·755485 | 5·94E-05 | 5·94E-05 | 1·32E-06 | 3·857980995 | 0·01 | RT-PCR |
| 39936_at | CCR2** | 1·44088 | 0·00136705 | 0·00136705 | 0·00120518 | 3·182692298 | 0·04 | RT-PCR |
MS versus Controls;
SLE versus Controls
DISCUSSION
In the current study we found that gene expression profiles of PBMC from MS and SLE patients differ from healthy subjects. This suggests that MS and SLE are highly related and represent a common autoimmune portrait that is distinct from healthy individuals. These results have a clinical ground, taking into consideration the similar clinical course with exacerbations and remissions, the increased incidence among women, the familial occurrence and the favourable effect of steroid or cytotoxic treatments in both diseases. The most notable findings in the common autoimmunity signature were genes relevant to basic autoimmune mechanisms, impaired apoptosis and proliferative advantage of immune activated cells. Imbalance of pro- and anti-apoptosis signals was demonstrated by up regulation of TRAF5 and CASP8 and down-expression of BCL2A, IER3 and nuclear receptor subfamily 4, group A, 1 and 2 (NR4A1 and NR4A2). The last group of genes is known to mediate T cell receptor-induced apoptosis through Fas ligand and TNFα or through early response of TCR-induced apoptosis of thymocytes [11]. Over expressed genes included immune cell receptors like interleukin 11 receptor alpha (IL11RA) which acts as an antagonist of IL11 mediated anti-inflammatory effects [12] and CD19 that is known to lower the threshold for antigen receptor stimulation on B lymphocytes and influence the balance between immunity and autoimmunity [13–16]. Transgenic mice that over express CD19, generate spontaneous antinuclear antibodies, rheumatoid factor and autoantibodies for ssDNA, dsDNA and histone by augmenting antigen receptor signalling [16]. The common autoimmunity signature also demonstrated over expression of adhesion molecules like PKP4, SCYE1 and PNN which may be associated with facilitation of the on going inflammation of the autoimmune process.
Down expressed genes within the autoimmunity signature were found to be associated with negative regulation of proliferation, and include TARBP1, CDKN1A (p21), GADD45B and BTG1. Transfection experiments using NIH 3T3 cells indicated that BTG1 negatively regulates cell proliferation [17].
One of the interesting finding in the autoimmunity signature was the aberrant expression of MMPs pathways. These pathways enhance trafficking of activated lymphocytes through the endothelial barrier and the extracellular matrix to damage the end organ in each disease. MMPs activity is controlled by specific tissue inhibitors (TIMPs), and especially TIMP1 inhibits the activity of most known MMPs. Inhibition of TIMP1 enhances cellular migration through collagen I in endothelial cells and affects programmed cell death [18]. Our findings of decreased TIMP1 expression in the autoimmunity signature, suggest increased MMP activity in target tissues as a result of lack of feedback mechanism. Similarly to our findings, reduced TIMP1 expression was also recently reported in MS patients using gene microarray of PBMC [19]. The lack of feedback inhibition by TIMP1 is supported by additional studies that demonstrated increased levels of MMP-9 and MMP-2 in both SLE and MS patients [20,21]. Moreover, the expression of MMPs and TIMPs is regulated by many cytokines particularly IL-1β and TGF-β, and TIMP-1 specifically is regulated by cytokines such as IL-6, VEGF and oncostatin M (OSM). Members of the proto-oncogenes JUN and FOS are also involved the activation/regulation of TIMP1 [22,23]. All these genes were found to be involved in the MMPs pathways within the autoimmunity portrait (Table 2). Recently, it was demonstrated that TIMP-1 protein level was augmented during IFN-β therapy in MS patients [24]. Taken together with our findings, we suggest that targeted therapy of enhanced TIMP-1 activity could shift the balance of MMPs/TIMP-1, in favour of reduced damage within the inflamed tissues.
In addition to the distinct autoimmune signature, we found a different specific gene expression profile which is unique to each disease, SLE or MS.
The MS specific disease signature was characterized by impairment in apoptosis related pathways, demonstrated by aberrant expression of NFKB and BIRC group of genes, over expression of adhesion related genes (ITGAL, ITGA6 and LY75), and of antigen presenting molecule (CD1D). Down expression of transcripts for the heat shock protein 70 (HSP70) including HSP-A1A, HSP-A1B and HSPA5 was unique to the MS signature. HSP70 was proved to be the most significant discriminator of MS versus controls by Bomprezzi et al. [19]. HSPs are involved in the presentation of endogenous antigens by MHC class I molecules and required for efficient antigenic epitope processing [25]. It is conceivable that a lower expression of MHC class-I on autoreactive T cells enable these cells to escape the censured regulation by CD8 cells that recognize autoimmune idiotypes [26]. Another specific finding in MS was the elevated expression of IL15. Over expression of IL15 affects T cells by up regulation of natural killer cells transcript 4 (NK4), and is required for memory T cells division [27]. The finding of over-expression of CD24 within the MS specific expression signature is in accordance with the fact that CD24 has been proven to be essential for the induction of experimental autoimmune encephalomyelitis (EAE) in mice [28] and with a recent report suggesting that CD24 is a possible genetic modifier for the risk and progression of MS [29].The current findings are in agreement with our previous results in MS patients [7]. We identified a unique transcriptional signature of PBMC from MS patients using oligonucleotide microarrays, irrespective of disease activation state or immunomodulatory treatment.
SLE patients are characterized by the constant presence of autoimmunity to nucleoproteins, particularly to chromatin and small nuclear RNA-protein that mediate premessenger RNA processing. Compatible with these features, we found that the SLE specific signature is characterized by aberrant expression of genes associated with replication, elongation, and maintenance and regulation of proliferation of nucleic acids. Specifically, DNA damage or repair inducible genes such as GADD45A, POLS, MBD4, ERCC2, MSH3, and FANCC were dysregulated. The FANCC gene (Fanconi anaemia complementation group C), which was found to be down-expressed, is known to be mutated in patients suffering from Fanconi anaemia, a congenital disease with impairments in DNA repair [30]. As FANCC gene is responsible for DNA repair, its under expression may lead to abnormal DNA fragments and result in appearance of antinuclear antibodies characterizing SLE. Similarly, the specific SLE signature included over expression of genes known to induce additional autoantibodies production against nuclear antigens in SLE patients like TOPBP1, NXP2 and IFI16 [31–33].
Other interesting genes found in the specific SLE signature included adhesion molecules (NELL2, EED, CCR2 and TSCI), different from those found in the MS specific signature (mainly characterized by integrins); and down expression of the G protein coupled receptor 65 (GPR65) also known as T cells death-associated gene 8 (TDAG8), involved in activation of caspase-9, -8, -3 and its low transcriptional expression may lead to prolonged survival of self reactive clones of T- lymphocytes [34,35].
The SLE signature mainly involved DNA damage/repair pathways. The impairment in these pathways may result in the exsistence of abnormal DNA fragments that will initiate production of nuclear autoatibodies. Similarly to our findings Baechler and his colleagues [36], found gene expression patterns that differentiated SLE patients from healthy subjects. Their main observation was related to dysregulation of genes related to IFN pathway, as the IFN signature also served as a marker for a more severe disease.
To summarize, identification of common and specific disease associated gene expression signatures is of importance to better understand the pathogenic mechanisms involved in autoimmune diseases and to enable tailoring of different therapeutic strategies. These strategies will target the common functional groups of genes contributing to the development of autoimmunity as well as the disease related distinct specific genes, as the common autoimmune signature reflects genetic susceptibility for impaired response to self-antigens and the unique part of gene expression reflects specific end-organ vulnerability.
REFERENCES
- 1.Jacobson DL, Gange SJ, Rose NR, Graham NM. Epidemiology and estimated population burden of selected autoimmune diseases in the United States. Clin Immunol Immunopathol. 1997;84:223–43. doi: 10.1006/clin.1997.4412. [DOI] [PubMed] [Google Scholar]
- 2.Marrack P, Kappler J, Kotzin BL. Autoimmune disease: why and where it occurs. Nat Med. 2001;7:899–905. doi: 10.1038/90935. [DOI] [PubMed] [Google Scholar]
- 3.Salaman MR. A two-step hypothesis for the appearance of autoimmune disease. Autoimmunity. 2003;36:57–61. doi: 10.1080/0891693031000068637. [DOI] [PubMed] [Google Scholar]
- 4.Prat A, Biernacki K, Lavoie JF, Poirier J, Duquette P, Antel JP. Migration of multiple sclerosis lymphocytes through brain endothelium. Arch Neurol. 2002;59:391–7. doi: 10.1001/archneur.59.3.391. [DOI] [PubMed] [Google Scholar]
- 5.Yasutomo K. Pathological lymphocyte activation by defective clearance of self-ligands in systemic lupus erythematosus. Rheumatology. 2003;42:214–22. doi: 10.1093/rheumatology/keg081. [DOI] [PubMed] [Google Scholar]
- 6.Maas K, Chan S, Parker J, Slater A, Moore J, Olsen N, Aune TM. Cutting edge: molecular portrait of human autoimmune disease. J Immunol. 2002;169:5–9. doi: 10.4049/jimmunol.169.1.5. [DOI] [PubMed] [Google Scholar]
- 7.Achiron A, Gurevich M, Friedman N, Kaminski N, Mandel M. Blood Transcriptional Signatures of Multiple Sclerosis: Unique gene expression of disease activity. Ann Neurol. 2004;55:410–7. doi: 10.1002/ana.20008. [DOI] [PubMed] [Google Scholar]
- 8.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaminski N, Friedman N. Practical approaches to analyzing results of microarray experiments. Am J Respir Cell Mol Biol. 2002;27:125–32. doi: 10.1165/ajrcmb.27.2.f247. [DOI] [PubMed] [Google Scholar]
- 10.Ben Dor L, Bruhn L, Friedman N, Nachman I, Schummer M, Yakhini Z. Tissue classification with gene expression profiles. J Comput Biol. 2000;7:559–83. doi: 10.1089/106652700750050943. [DOI] [PubMed] [Google Scholar]
- 11.Rathmell JC, Thompson CB. Pathways of apoptosis in lymphocyte development, homeostasis, and disease. Cell. 2002;109(Suppl.):97–107. doi: 10.1016/s0092-8674(02)00704-3. [DOI] [PubMed] [Google Scholar]
- 12.Curtis DJ, Hilton DJ, Roberts B, Murray L, Nicola N, Begley CG. Recombinant soluble interleukin-11 (IL-11) receptor alpha-chain can act as an IL-11 antagonist. Blood. 1997;90:4403–12. [PubMed] [Google Scholar]
- 13.Carter RH, Fearon DT. CD19: lowering the threshold for antigen receptor stimulation of B lymphocytes. Science. 1992;256:105–7. doi: 10.1126/science.1373518. [DOI] [PubMed] [Google Scholar]
- 14.Inaoki M, Sato S, Weintraub BC, Goodnow CC, Tedder TF. CD19-regulated signaling thresholds control peripheral tolerance and autoantibody production in B lymphocytes. J Exp Med. 1997;186:1923–31. doi: 10.1084/jem.186.11.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sato S, Ono N, Steeber DA, Pisetsky DS, Tedder TF. CD19 regulates B lymphocyte signaling thresholds critical for the development of B-1 lineage cells and autoimmunity. J Immunol. 1996;157:4371–8. [PubMed] [Google Scholar]
- 16.Sato S, Hasegawa M, Fujimoto M, Tedder TF, Takehara K. Quantitative genetic variation in CD19 expression correlates with autoimmunity. J Immunol. 2000;165:6635–43. doi: 10.4049/jimmunol.165.11.6635. [DOI] [PubMed] [Google Scholar]
- 17.Rouault JP, Rimokh R, Tessa C, et al. BTG1, a member of a new family of antiproliferative genes. EMBO J. 1992;11:1663–70. doi: 10.1002/j.1460-2075.1992.tb05213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zucker S, Hymowitz M, Conner C, et al. Measurement of matrix metalloproteinases and tissue inhibitors of metalloproteinases in blood and tissues. Clinical and experimental applications. Ann N Y Acad Sci. 1999;878:212–27. doi: 10.1111/j.1749-6632.1999.tb07687.x. [DOI] [PubMed] [Google Scholar]
- 19.Bomprezzi R, Ringnér M, Kim S, et al. Gene expression profile in multiple sclerosis patients and healthy controls: identifying pathways relevant to disease. Hum Mol Genet. 2003;12:2191–9. doi: 10.1093/hmg/ddg221. [DOI] [PubMed] [Google Scholar]
- 20.Chandler S, Miller KM, Clements JM, Lury J, Corkill D, Anthony DC, Adams SE, Gearing AJ. Matrix metalloproteinases, tumor necrosis factor and multiple sclerosis: an overview. J Neuroimmunol. 1997;72:155–61. doi: 10.1016/s0165-5728(96)00179-8. [DOI] [PubMed] [Google Scholar]
- 21.Faber E, Sthoeger Z, Tcherniack A, Dayan M, Mozes E. Activity of matrix metalloproteinase-9 is elevated in sera of patients with systemic lupus erythematosus. Clin Exp Immunol. 2002;127:393–8. doi: 10.1046/j.1365-2249.2002.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Botelho FM, Edwards DR, Richards CD. Oncostatin M stimulates c-Fos to bind a transcriptionally responsive AP-1 element within the tissue inhibitor of metalloproteinase-1 promoter. J Biol Chem. 1998;273:5211–8. doi: 10.1074/jbc.273.9.5211. [DOI] [PubMed] [Google Scholar]
- 23.Zeng G, McCue HM, Mastrangelo L, Millis AJ. Endogenous TGF-beta activity is modified during cellular aging. effects on metalloproteinase and TIMP-1 expression. Exp Cell Res. 1996;228:271–6. doi: 10.1006/excr.1996.0326. [DOI] [PubMed] [Google Scholar]
- 24.Waubant E, Gee L, Miller K, Stabler G, Goodkin D. IFN-beta1a may increase serum levels of TIMP-1 in patients with relapsing-remitting multiple sclerosis. J Interferon Cytokine Res. 2001;21:181–5. doi: 10.1089/107999001750133230. [DOI] [PubMed] [Google Scholar]
- 25.Rechsteiner M, Realini C, Ustrell V. The proteasome activator 11 S REG (PA28) and class I antigen presentation. Biochem J. 2000;345 Part 1:1–15. [PMC free article] [PubMed] [Google Scholar]
- 26.Zang YC, Hong J, Rivera VM, Killian J, Zhang JZ. Preferential recognition of TCR hypervariable regions by human anti-idiotypic T cells induced by T cell vaccination. J Immunol. 2000;164:4011–7. doi: 10.4049/jimmunol.164.8.4011. [DOI] [PubMed] [Google Scholar]
- 27.Ku CC, Murakami M, Sakamoto A, Kappler J, Marrack P. Control of homeostasis of CD8+ memory T cells by opposing cytokines. Science. 2000;288:675–8. doi: 10.1126/science.288.5466.675. [DOI] [PubMed] [Google Scholar]
- 28.Bai XF, Liu JQ, Liu X, Guo Y, Cox K, Wen J, Zheng P, Liu Y. The heat-stable antigen determines pathogenicity of self-reactive T cells in experimental autoimmune encephalomyelitis. J Clin Invest. 2000;105:1227–32. doi: 10.1172/JCI9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Q, Rammohan K, Lin S, et al. CD24 is a genetic modifier for risk and progression of multiple sclerosis. PNAS. 2003;100:15041–6. doi: 10.1073/pnas.2533866100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strathdee CA, Duncan AM, Buchwald M. Evidence for at least four Fanconi anaemia genes including FANCC on chromosome 9. Nat Genet. 1992;1:196–8. doi: 10.1038/ng0692-196. [DOI] [PubMed] [Google Scholar]
- 31.Larsen AK, Escargueil AE, Skladanowski A. From DNA damage to G2 arrest: the many roles of topoisomerase II. Prog Cell Cycle Res. 2003;5:295–300. [PubMed] [Google Scholar]
- 32.Dawson MJ, Trapani JA. The interferon-inducible autoantigen, IFI 16. localization to the nucleolus and identification of a DNA-binding domain. Biochem Biophys Res Commun. 1995;214:152–62. doi: 10.1006/bbrc.1995.2269. [DOI] [PubMed] [Google Scholar]
- 33.Seelig HP, Ehrfeld H, Renz M. Interferon-gamma-inducible protein p16. A new target of antinuclear antibodies in patients with systemic lupus erythematosus. Arthritis Rheum. 1994;37:1672–83. doi: 10.1002/art.1780371117. [DOI] [PubMed] [Google Scholar]
- 34.Kyaw H, Zeng Z, Su K, Fan P, Shell BK, Carter KC, Li Y. Cloning, characterization, and mapping of human homolog of mouse T-cell death-associated gene. DNA Cell Biol. 1998;17:493–500. doi: 10.1089/dna.1998.17.493. [DOI] [PubMed] [Google Scholar]
- 35.Tosa N, Murakami M, Jia WY, et al. Critical function of T cell death-associated gene 8 in glucocorticoid-induced thymocyte apoptosis. Int Immunol. 2003;15:741–9. doi: 10.1093/intimm/dxg070. [DOI] [PubMed] [Google Scholar]
- 36.Baechler EC, Batliwalla FM, Karypis G, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100:2610–5. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]