Abstract
Yaf9 is one of three proteins in budding yeast containing a YEATS domain. We show that Yaf9 is part of a large complex and that it coprecipitates with three known subunits of the NuA4 histone acetyltransferase. Although Esa1, the catalytic subunit of NuA4, is essential for viability, we found that yaf9Δ mutants are viable but hypersensitive to microtubule depolymerizing agents and synthetically lethal with two different mutants of the mitotic apparatus. Microtubules depolymerized more readily in the yaf9Δ mutant compared to the wild type in the presence of nocodazole, and recovery of microtubule polymerization and cell division from limiting concentrations of nocodazole was inhibited. Two other NuA4 mutants (esa1-1851 and yng2Δ) and nonacetylatable histone H4 mutants were also sensitive to benomyl. Furthermore, wild-type budding yeast were more resistant to benomyl when grown in the presence of trichostatin A, a histone deacetylase inhibitor. These results strongly suggest that acetylation of histone H4 by NuA4 is required for the cellular resistance to spindle stress.
The great majority of solid tumors are composed of aneuploid cancer cells (29, 38). This aneuploidy is thought to contribute to defects in gene expression and genome stability that fuel the oncogenic process. Aneuploidy is caused by chromosome mis-segregation due to defects in the mitotic apparatus and/or checkpoint pathways that verify the faithful assembly of the spindle. Several mitotic checkpoint pathways have been described (48). These pathways prevent anaphase entry in cells containing unattached kinetochores, in the absence of tension at kinetochores containing attached microtubules (MTs), or in cells containing incorrectly oriented spindles. Recently, a pathway involving the Chfr ubiquitin ligase has also been suggested to respond to spindle stress by delaying the entry into metaphase (61). These mitotic checkpoint pathways prevent aneuploidy by inhibiting cell cycle progression, and they may also contribute, along with still other checkpoints, to inducing apoptosis in response to spindle stress (59, 68). In this respect, they resemble DNA damage checkpoints. However, in addition to inhibiting cell cycle progression and inducing apoptosis, DNA damage checkpoints also have key roles in the cellular response to DNA damage by activating the transcription of DNA repair genes and by phosphorylating DNA replication, repair, and recombination proteins to facilitate the repair of DNA damage (16). Similarly, it seems likely that cells have evolved pathways to facilitate the repair of, and recovery from, spindle stress, although such pathways have not yet been described. We show here that such a pathway may exist in budding yeast and that it involves the acetylation of histone H4 by the NuA4 histone acetyltransferase (HAT) complex.
The N-terminal lysines of the histones are crucial functional targets of HATs (56, 63). Histone acetylation is thought to affect chromatin structure and gene expression by neutralizing the highly basic N-terminal tails of the histones and thereby decompacting chromatin (14). Specific patterns of histone acetylation, along with other histone modifications, also serve as recognition sites for the recruitment of particular proteins to chromatin (31). Histone acetylation is important in gene expression, DNA repair (24), and kinetochore function (15, 67). In budding yeast, the NuA4 HAT complex is responsible for the bulk of histone H4 acetylation on lysines 5, 8, and 12 (1, 12, 39, 54), whereas lysine-16 is mainly acetylated by the Sas2 HAT complex (34, 66). The NuA4 HAT complex is composed of ca. 12 subunits (1, 19, 50). Nine of these have been identified: Esa1 is the catalytic subunit of the complex; Tra1 belongs to the phosphatidylinositol kinase family; Act1 is the conventional yeast actin; Arp4 is an actin-like protein; and Epl1, Yng2, Eaf2, Eaf3, and Eaf4 are subunits with unknown biochemical functions. NuA4 has been implicated in transcriptional activation (19, 54) and DNA double-strand break repair (4). Ynl107/Yaf9, a novel subunit of NuA4 (see below), has also been shown to be required for UV resistance (5). In the present study, we show that Ynl107/Yaf9, Yng2, and acetylated histone H4 are required for the cellular resistance to spindle stress. Complexes resembling NuA4 are found in animal cells, and they have been implicated in transcriptional regulation and DNA repair (28, 56, 63). Given these similarities, it is possible that these complexes also participate in the response of animal cells to spindle stress.
MATERIALS AND METHODS
Plasmids, strains, media, and inhibitors.
Plasmids and yeast strains used in the present study are listed in Tables 1 and 2, respectively. Standard yeast genetic techniques and media were used (62). Yeast strains were made by using PCR-amplified cassettes (40). Constructions were verified by PCR and Western blot analysis for tagged proteins. Benomyl, nocodazole, and trichostatin A (TSA) were purchased from Sigma-Aldrich.
TABLE 1.
Plasmids used in this study
TABLE 2.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| CMY1228 | MATaura3-52::pAFS91 his3Δ200 trp1Δ63 leu2Δ1 ade2-101 lys2-801 | This study |
| CMY1229 | MATaura3-52::pAFS91 yaf9::KanMX his3Δ200 trp1Δ63 leu2Δ1 ade2-101 lys2-801 | This study |
| CMY1237 | MAT? yng2::KanMX6 leu2-3,112 his3-11,15 ura3-1 trp1-1 ade2-1 | This study |
| ILM49 | MATaYAF9-3HA leu2-3,112 his3-11,15 ura3-1 trp1-1 ade2-1 | This study |
| ILM63 | MATaleu2-3,112 his3-11,15 ura3-1 trp1-1 ade2-11 yaf9::KanMX6 | This study |
| ILM74 | MATamps2-1 ura3-52 his3Δ200 trp1Δ63 leu2Δ1 ade2-101 lys2-801 YAF9-GFP::KanMX6 | This study |
| ILM113 | MATaura3-52 his3Δ200 TRP1::pCS51 leu2Δ1 ade2-101 lys2-801 yaf9::KanMX6 bbp1::HIS3MX4/pli36 | This study |
| ILM128 | MATaspc24-11 ura3-52 his3Δ200 trp1Δ63 leu2Δ1 ade2-101 lys2-801 | This study |
| ILM140 | MATaspc24-11 ura3-52 his3Δ200 trp1Δ63 leu2Δ1 ade2-101 lys2-801 yaf9::KanMX6/pli39 | This study |
| ILM162 | MATaura3-52 his3Δ200 trp1Δ63 leu2Δ1 ade2-101 lys2-801 yaf9::KanMX6 | This study |
| JCY1258 | MATaYNG2-13myc leu2-3,112 his3-11,15 ura3-1 trp1-1 ade2-1 | 11 |
| JCY1259 | MATaEPL1-3HA leu2-3,112 his3-11,15 ura3-1 trp1-1 ade2-1 | 11 |
| MSY534 | MATahhf1-22 ura3-52 trp1-289 leu2-3,112 Δ(hht2 hhf2) | 42 |
| MSY535 | MATahhf1-10 ura3-52 lys2-Δ201 leu2-3,112 Δ(hht2 hhf2) | 42 |
| MSY538 | MATahhf1-28 ura3-52 lys2-Δ201 leu2-3,112 Δ(hht2 hhf2) | 42 |
| MSY568 | MATahhf1-23 ura3-52 lys2-Δ201 leu2-3,112 Δ(hht2 hhf2) | 42 |
| MSY569 | MATahhf1-24 ura3-52 lys2-Δ201 leu2-3,112 Δ(hht2 hhf2) | 42 |
| MSY570 | MATahhf1-25 ura3-52 lys2-Δ201 leu2-3,112 Δ(hht2 hhf2) | 42 |
| MSY571 | MATahhf1-32 ura3-52 lys2-Δ201 leu2-3,112 Δ(hht2 hhf2) | 42 |
| MSY572 | MATahhf1-27 ura3-52 lys2-Δ201 leu2-3,112 Δ(hht2 hhf2) | 42 |
| MSY581 | MATahhf1-30 ura3-52 lys2-Δ201 leu2-3,112 Δ(hht2 hhf2) | 42 |
| MSY605 | MATahhf1-34 ura3-52 lys2-Δ201 leu2-3,112 Δ(hht2 hhf2) | 42 |
| MSY2431 | MATaleu2-3,112 his3-11,15 ura3-1 trp1-1 ade2-11 esa1-1851::K.l.-URA3 | 4 |
| MSY2432 | MATaleu2-3,112 his3-11,15 ura3-1 trp1-1 ade2-11 esa1-L357H::K.l.-URA3 | 4 |
| W303-1a | MATaleu2-3,112 his3-11,15 ura3-1 trp1-1 ade2-11 | R. Rothstein |
| YCS64 | MATaura3-52 his3Δ200 TRP1::pCS51 leu2Δ1 ade2-101 lys2-801 bbp1::HIS3MX4 | 60 |
| YPH499 | MATaura3-52 his3Δ200 trp1Δ63 leu2Δ1 ade2-101 lys2-801 | Sikorski and Hieter |
Microscopy.
Indirect immunofluorescence was performed by using published protocols (33). For in vivo visualization of green fluorescent protein (GFP) fusions, yeast strains were visualized directly or after incubation with 25 μg of 1,4-diazabicyclo(2.2.2)octane (DABCO; Sigma)/ml plus 0.5 μg of DAPI (4′,6′-diamidino-2-phenylindole)/ml in 25% phosphate-buffered saline (PBS) plus 75% glycerol. All observations were made with a Leica DMRXA microscope equipped with a Roper Scientific MicroMax cooled charge-coupled device camera and METAMORPH software (Universal Imaging).
Synthetic lethality and complementation.
Thermosensitive bbp1-1 and spc24-11 mutants were transformed by URA3 plasmids containing the wild-type alleles of each gene (pli36 and pli39, respectively). The entire YAF9 coding sequence was then replaced with the KanMX6 cassette (40) in each of the complemented strains. The double-mutant strains containing the wild-type allele of the thermosensitive mutants on a YCp-URA3 plasmid were then plated on medium lacking uracil or containing 5-fluoroorotic acid (5-FOA) (6) at the permissive temperature of 24°C in order to test for the ability of strains containing the double mutants to survive in the absence of a plasmid complementing the temperature-sensitive mutation.
Spindle sensitivity.
Exponentially growing wild-type (YPH499) and ILM162 (yaf9Δ) strains in yeast extract-peptone-dextrose (YPD) medium containing 50 μg of adenine/ml were treated with nocodazole at 7.5 or 15 μg/ml. Once per hour for 6 h, aliquots were taken and analyzed to determine the percentage of rebudded cells and the cell viability. An aliquot of cells was also fixed with 70% ethanol for fluorescence-activated cell-sorting analysis (27).
Gel filtration analysis.
A total of 50 ml of exponentially growing YPH499 cells at an optical density at 600 nm of 0.5 in YPD was washed with PBS, resuspended in 4 ml of PBS plus Complete protease inhibitor cocktail (Roche Biochemicals), and broken in an Eaton press. The lysate was then centrifuged for 15 min at 4°C in an Eppendorf centrifuge, and the supernatant was passed through a 0.45-μm (pore-size) Nalgene filter. Then, 100 μl of filtered protein extract containing 300 μg of protein was applied to a Superdex 200 gel filtration column on a Pharmacia SMART analytical chromatography apparatus. Next, 50-μl fractions were collected, and the elution positions of Yaf9 and Esa1 were determined by immunoblotting and compared to the elution positions of a standard set of marker proteins with known molecular masses.
Immunoblots with antibodies to acetyl-lysine-histone H4.
Cultures (10 ml) of yeast cells were centrifuged and washed once with water, and the pellets were then resuspended in 300 μl of HSB lysis buffer (45 mM HEPES-KOH [pH 7.4], 150 mM NaCl, 10% glycerol, 1 mM EDTA, 0.5% NP-40, Complete protease inhibitor cocktail). Then, 250 μl of acid-washed glass beads was added, and the cells were lysed with a Bead-Beater. The extract was spun in an Eppendorf centrifuge for 10 min at 4°C, and the supernatant was recovered. Proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12% polyacrylamide gels and transferred to nitrocellulose membranes. Membranes were blocked with Tris-buffered saline plus 0.1% Tween (TBST) containing 5% nonfat powdered milk and then incubated with a 1/1,000 dilution of anti-hyperacetylated histone H4 (Penta) rabbit antibodies from Upstate Biotechnology (catalog no. 06-946) for 1 h at room temperature. Membranes were washed with TBST and then incubated with a 1/3,000 dilution of a goat anti-rabbit immunoglobulin G (IgG) conjugated with horseradish peroxidase in TBS-5% milk for 1 h at room temperature. After a wash with TBST, the immunoblots were visualized with enhanced chemiluminescence (Amersham-Pharmacia).
Coimmunoprecipitation analyses.
Exponentially growing yeast cell cultures (400 ml) in YPD at an optical density at 600 nm of 0.6 were centrifuged, washed once with cold water, and resuspended in 0.7 ml of immunoprecipitation (IP) buffer (50 mM Tris-Cl [pH 7.5], 100 mM NaCl, 10 mM EDTA, 15% glycerol, Complete protease inhibitor cocktail), and the cells were broken in an Eaton press. Triton X-100 was then added to 0.1%, and the lysate was centrifuged for 15 min at 4°C in an Eppendorf centrifuge. Next, 360 μg of protein extract in 100 μl of IP buffer plus 0.1% Triton X-100 were mixed with 40 μl of Dynal magnetic Dynabeads Pan-Mouse IgG saturated with either 12CA5 anti-HA monoclonal antibodies, 9E10 anti-myc monoclonal antibodies, or no antibody, followed by incubation overnight at 4°C. The beads were washed three times with 1 ml of IP buffer plus 0.1% Trition X-100 and then heated at 94°C for 5 min in SDS-PAGE sample buffer to release immunoprecipitated proteins.
Quantification of Western blots.
Proteins were separated by SDS-PAGE and then transferred to nitrocellulose membranes. After incubation with appropriate antibodies, the Western blots were developed by using enhanced chemiluminescence plus reagents (Amersham-Pharmacia); the chemiluminescent images were acquired and quantified with a Fluorochem Imager from Alpha Innotech, Inc. This imager uses a photographic lens and a cooled charge-coupled device camera to quantitatively capture and digitize the chemiluminescent signal.
Microarray analyses.
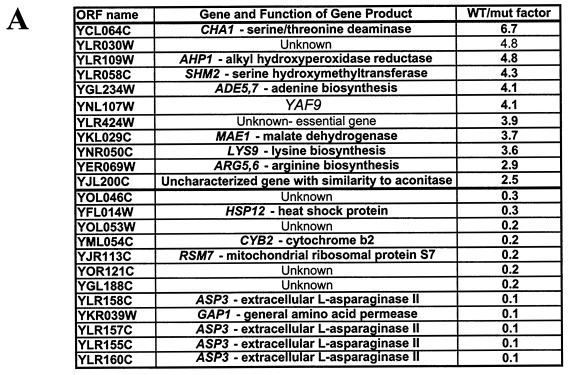
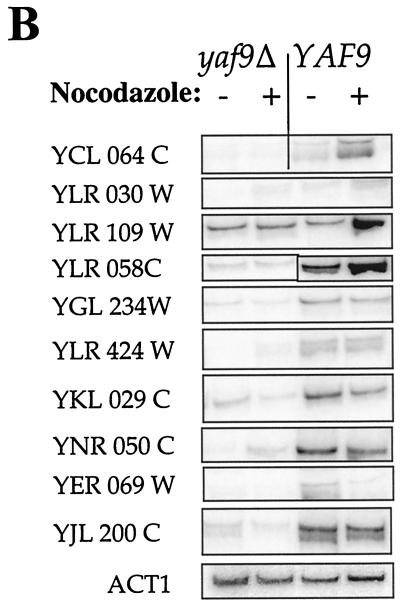
Exponentially growing wild-type (YPH499) and ILM162 (yaf9Δ) strains in YPD media containing 50 μg of adenine/ml were treated with 15 μg of nocodazole/ml until >90% of cells accumulated with a large bud (ca. 2.5 h). Cells were then harvested, and RNA was prepared by using a Qiagen RNeasy kit after the cells were broken with an Eaton press. A 12.5-mg portion of total RNA was used as a template for double-stranded cDNA synthesis. Direct labeling of cDNA was performed during the synthesis as previously described (17). Labeled cDNA was hybridized to yeast genome arrays produced by the Service de Génomique Fonctionnelle-CEA/Evry. Hybridized arrays were scanned with a GenePix 4000A scanner (Axon Instruments, Inc.), and fluorescence ratio measurements were determined with the GenePix Pro 3.0 software (Axon Instruments). From five independent arrays, we chose open reading frames (ORFs) showing a modified signal on at least three of the five arrays, in the same direction and with a wild-type/mutant ratio differing by a factor of 2.5 or more. (These ORFs are listed in Fig. 8A.) The microarray results were confirmed by quantitative reverse transcription-PCR (RT-PCR) analysis of total RNA as described previously (22). The ACT1 mRNA was used as a normalization standard for each amplification reaction. Oligonucleotides were chosen for each ORF to give an amplification product of ca. 400 bp.
FIG. 8.
Identification of genes whose expression is altered in the yaf9Δ mutant (ILM162) relative to the wild type (YPH499) by whole-genome microarray analysis. (A) List of genes whose expression is increased at least 2.5-fold or decreased at least 3-fold in the wild type versus yaf9Δ for cells treated with 15 μg of nocodazole/ml for 3 h. (B) Verification by RT-PCR of genes whose expression is inhibited in the yaf9Δ mutant versus the wild type for cells growing exponentially in YPD or after treatment with 15 μg of nocodazole/ml for 3 h.
Antibodies.
Rabbit polyclonal antibodies to Yaf9 were produced by immunizing rabbits with a maltose-binding protein (MBP)-Yaf9 fusion protein purified from Escherichia coli. Antibodies were affinity purified on an MBP-Yaf9 column. These antibodies were then passed over an MBP column to deplete antibodies that bind MBP. A goat polyclonal antibody directed against Esa1 was purchased from Santa Cruz Biotechnology (sc-12155). An anti-hyperacetylated histone H4 (Penta) rabbit antibody (catalog no. 06-946) was purchased from Upstate Biotechnology. A rabbit antibody against glucose-6-phosphate dehydrogenase was purchased from Sigma (catalog no. A9521).
RESULTS
Ynl107/Yaf9 interacts with Mps2 and is a member of a family of proteins containing YEATS domains that is involved in chromatin modification and transcriptional regulation.
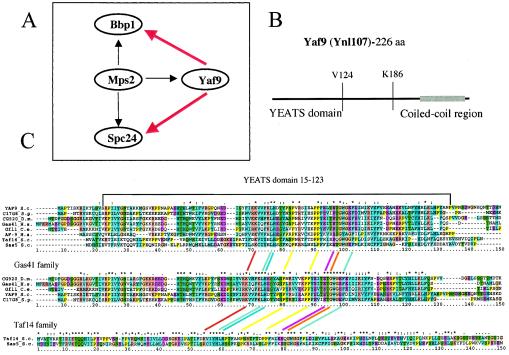
Mps2 (for monopolar spindle 2) is a spindle pole body (SPB) and nuclear envelope protein that is required for the insertion of the duplicated SPB into the nuclear envelope (47). We identified YNL107w as one of three genes coding for potential interacting partners of the Mps2 protein (36), the other two genes being BBP1 and SPC24 (Fig. 1A). YNL107w encodes a 226-amino-acid protein containing an amino-terminal 124-amino-acid YEATS domain and a predicted carboxy-terminal coiled-coil sequence (Fig. 1B). The C-terminal 40 amino acids containing the coiled-coil sequence was the minimal sequence interacting with Mps2 in our two-hybrid screen. The YEATS (Ynl107, ENL, AF9, and TFIIF small subunit) domain is found in many proteins implicated in chromatin modification and transcriptional regulation, but its structure and function are not yet determined (Pfam accession no. PF003366). Ynl107 is similar to a large number of proteins containing a YEATS domain including the yeast Taf14 (also known as Anc1, Tfg3, and Taf30) (8, 25) and Sas5 proteins (32, 43, 51) and the human proteins AF9 (13) and Gas41 (18, 46) implicated in oncogenesis. Due to its similarity to human AF9, the YNL107w ORF was named YAF9 (Yeast AF-9) in the Saccharomyces Genome Database (http://genome-www4.stanford.edu/cgi-bin/SGD/locus.pl?locus =yaf9). However, the similarity of Yaf9 to AF9 and most other YEATS proteins is restricted to the amino-terminal YEATS domain, and CLUSTALW alignments (26) show that the Yaf9 YEATS domain is most similar to that found in the Gas41 family (Fig. 1C). Likewise, the yeast Taf14 and Sas5 YEATS domains are more similar to each other than to the other YEATS domains, whereas the human AF9 YEATS domain is about equally distant from the Gas41 and Taf14/Sas5 families. Furthermore, hydrophobic cluster analysis (HCA) (9, 37) showed that Yaf9 has a distribution of hydrophobic and charged amino acids in its carboxy-terminal domain that is most similar to that of the Gas41 sequence (Fig. 2) Finally, Yaf9 and Gas41 are similar in size (226 and 223 amino acids, respectively), whereas the AF9 protein is much larger (568 amino acids). Gas41 thus seems to be the human protein that is most similar to Yaf9.
FIG. 1.
Yaf9(Ynl107) interacts with Mps2 in a two-hybrid screen, contains a YEATS domain, and shows most similarity to the family of Gas41-like sequences in metazoans. (A) Yaf9, Spc24, and Bbp1 interact with Mps2 in a two-hybrid screen. The black arrows indicate two-hybrid interactions, and the red arrows indicate synthetic lethality between yaf9Δ and either bbp1-1 or spc24-11 mutants (see Fig. 3A). (B) Schematic representation of the Yaf9 sequence showing the location of the N-terminal YEATS domain and the minimal C-terminal 40-amino-acid sequence interacting with Mps2 in the two-hybrid screen. This C-terminal region contains a predicted 30-amino-acid coiled-coil sequence (gray bar) as detected by the COILS program (41) (http://www.ch.embnet.org/software/COILS_form.html). V124 and K186 indicate valine-124 and lysine-186 of the Yaf9 sequence. (C) CLUSTALW alignments (26) of YEATS domains found in proteins similar to Yaf9. Shown are YEATS domains from Saccharomyces cerevisiae (S.c.) Yaf9 (Ynl107), Sas5, and Taf14, Schizosaccharomyces pombe (S.p.) SPAC17G8.07, Drosophila melanogaster (D.m.) CG9207, Caenorhabditis elegans (C.e.) Gfl-1, and Homo sapiens (H.s.) Gas41 and AF9. This analysis shows that Yaf9 and the Gas41 family of proteins contain YEATS domains that are more similar to each other than the YEATS domains found in the human AF9 and the yeast Taf14 and Sas5 proteins. The amino acid color code is as follows: yellow for proline, beige for glycine, purple for acidic residues, orange for basic residues, green for polar residues, light blue for hydrophobic residues, and dark blue for aromatic residues. An asterisk above the column of aligned amino acids indicates 100% identity, two dots indicate highly conserved amino acids, and a single dot indicates similar amino acids.
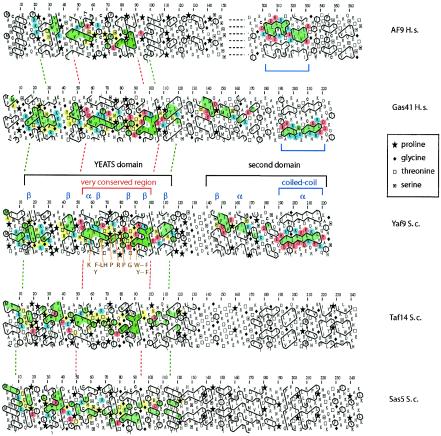
FIG. 2.
HCA indicates that Yaf9 is more similar to human (H.s.) Gas41 than to human AF9 or two other YEATS-domain proteins in S. cerevisiae (Taf14 and Sas5). HCA is a visual method that predicts protein secondary structure (9, 37). The linear one-dimensional sequence of the protein is written on an alpha-helix displayed along a cylinder. The cylinder is then cut parallel to its axis and unrolled in a bidimensional diagram. This diagram is duplicated in order to restore the full environment of each amino acid. The contours of clusters of hydrophobic residues are then drawn. This process has been automated by an online server (http://smi.snv.jussieu.fr/hca/hca-form.html). The alpha-helical net offers the best correspondence between the positions of hydrophobic clusters and regular secondary structures (alpha-helix and β strand). The HCA program uses the standard one-letter code for amino acids except for proline (regular secondary structure breaker), glycine (the least constrained amino acid), and serine and threonine (which can be accommodated in either hydrophilic or hydrophobic environments). Sequence features that were found in similar positions in Yaf9 and the other YEATS domain proteins were colored as follows: common hydrophobic clusters are in green, common nonhydrophobic amino acids are in yellow, positively charged amino” acids are in blue, and negatively charged amino acids are in red. “α” and “β” indicate the position of alpha-helical and β-sheet secondary structures for Yaf9 as predicted by the HCA analysis. Blue brackets show the positions of predicted coiled-coil sequences in the C-terminal regions of Yaf9, Gas41, and AF9. The global analysis of the sequences shows that these proteins are composed of two domains: a similar N-terminal region that corresponds to the YEATS domain and a C-terminal region that is more divergent. For Yaf9, Taf14, and AF9, a linker region could be clearly seen between these two domains. Examination of the HCA plots of human Gas41, AF9 and S. cerevisiae Yaf9, Taf14, and Sas5 sequences reveals the presence of similar hydrophobic motifs in the YEATS domain. The boundaries of this domain in the different proteins are shown with a dotted green line. Residues 55 to 100 of the Yaf9 sequence indicate an extremely conserved region shown within the dotted red lines. This central part of the first domain of the proteins was found to be characteristic of the YEATS domain family. The CLUSTALW (Fig. 1C) and HCA analyses show that the yeast Yaf9 and the human Gas41 YEATS domains are most similar to each other, whereas the yeast Taf14 and Sas5 YEATS domains are more similar to each other than either is to the remaining proteins. The second part of these proteins is more divergent and no similarity (>30%) in primary sequence could be found. Furthermore, the HCA profiles of Sas5, Taf14, and Yaf9 are different. On the other hand, similar hydrophobic motifs are found for the second domain of Yaf9 and Gas41. First, there are two hydrophobic clusters (green) separated by a conserved proline. Some amino acids, in particular charged residues, around this motif are well conserved. Second, a long alpha-helix predicted to form a coiled coil (blue brackets) is found at the C terminus of both Yaf9 and Gas41. We noted, however, that the charge distributions around these coiled coils are different, suggesting that their binding specificity may have diverged. Human AF9, both by its length (more than twice the length of the other YEATS proteins shown here) and by the profiling of its domains, seems the least similar of all of the YEATS proteins shown here.
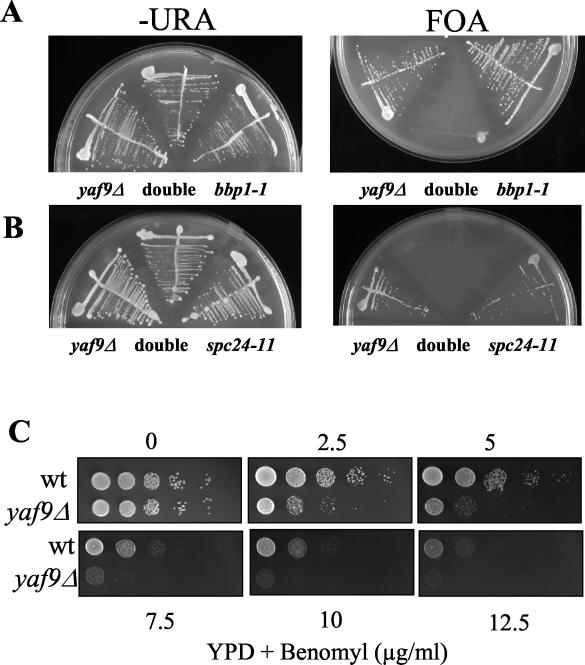
The yaf9Δ mutant is synthetically lethal with mutants of the mitotic apparatus and is hypersensitive to MT depolymerizing agents.
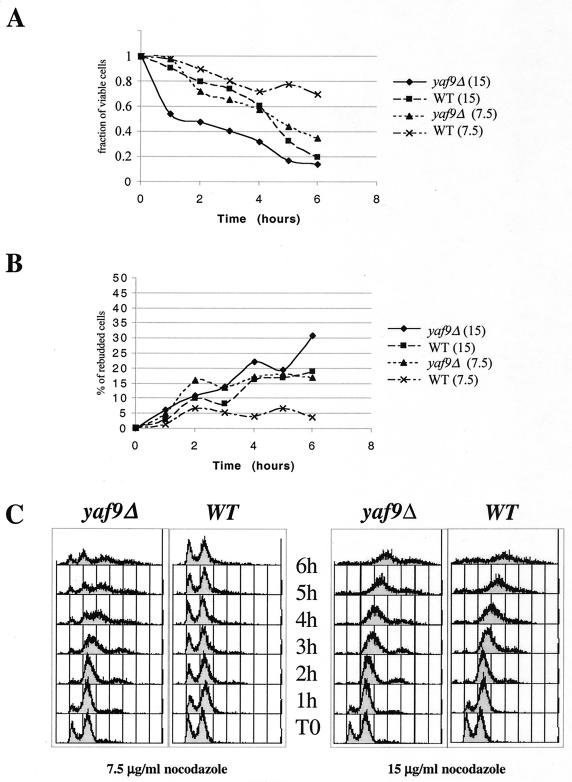
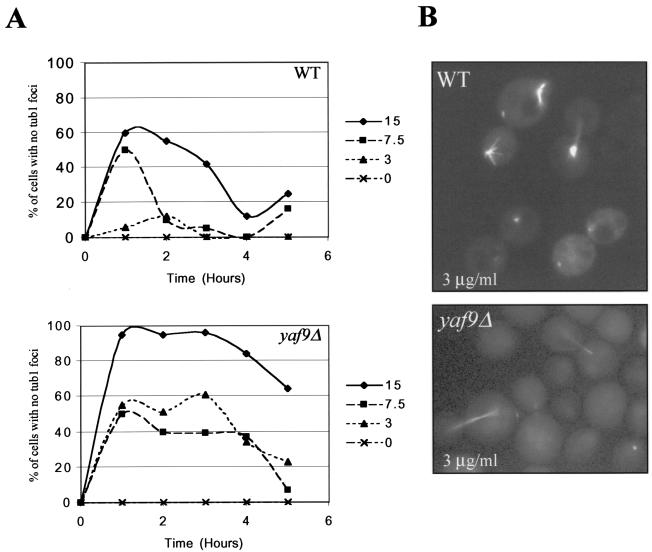
YAF9 is a nonessential yeast gene, and the yaf9Δ mutant showed little or no growth defect relative to the wild type at 24 to 30°C but did grow more slowly than the wild type at 16°C and was temperature sensitive for growth at 37°C in the W303 strain background but not in the YPH or BY strain backgrounds (data not shown). Since Yaf9 interacted with Mps2 in our two-hybrid screen, we sought genetic interactions between yaf9Δ and mutants of the mitotic apparatus. The bbp1-1 mutant is unable to insert a newly synthesized SPB into the nuclear envelope at 37°C (60), whereas the spc24-11 mutant has a kinetochore defect at 37°C (36). The yaf9Δ mutant was found to be synthetically lethal when combined at the permissive temperature of 24°C with either the bbp1-1 mutant or the spc24-11 mutant (Fig. 3A and B) Synthetic lethality was thus observed with mutants affecting the function of both the SPB and the kinetochore. This result suggested that Yaf9 might be required for viability in conditions in which the structural integrity of the spindle was compromised. In support of this notion, we found that the yaf9Δ mutant was hypersensitive to growth on media containing the MT depolymerizing agent benomyl (Fig. 3C). We studied the response of the mutant in liquid cultures to another MT depolymerizing agent, nocodazole, in order to better characterize its defect. yaf9Δ cells lost viability more rapidly than the congenic wild type in the presence of nocodazole (Fig. 4A) and showed a slight increase in the frequency of multibudded cells (Fig. 4B). However, the loss of viability and the frequency of rebudding were less elevated than for spindle checkpoint mutants (data not shown) (64, 69). Flow cytometric analyses showed that the haploid yaf9Δ mutant accumulated with a 2C DNA content after 1 h of treatment with 7.5 μg of nocodazole/ml, whereas the wild-type strain showed only a slight and transient response at this concentration (Fig. 4C). The yaf9Δ mutant is thus hypersensitive to both nocodazole and benomyl. After 4 h of incubation in the presence of 7.5 μg of nocodazole/ml, the yaf9Δ cells escaped from the mitotic arrest, and the flow cytometry profiles suggested that some chromosome mis-segregation might have occurred to give rise to a small fraction of cells with a less than 1C and greater than 2C DNA content. When treated with 15 μg of nocodazole/ml, both the yaf9Δ mutant and the wild type remained arrested with a 2C DNA content over the 6-h period of the experiment (Fig. 4C). This result indicated that the nocodazole sensitivity of the yaf9Δ mutant was not due to a defect in the cell cycle arrest mediated by the spindle checkpoint but was likely due to a defect in the cellular response to MT depolymerization at the level of recovery from spindle stress. Further support of this interpretation was obtained by visualizing MTs in living cells expressing a GFP-Tub1 fusion protein that has previously been used to characterize spindle dynamics in living cells (65). Expression of GFP-Tub1 did not adversely affect the growth of the yaf9Δ mutant. Wild-type and yaf9Δ cells expressing GFP-Tub1 were treated with 3, 7.5, and 15 μg of nocodazole/ml for 5 h. At each hour, an aliquot of living cells was observed, and the percentage of large-budded cells with depolymerized MTs and no visible focus of tubulin staining at the spindle pole bodies was quantified (Fig. 5A) An example of wild-type and yaf9Δ cells expressing GFP-Tub1 and treated with 3 μg of nocodazole/ml for 3 h is shown in Fig. 5B. At each concentration of nocodazole, a higher percentage of the yaf9Δ cells contained disassembled spindles compared to the wild type. Moreover, at lower concentrations of nocodazole, a transient depolymerization of MTs was seen in the wild type, whereas the recovery of MTs was inhibited in the yaf9Δ mutant. Yaf9 is thus required for a normal level of resistance of yeast cells to MT depolymerizing agents.
FIG. 3.
yaf9Δ is synthetically lethal with SPB and kinetochore mutants and it is hyper-sensitive to benomyl. (A) yaf9Δ is synthetically lethal with the bbp1-1 mutant. yaf9Δ (ILM162), bbp1-1 (YCS64), and yaf9Δ bbp1-1 (ILM113) cells containing the BBP1 URA3 plasmid (pli36) were streaked at 24°C, the permissive temperature for the bbp1-1 mutant, onto plates without uracil (−URA) or onto plates containing 5-FOA to counterselect the URA3 plasmid. The absence of growth on the 5-FOA plate of the yaf9Δ bbp1-1 (ILM113) cells (double in the figure) shows that the yaf9Δ bbp1-1 double mutant cannot grow in the absence of the BBP1 URA3 plasmid. (B) yaf9Δ is synthetically lethal with the spc24-11 mutant. yaf9Δ (ILM162), spc24-11 (ILM128), and yaf9Δ spc24-11 (ILM140) cells containing the SPC24 URA3 plasmid (pli39) were streaked at 24°C, the permissive temperature for the spc24-11 mutant, onto plates without uracil (−URA) or onto plates containing 5-FOA to counterselect the URA3 plasmid. The absence of growth on the 5-FOA plate of the yaf9Δ spc24-11 (ILM140) cells (double in the figure) shows that the yaf9Δ spc24-11 double mutant cannot grow in the absence of the SPC24 URA3 plasmid. (C) The yaf9Δ mutant is hypersensitive to benomyl. Wild-type (YPH499) and yaf9Δ (ILM162) cells at the same cell density in YPD were spotted in 10-fold serial dilutions onto YPD plates containing the indicated concentrations of benomyl, and the plates were subsequently incubated at 24°C for 3 days. WT, wild type.
FIG. 4.
yaf9Δ mutants are hypersensitive to nocodazole, but they do not show significant defects in cell cycle arrest. (A to C) Wild-type (YPH499) and yaf9Δ (ILM162) mutants growing exponentially in YPD were treated with 7.5 or 15 μg of nocodazole/ml for the indicated periods of time at 30°C, and the fraction of viable cells (A) and the percentage of rebudded cells (cells with two buds rather than one) (B) were determined at each of the indicated times. (C) An aliquot of cells was also fixed for flow cytometry to determine their DNA content. WT, wild type.
FIG. 5.
MTs depolymerize more readily and recover more slowly from limiting nocodazole in the yaf9Δ mutant compared to the wild type. Wild-type (CMY1228) and yaf9Δ cells (CMY1229) expressing GFP-Tub1 were grown in YPD at 30°C and treated with nocodazole at the concentrations and for the time periods indicated in the figure. (A) The percentage of wild-type and yaf9Δ cells with no detectable GFP-Tub1 foci after treatment with the indicated concentrations of nocodazole was quantified. A fluorescent GFP-Tub1 focus is found at the SPB of cells containing polymerized tubulin. The absence of such foci is thus an indication of an essentially complete depolymerization of MTs. (B) Images illustrating the different responses of wild-type and yaf9Δ cells to incubation with 3 μg of nocodazole/ml for 3 h. WT, wild type.
Yaf9 is a subunit of the NuA4 HAT complex.
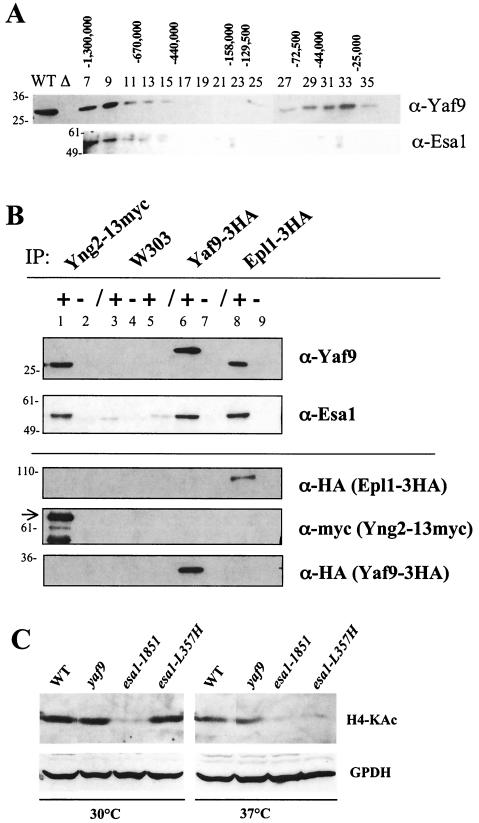
The Yaf9 sequence contains a YEATS domain that is often found in subunits of complexes involved in chromatin modification and transcriptional regulation. A systematic analysis of protein complexes in yeast showed that Yaf9/Ynl107 could be purified in association with some known subunits of the NuA4 HAT complex (20). To test for the presence of Yaf9 in protein complexes, we fractionated whole-cell protein extracts of wild-type yeast cells on a Superdex 200 gel filtration column. Yaf9 was found both in high-molecular-weight fractions corresponding to the size of an ∼1-MDa complex and in low-molecular-weight fractions that may correspond to the monomeric protein (Fig. 6A). Esa1, the catalytic subunit of the NuA4 HAT complex, cofractionated with Yaf9 in the high-molecular-weight fractions. This result was consistent with Yaf9 being a subunit of the NuA4 complex with Esa1. We further verified the presence of Yaf9 in the NuA4 complex by testing for the coimmunoprecipitation of Yaf9 with Esa1, the catalytic subunit of the NuA4 complex, as well as with two other subunits of the complex: Yng2 (11, 39, 50) and Epl1 (19). Western blotting showed that both Yaf9 and Esa1 coprecipitated with Yng2-13myc and with Epl1-3HA (Fig. 6B). Likewise, Esa1 was found to coprecipitate with Yaf9-3HA. The combined data strongly suggest that Yaf9 is a previously unidentified subunit of NuA4.
FIG. 6.
Yaf9 is in a large-molecular-weight complex, coprecipitates with the Esa1, Epl1, and Yng2 subunits of the NuA4 HAT complex but is not required for global histone H4 acetylation by NuA4. (A) A whole-cell protein extract from the wild-type strain YPH499 was chromatographed on a Superdex-200 gel filtration column. Fractions were collected and analyzed by SDS-PAGE and immunoblotting with anti-Yaf9 and anti-Esa1 antibodies. The numbers above each lane indicate the fractions and the molecular weight of marker proteins eluting in those fractions. Thenumbers to the left of the immunoblot indicate the molecular weight of precolored marker proteins. WT indicates a lane containing 40 μg of protein extract from the wild-type strain, and Δ indicates a lane containing 40 μg of protein extract from a yaf9Δ mutant. (B) Yaf9 coprecipitates with Esa1, Epl1, and Yng2. Extracts prepared from the wild-type W303-1a and from strains expressing Yng2-13myc (JCY1258), Yaf9-3HA (ILM49), or Epl1-3HA (JCY1259) were used for IPs with anti-myc or anti-HA monoclonal antibodies. The immunoprecipitates were electrophoresed on SDS-polyacrylamide gels, transferred to membranes, and probed consecutively with anti-Yaf9, anti-Esa1, anti-HA, and anti-myc antibodies in order to test for coprecipitation of NuA4 subunits and to verify the efficiency of IP of each tagged protein. Lane 1, anti-myc IP from extract of JCY1258 (Yng2-13myc) showing coimmunoprecipitation of Yaf9 and Esa1 with Yng2-13myc; lane 2, mock IP without primary antibody from JCY1258 (Yng2-13myc); lane 3, anti-myc IP from extract of untagged W303-1a; lane 4, mock IP without primary antibody from W303-1a; lane 5, anti-HA IP from extract of untagged W303-1a; lane 6, anti-HA IP from extract of ILM49 (Yaf9-3HA) showing coimmunoprecipitation of Esa1 with Yaf9-3HA; lane 7, mock IP without primary antibody from ILM49 (Yaf9-3HA); lane 8, anti-HA IP from extract of JCY1259 (Epl1-3HA) showing coimmunoprecipitation of Yaf9 and Esa1 with Epl1-3HA; lane 9, mock IP without primary antibody from JCY1259 (Epl1-3HA). The numbers to the left of each membrane show the molecular masses (in kilodaltons) of precolored protein markers. The arrow indicates Yng2-13myc. The two faster-migrating bands are proteolytic fragments produced during the IP incubations. (C) Immunoblots of whole-cell protein extracts from wild-type (W303-1a), yaf9Δ (ILM63), esa1-1851, and esa1-L357H cells grown at log phase in YPD at 30°C or transferred from 30 to 37°C for 4 h. The membranes were probed with anti-hyperacetylated histone H4 (Penta) rabbit antibodies (H4-Kac) from Upstate Biotechnology (catalog no. 06-946) and then reprobed with antibodies to glucose-6-phosphate dehydrogenase (GPDH) to verify the loading of equal amounts of protein. WT, wild type.
The yaf9Δ mutant does not show a global reduction of acetylated histone H4.
We tested the effect of the yaf9Δ mutant on NuA4 function by immunoblotting protein extracts from wild-type and mutant strains with an antibody that recognizes hyperacetylated histone H4. This antibody recognizes tetra-acetylated histone H4, and it may recognize triacetylated forms. The yaf9Δ mutant did not show significantly reduced amounts of hyperacetylated histone H4 compared to the wild type at either 30°C or 37°C (Fig. 6C). In contrast, two temperature-sensitive mutants of the Esa1 catalytic subunit of NuA4 showed strong defects, as previously reported (4). The esa1-L357H mutant had normal levels of hyperacetylated histone H4 at 30°C but much less than the wild type after transfer to 37°C. The esa1-1851 mutant showed drastically reduced levels of hyperacetylated histone H4 at both 30 and 37°C. These results indicated that Yaf9 is not required for global H4 acetylation, but they did not exclude the possibility that it might be required to target the NuA4 complex to some specific genomic locations or for the acetylation of some other substrate.
Yaf9 is a nuclear protein whose levels are increased when cells are treated with nocodazole.
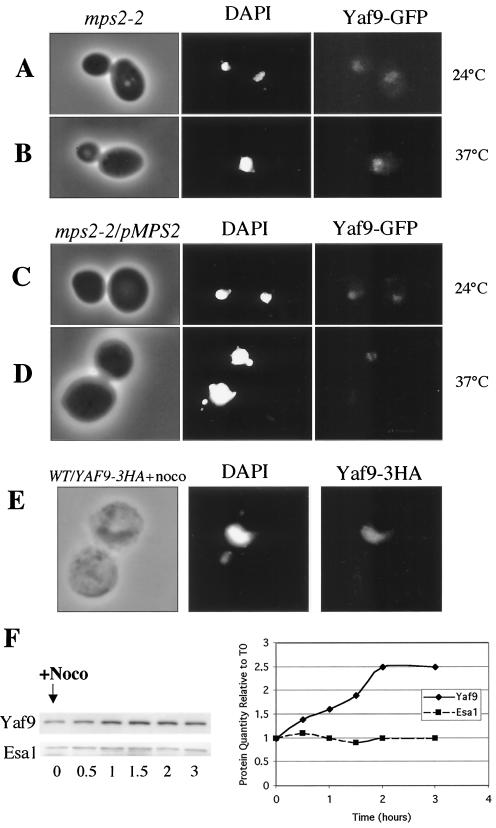
The intracellular localization of Yaf9 was determined in living cells by using a Yaf9-GFP fusion protein and in fixed cells by immunofluorescence by using a Yaf9-3HA epitope-tagged protein. The Yaf9-GFP and Yaf9-3HA proteins appeared to be functionally normal in that cells expressing these fusion proteins in place of the wild-type Yaf9 had the same resistance to benomyl as had the wild type (data not shown). Localization was observed in wild-type cells in the absence or the presence of nocodazole and in an mps2-2 mutant at the permissive and restrictive temperatures. The mps2-2 mutant is unable to insert a duplicated SPB into the nuclear envelope and thus gives rise to cells with monopolar spindles at 37°C (47). A weak intranuclear localization was seen for Yaf9-GFP in an mps2-2 mutant at the permissive temperature of 24°C (Fig. 7A) as well as in the isogenic wild type created by transforming the mps2-2 mutant with a plasmid containing the wild-type MPS2 gene (Fig. 7C). Yaf9-GFP remained nuclear in these cells at the restrictive temperature of 37°C; however, the intensity of the Yaf9-GFP signal increased in the mps2-2 mutant after incubation at 37°C for 3 h (Fig. 7B) but not in the isogenic wild type (Fig. 7D). The immunofluorescence localization of Yaf9-3HA was only weakly visible in wild-type cells (data not shown). However, when these cells were incubated in nocodazole for 3 h, the nuclear localization of Yaf9-3HA was readily apparent (Fig. 7E). The combined results on the intracellular localization of Yaf9 suggested that it was a nuclear protein whose levels increased in conditions in which the integrity of the mitotic spindle was affected. We determined more precisely the quantity of Yaf9 in nocodazole-treated cells by immunoblotting. Yaf9 levels increased 2.5-fold within 2 h of treatment with nocodazole (Fig. 7F). In contrast, the levels of the Esa1 catalytic subunit of the NuA4 complex remained constant in the presence of nocodazole (Fig. 7F). Thus, Yaf9 protein levels seem to be increased in response to spindle stress.
FIG. 7.
Yaf9 is a nuclear protein whose levels increase in response to spindle stress. (A to D) Yeast cells were observed by fluorescence microscopy to localize Yaf9-GFP with regard to nuclear DNA stained with DAPI. No signal was detectable with the GFP filter set for the parental control strain that did not express Yaf9-GFP. (A and B) mps2-2 YAF9-GFP (ILM74) cells grown at 24°C (A) or at 37°C (B) for 3 h; (C and D)mps2-2 YAF9-GFP/pMPS2 (ILM74/pli23) cells grown at 24°C (C) or at 37°C (D) for 3 h. (E) Wild-type/Yaf9-3HA cells (ILM49) growing in YPD at 30°C were treated with 15 μg of nocodazole/ml for 3 h. Cells were then fixed for immunofluorescence with anti-HA monoclonal antibodies, and nuclear DNA was stained with DAPI. No immunofluorescent signal was visible for the parental control strain that did not express Yaf9-3HA. (F) Western blot showing the levels of Yaf9 and Esa1 in whole-cell protein extracts prepared from wild-type cells (YPH499) treated with 15 μg of nocodazole/ml in YPD at 30°C for the indicated periods of time. The Yaf9 and Esa1 protein bands were quantified with a Fluorochem Imager. WT, wild type.
Altered gene regulation in the yaf9Δ mutant.
ESA1, the gene encoding the catalytic acetyltransferase subunit of NuA4, is essential for yeast cell viability (12). Analysis of esa1 conditional mutants has shown that it is required for the bulk of histone H4 N-terminal acetylation and for the correct expression of a subset of yeast genes (19, 39, 54). Since Yaf9 is a nonessential subunit of NuA4, it cannot be required for all NuA4 functions. We used yeast genome microarrays to explore any differences in the transcriptomes of yaf9Δ and wild-type cells that were treated with nocodazole for 3 h, since perturbations in gene expression could contribute to the phenotypes of the yaf9Δ mutant. This analysis identified 11 genes whose expression was reduced at least 2.5-fold in the yaf9Δ mutant compared to the wild type (including the YAF9 gene itself, as is expected since this sequence is deleted in the yaf9Δ strain) and 12 genes whose expression was elevated at least threefold in the yaf9Δ mutant compared to the wild type (Fig. 8A). We used the RT-PCR to verify the microarray results for 10 genes that were hypoexpressed in the yaf9Δ mutant relative to the wild type (Fig. 8B). RT-PCR analysis confirmed the results of the microarray results and showed that most of the genes that were underexpressed in yaf9Δ in the presence of nocodazole were also underexpressed in the mutant in the absence of nocodazole. However, one gene (AHP1/YLR109w encoding alkyl hydroxyperoxidase reductase) seemed to be expressed normally in the yaf9Δ mutant in the absence of nocodazole but was underexpressed in the presence of nocodazole. AHP1 thus seems to be induced in the wild type in the presence of nocodazole in a manner dependent on Yaf9.
None of the genes whose expression is affected in the yaf9Δ mutant encode proteins that are obviously related to MT stability. Nine of the ten genes that are hypoexpressed in the yaf9Δ mutant relative to the wild type are nonessential for viability, including AHP1. We thus tested the benomyl sensitivity of individual yeast strains from which each of these genes had been deleted. All nine mutant strains had a wild-type level of benomyl resistance (data not shown). Individual underexpression of these genes in the yaf9Δ mutant thus cannot be responsible for its benomyl sensitivity. The essential YLR424W gene that is also underexpressed in the yaf9Δ mutant encodes a protein that copurifies with a large mRNA splicing complex that contains the Cef1 protein (20). Interestingly, cef1 mutants are hypersensitive to benomyl at their permissive temperature and arrest preferentially in mitosis at their restrictive temperature principally due to defects in the splicing of the intron from the TUB1 gene encoding the major isoform of alpha tubulin (7). Replacing the wild-type TUB1 gene with an intronless version of the gene suppressed these phenotypes in the cef1 mutant. However, replacing the wild-type TUB1 gene with an intronless version of the gene did not suppress the benomyl sensitivity of the yaf9Δ mutant (data not shown). Thus, gene expression defects may not be responsible for this phenotype.
The yng2Δ mutant is sensitive to benomyl, and the benomyl resistance of the wild-type strain is increased by TSA, an inhibitor of HDACs.
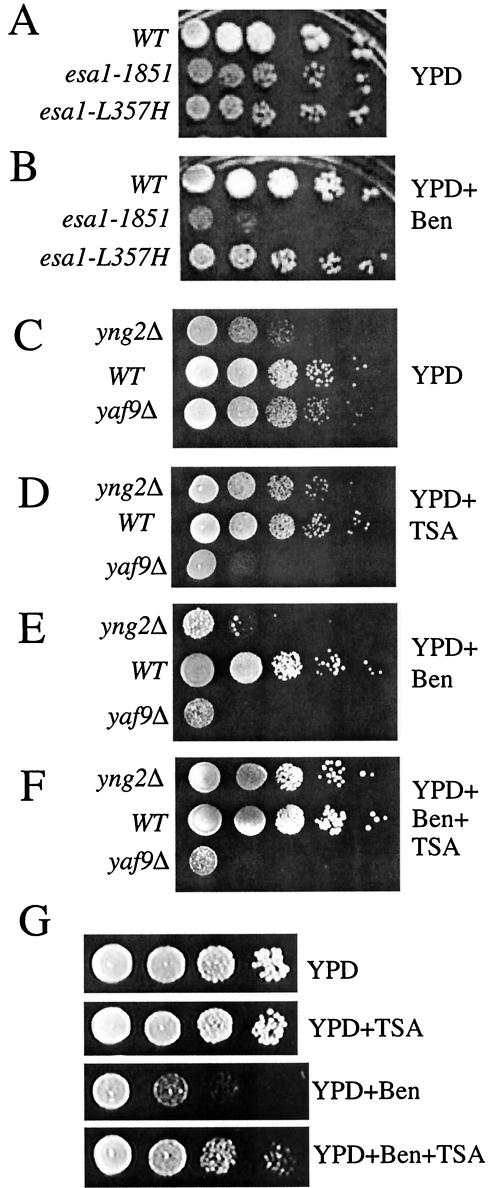
The sensitivity of the yaf9Δ mutant might be due to a defect in NuA4 complex function, or it might be due to a function that is specific to Yaf9 and distinct from any possible function it might have as part of the NuA4 complex. We examined three other NuA4 mutants—two different esa1 temperature-sensitive mutants (4) and a yng2Δ mutant (11)—for their sensitivity to benomyl in order to test for a possible function of NuA4 in the cellular response to MT depolymerization. The esa1-1851 mutant was hypersensitive to benomyl at 30°C, whereas the esa1-L357H mutant was not (Fig. 9A and B). Thus, reduced acetylated histone H4 levels in the esa1-1851 mutant (Fig. 6C) is correlated with hypersensitivity to benomyl. The yng2Δ mutant was also sensitive to benomyl (Fig. 9C and E). Global histone H4 acetylation is reduced in the yng2Δ mutant, and these cells grow more poorly than in the wild type, but Yng2 is not required for viability (11, 39, 50). The sensitivity of the esa1-1851 and yng2Δ mutants to benomyl suggests that NuA4 acetylation of some substrate is required for normal cellular resistance to benomyl. This interpretation is supported by the effects of TSA, an inhibitor of histone deacetylases (HDACs) (71), on the resistance of yeast cells to benomyl. The resistance of wild-type yeast cells to benomyl was increased when cells were grown in the presence of 30 μg of TSA/ml (Fig. 9E to G). This result suggests that the hyperacetylation of some cellular protein or proteins increases the resistance of wild-type yeast cells to benomyl. TSA has been shown to partially suppress the slow growth phenotype, the G2/M accumulation, and the histone H4 hypoacetylation of the yng2Δ mutant (11). We thus examined the effect of TSA on the benomyl sensitivity of the yng2Δ and yaf9Δ mutants. As for its other known phenotypes, TSA suppressed the benomyl hypersensitivity of the yng2Δ mutant (Fig. 9E and F). Remarkably, however, the opposite effect was observed with regards to the yaf9Δ mutant: TSA inhibited its growth in the absence of benomyl and did not suppress its sensitivity to benomyl (Fig. 9C to F). The benomyl sensitivity of the yaf9Δ, yng2Δ, and esa1-1851 mutants suggests that they have common defects that cause this phenotype, but the opposing response of the yaf9Δ and yng2Δ mutants to TSA indicates that they must also have some distinct biochemical functions.
FIG. 9.
The esa1-1851 and yng2Δ mutants are hypersensitive to benomyl, and the HDAC inhibitor TSA differentially affects the growth and benomyl resistance of wild type and NuA4 HAT mutants. (A and B) Wild type (W303-1a) and esa1-1851 (MSY2431) and esa1-L357H (MSY2432) mutants were spotted onto either YPD alone or YPD plus 15 μg of benomyl/ml at 30°C for 3 days. (C to F) Tenfold serial dilutions of exponentially growing yng2Δ (CMY1237), wild-type (W303-1a), and yaf9Δ (ILM63) cells in YPD at the same cell density were spotted onto plates containing the indicated media and incubatedat 24°C for 1 day (A and B) or 4 days (C and D). YPD+TSA is YPD containing TSA at 30 μg/ml, YPD+Ben is YPD containing benomyl at 10 μg/ml, YPD+Ben+TSA is YPD containing benomyl at 10 μg/ml and TSA at 30 μg/ml. (G) Wild-type (W303-1a) cells were spotted on the indicated media and incubated at 24°C for 2 days. WT, wild type.
Acetylation of histone H4 is required for benomyl resistance.
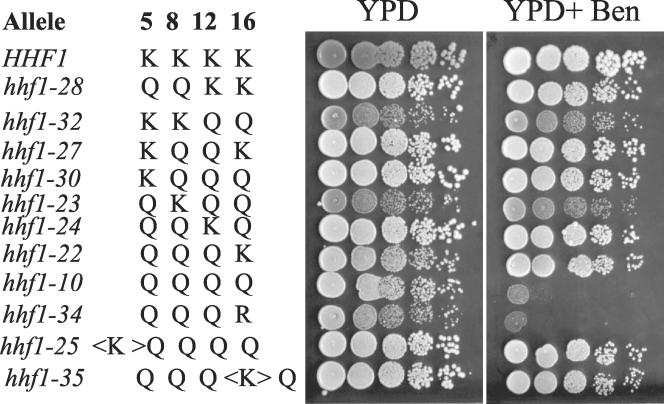
The N-terminal lysines of histone H4 are the best-characterized substrates of the NuA4 HAT. We thus tested nonacetylatable mutants of histone H4 (42) for their benomyl sensitivity (Fig. 10) Mutants containing at least one of four N-terminal lysines were nearly as resistant to benomyl as the wild type. Strikingly, nonacetylatable histone H4 mutants in which all four N-terminal lysine residues (K5, K8, K12, and K16) of histone H4 are mutated to glutamine (hhf1-10) or glutamine plus arginine (hhf1-34) were hypersensitive to benomyl. Insertion of an ectopic lysine to the quadruple lysine-to-glutamine mutant (hhf1-25 and hhf1-35) restored resistance to benomyl. These results strongly suggest that the acetylation of one or more N-terminal lysines of histone H4 by the NuA4 HAT is required for cellular resistance to benomyl.
FIG. 10.
Nonacetylatable histone H4 mutants are hypersensitive to benomyl. Wild-type and mutant cells at the same cell density in YPD were spotted in 10-fold serial dilutions onto YPD plates without benomyl (YPD) or with 15 μg of benomyl/ml (Ben), followed by incubation at 28°C for 3 days. For each mutant allele, the amino acid substitutions at positions 5, 8, 12, and 16 or the N-terminal tail are indicated as K (lysine), Q (glutamine), or R (arginine). Allele hhf1-25 contains an insertion of G-K-G at position 3 of the hhf1-10 tail (designated <K>QQQQ), whereas allele hhf1-35 contains an insertion of G-K-G at position 12 of the hhf1-10 tail (designated QQQ<K>Q). Both hhf1-10 and hhf1-34 are roughly 5 orders of magnitude hypersensitive to benomyl and are the only mutants that lack any acetylatable lysine in their histone H4 tail.
DISCUSSION
Ynl107w/Yaf9 is a novel subunit of the NuA4 HAT complex containing a YEATS domain.
A recent systematic characterization of protein complexes in yeast indicated that Ynl107/Yaf9 copurifies with some known subunits of the NuA4 HAT complex (20). In agreement with that study and with unpublished work from other laboratories (B. Cairns, unpublished data), we found that Ynl107w/Yaf9 is in a large-molecular-weight complex and that it coprecipitates with three known subunits of NuA4: Esa1, Epl1, and Yng2. Analysis of the amino acid sequence of Ynl107/Yaf9 indicates that it contains an N-terminal YEATS domain and a predicted C-terminal coiled-coil sequence. Interestingly, two other yeast proteins, Sas5 and Taf14 (Anc1/Tfg3/Taf30), also contain YEATS domains and are associated with histone acetylation complexes. Sas5 is a subunit of the Sas2 HAT complex (43, 51) and Taf14 (Anc1/Tfg3/Taf30) is a component of the NuA3 (Sas3) HAT complex (32). However, Taf14 (Anc1/Tfg3/Taf30) is also associated with the Swi/Snf ATPase remodeling complex (8) and the TFIIF (25) and TFIID (58) RNA polymerase II transcription complexes. Unfortunately, the structure and the function of the YEATS domain have not yet been determined, and the precise function of these proteins within their protein complexes is not known. Esa1, the catalytic acetyltransferase subunit of NuA4 is essential for yeast cell viability, but the ynl107wΔ/yaf9Δ mutant has little or no growth defect at 24 to 30°C. Thus, Ynl107/Yaf9 is not necessary for the essential functions of NuA4. Moreover, the global levels of histone H4 acetylation were not significantly diminished in the yaf9Δ mutant (Fig. 6C). One interesting possibility for Yaf9 function is that it may target the NuA4 complex to acetylate histone H4 at some specific genomic locations. This idea could by tested by genome-wide chromatin IP experiments (55).
Ynl107 was named Yaf9 because of its similarity to human AF9, a protein that induces acute leukemia when fused to the N-terminal portion of the MLL gene product (13). However, this similarity is restricted to the YEATS domain of the two proteins, and our sequence analysis and the predicted secondary structure of Ynl107 indicates that the human Gas41 and its metazoan homologs are more similar to Ynl107 than are the other YEATS domain proteins. Gas41 is a sequence that was identified by its amplification in human gliomas (18, 46). It will be of great interest to determine whether Gas41 is a subunit of a NuA4-like HAT complex in animal cells and whether YEATS domain proteins are generally associated with HAT complexes.
Yaf9, the NuA4 HAT complex, and histone H4 acetylation are required for normal cellular resistance to spindle stress.
We isolated Yaf9 in a two-hybrid screen with Mps2 as bait (36). Mps2 is a component of the SPB and the nuclear envelope that is required for insertion of the newly duplicated SPB into the nuclear envelope (47). It is thus necessary for establishing a normal bipolar spindle. We sought to test the physiological significance of this interaction and discovered that the yaf9Δ mutant is synthetically lethal with two mitotic mutants that we tested, i.e., the bbp1-1 and spc24-11 mutants. Remarkably, these mutants affect two distinct mitotic functions: Bbp1 is required along with Mps2 for insertion of the SPB into the nuclear envelope (60), whereas Spc24 is a kinetochore subunit that is required for stable binding of kinetochores to spindle MTs (30, 36, 70). Furthermore, we discovered that the yaf9Δ mutant is hypersensitive to MT depolymerizing agents such as nocodazole and benomyl. These results show that Yaf9 is required for viability under conditions in which the integrity of the mitotic spindle is compromised.
Interestingly, Gas41, the human protein that is most similar to Yaf9, has been found to interact with the NumA (23) and Tacc1 (35) proteins. In animal cells in mitosis, NumA is concentrated near centrosomes and helps organize the minus ends of spindle MTs (44). Tacc1 was also implicated in spindle structure (21). Given the intriguing similarities in the interactions between Yaf9-Mps2 and Gas41-NumA+Tacc1, it would be worthwhile to test for a possible role for Gas41 in responding to spindle stress in animal cells.
Phenotypic analysis of the yaf9Δ mutant indicates that its sensitivity to MT depolymerizing agents is not due to an obvious defect in the mitotic spindle checkpoint pathways. The yaf9Δ mutant does not exit mitosis prematurely in the presence of nocodazole as do the spindle checkpoint mutants. Instead, the MTs of the yaf9Δ mutant disassemble more readily than those of the wild type at the same concentration of nocodazole, and reassembly of MTs in the presence of limiting concentrations of nocodazole is inhibited in the yaf9Δ mutant. Furthermore, the yaf9Δ mutant is blocked in mitosis at lower concentrations of nocodazole than the wild type and recovers from this mitotic arrest less efficiently. Since Yaf9 is a subunit of the NuA4 histone acetylase complex, we tested the need of this complex for benomyl resistance by examining other NuA4 mutants. We found that the esa1-1851 and yng2Δ mutants are also hypersensitive to benomyl, and these mutants are known to have reduced levels of histone H4 acetylation (4, 11, 39, 50). Furthermore, we found that nonacetylatable histone H4 mutants were also hypersensitive to benomyl. In these mutants, the four N-terminal lysines of histone H4 were mutated to glutamine or to glutamine plus arginine. The similarity of these mutants to the NuA4 mutants strongly suggests that histone H4 is the relevant substrate that must be acetylated by NuA4 in order to confer a cellular resistance to benomyl. Yaf9 may target the NuA4 complex to specific genomic sites where histone H4 acetylation is important for responding to mitotic stress.
There are several possible ways to explain the sensitivity of NuA4 mutants and nonacetylatable histone H4 mutants to spindle stress. One possibility is through defects in the expression of genes encoding proteins involved in MT assembly or structure, since histone H4 acetylation is implicated in transcriptional regulation (56, 63). We identified 10 genes that were underexpressed in the yaf9Δ mutant relative to the wild type and 12 genes that were overexpressed. None of these genes are obviously involved in spindle functions, and we directly tested and ruled out a phenotypic contribution for the 10 genes that are hypoexpressed in the yaf9Δ mutant. Although we cannot exclude an effect of perturbed gene expression on the benomyl hypersensitivity of yaf9Δ, our current data do not favor this model.
Another possibility is that the acetylation of histone H4 is required for the recruitment of specific proteins to chromatin (31), some of which may be necessary to form a structurally resistant spindle. Recruitment of the Ran GTPase and its GDP/GTP exchange factor Rcc1 to chromatin is important for proper mitotic spindle assembly in Xenopus extracts and human cells (10, 45). Ran and its regulators also influence spindle structure and function in budding yeast (52, 53), fission yeast (57), and worms (2). Ran binds chromatin directly through interactions with histones H3 and H4 and indirectly through binding to Rcc1 that interacts directly with histones H2a and H2b (3, 49). However, docking of Ran to chromatin does not appear to require the N-terminal extremities of histones H3 and H4 (3), which suggests that its chromatin binding might not be affected in NuA4 mutants or the nonacetylatable histone H4 mutant. Another possibility is that centromeric histone acetylation directly or indirectly affects kinetochore-MT interactions. In budding yeast, we found that treatment of wild-type yeast cells with TSA, an HDAC inhibitor, increases their resistance to benomyl. Thus, the degree of acetylation of some protein in budding yeast, presumably histone H4, is limiting its resistance to benomyl. This result is in contrast to the effect of TSA in fission yeast (15) and animal (67) cells. In the latter two cases, TSA treatment perturbs the heterochromatic centromeric chromatin and leads to increased rates of chromosome loss. Budding yeast centromeres are smaller and simpler than those of fission yeast and animal cells. The absence of extensive heterochromatin around the budding yeast centromeres may have allowed us to observe a positive effect of protein acetylation on spindle stability. In eukaryotes containing extensive centromeric heterochromatin, kinetochore perturbation by histone hyperacetylation of centromeric heterochromatin may be dominant to any potential beneficial effects of TSA treatment on spindle stability. Determining the exact mechanism by which protein acetylation increases spindle stability in budding yeast will allow us to test for its conservation in other eukaryotes.
Acknowledgments
We are grateful to Michel Werner for establishing the yeast microarray facilities at the SBGM and to Véronique Bordas-Lefloch for experimental advice with the microarrays. We thank Caroline Dubacq, Christophe Leroy, Marie-Claude Marsolier, and the Cell Cycle group for their invaluable discussions and support and Domenico Libri for his warm welcome, encouragement, and support. We thank John Choy, Steve Kron, Erik Spedale, Lorraine Pilus, and Kathy Gould for strains and advice.
I.L.M. was supported by an allocation from the French Ministère de l'Education Nationale, de la Recherche, et de la Technologie and by stipend support from the Association pour la Recherche sur le Cancer (ARC). M.M.S. was supported by NIH grant GM28920, and C.M. received funding from the Association pour la Recherche sur le Cancer (ARC 4470) and from a collaborative program between the CEA and the Curie Institute on epigenetic parameters in the response to genotoxic agents and the control of the cell cycle.
REFERENCES
- 1.Allard, S., R. T. Utley, J. Savard, A. Clarke, P. Grant, C. J. Brandl, L. Pillus, J. L. Workman, and J. Cote. 1999. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex containing Esa1p and the ATM-related cofactor Tra1p. EMBO J. 18:5108-5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Askjaer, P., V. Galy, E. Hannak, and I. W. Mattaj. 2002. Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol. Biol. Cell 13:4355-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bilbao-Cortes, D., M. Hetzer, G. Langst, P. B. Becker, and I. W. Mattaj. 2002. Ran binds to chromatin by two distinct mechanisms. Curr. Biol. 12:1151-1156. [DOI] [PubMed] [Google Scholar]
- 4.Bird, A. W., D. Y. Yu, M. G. Pray-Grant, Q. Qiu, K. E. Harmon, P. C. Megee, P. A. Grant, M. M. Smith, and M. F. Christman. 2002. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419:411-415. [DOI] [PubMed] [Google Scholar]
- 5.Birrell, G. W., G. Giaever, A. M. Chu, R. W. Davis, and J. M. Brown. 2001. A genome-wide screen in Saccharomyces cerevisiae for genes affecting UV radiation sensitivity. Proc. Natl. Acad. Sci. USA 98:12608-12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boeke, J. D., F. LaCroute, and G. R. Fink. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345-346. [DOI] [PubMed] [Google Scholar]
- 7.Burns, C. G., R. Ohi, S. Mehta, E. T. O'Toole, M. Winey, T. A. Clark, C. W. Sugnet, M. Ares, Jr., and K. L. Gould. 2002. Removal of a single alpha-tubulin gene intron suppresses cell cycle arrest phenotypes of splicing factor mutations in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:801-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns, B. R., N. L. Henry, and R. D. Kornberg. 1996. TFG/TAF30/ANC1, a component of the yeast SWI/SNF complex that is similar to the leukemogenic proteins ENL and AF-9. Mol. Cell. Biol. 16:3308-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Callebaut, I., G. Labesse, P. Durand, A. Poupon, L. Canard, J. Chomilier, B. Henrissat, and J. P. Mornon. 1997. Deciphering protein sequence information through hydrophobic cluster analysis (HCA): current status and perspectives. Cell Mol. Life Sci. 53:621-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carazo-Salas, R. E., G. Guarguaglini, O. J. Gruss, A. Segref, E. Karsenti, and I. W. Mattaj. 1999. Generation of GTP-bound Ran by RCC1 is required for chromatin-induced mitotic spindle formation. Nature 400:178-181. [DOI] [PubMed] [Google Scholar]
- 11.Choy, J. S., B. T. Tobe, J. H. Huh, and S. J. Kron. 2001. Yng2p-dependent NuA4 histone H4 acetylation activity is required for mitotic and meiotic progression. J. Biol. Chem. 276:43653-43662. [DOI] [PubMed] [Google Scholar]
- 12.Clarke, A. S., J. E. Lowell, S. J. Jacobson, and L. Pillus. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 19:2515-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corral, J., I. Lavenir, H. Impey, A. J. Warren, A. Forster, T. A. Larson, S. Bell, A. N. McKenzie, G. King, and T. H. Rabbitts. 1996. An Mll-AF9 fusion gene made by homologous recombination causes acute leukemia in chimeric mice: a method to create fusion oncogenes. Cell 85:853-861. [DOI] [PubMed] [Google Scholar]
- 14.Eberharter, A., and P. B. Becker. 2002. Histone acetylation: a switch between repressive and permissive chromatin. EMBO Rep. 3:224-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ekwall, K., T. Olsson, B. M. Turner, G. Cranston, and R. C. Allshire. 1997. Transient inhibition of histone deacetylation alters the structural and functional imprint at fission yeast centromeres. Cell 91:1021-1032. [DOI] [PubMed] [Google Scholar]
- 16.Elledge, S. J. 1996. Cell cycle checkpoints: preventing an identity crisis. Science 274:1664-1672. [DOI] [PubMed] [Google Scholar]
- 17.Fauchon, M., G. Lagniel, J. C. Aude, L. Lombardia, P. Soularue, C. Petat, G. Marguerie, A. Sentenac, M. Werner, and J. Labarre. 2002. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell 9:713-723. [DOI] [PubMed] [Google Scholar]
- 18.Fischer, U., D. Heckel, A. Michel, M. Janka, T. Hulsebos, and E. Meese. 1997. Cloning of a novel transcription factor-like gene amplified in human glioma including astrocytoma grade I. Hum. Mol. Genet. 6:1817-1822. [DOI] [PubMed] [Google Scholar]
- 19.Galarneau, L., A. Nourani, A. A. Boudreault, Y. Zhang, L. Heliot, S. Allard, J. Savard, W. S. Lane, D. J. Stillman, and J. Cote. 2000. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol. Cell 5:927-937. [DOI] [PubMed] [Google Scholar]
- 20.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 21.Gergely, F. 2002. Centrosomal TACCtics. Bioessays 24:915-925. [DOI] [PubMed] [Google Scholar]
- 22.Godon, C., G. Lagniel, J. Lee, J. M. Buhler, S. Kieffer, M. Perrot, H. Boucherie, M. B. Toledano, and J. Labarre. 1998. The H2O2 stimulon in Saccharomyces cerevisiae. J. Biol. Chem. 273:22480-22489. [DOI] [PubMed] [Google Scholar]
- 23.Harborth, J., K. Weber, and M. Osborn. 2000. GAS41, a highly conserved protein in eukaryotic nuclei, binds to NuMA. J. Biol. Chem. 275:31979-31985. [DOI] [PubMed] [Google Scholar]
- 24.Hasan, S., and M. O. Hottiger. 2002. Histone acetyl transferases: a role in DNA repair and DNA replication. J. Mol. Med. 80:463-474. [DOI] [PubMed] [Google Scholar]
- 25.Henry, N. L., A. M. Campbell, W. J. Feaver, D. Poon, P. A. Weil, and R. D. Kornberg. 1994. TFIIF-TAF-RNA polymerase II connection. Genes Dev. 8:2868-2878. [DOI] [PubMed] [Google Scholar]
- 26.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 27.Hutter, K. J., and H. E. Eipel. 1979. Microbial determinations by flow cytometry. J. Gen. Microbiol. 113:369-375. [DOI] [PubMed] [Google Scholar]
- 28.Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang, M. Horikoshi, R. Scully, J. Qin, and Y. Nakatani. 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102:463-473. [DOI] [PubMed] [Google Scholar]
- 29.Jallepalli, P. V., and C. Lengauer. 2001. Chromosome segregation and cancer: cutting through the mystery. Nat. Rev. Cancer. 1:109-117. [DOI] [PubMed] [Google Scholar]
- 30.Janke, C., J. Ortiz, J. Lechner, A. Shevchenko, M. M. Magiera, C. Schramm, and E. Schiebel. 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20:777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 32.John, S., L. Howe, S. T. Tafrov, P. A. Grant, R. Sternglanz, and J. L. Workman. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14:1196-1208. [PMC free article] [PubMed]
- 33.Kilmartin, J. V., and A. E. Adams. 1984. Structural rearrangements of tubulin and actin during the cell cycle of the yeast Saccharomyces. J. Cell Biol. 98:922-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura, A., T. Umehara, and M. Horikoshi. 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32:370-377. [DOI] [PubMed] [Google Scholar]
- 35.Lauffart, B., S. J. Howell, J. E. Tasch, J. K. Cowell, and I. H. Still. 2002. Interaction of the transforming acidic coiled-coil 1 (TACC1) protein with ch-TOG and GAS41/NuBI1 suggests multiple TACC1-containing protein complexes in human cells. Biochem. J. 363:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Masson, I., C. Saveanu, A. Chevalier, A. Namane, R. Gobin, M. Fromont-Racine, A. Jacquier, and C. Mann. 2002. Spc24 interacts with Mps2 and is required for chromosome segregation, but is not implicated in spindle pole body duplication. Mol. Microbiol. 43:1431-1443. [DOI] [PubMed] [Google Scholar]
- 37.Lemesle-Varloot, L., B. Henrissat, C. Gaboriaud, V. Bissery, A. Morgat, and J. P. Mornon. 1990. Hydrophobic cluster analysis: procedures to derive structural and functional information from 2-D representation of protein sequences. Biochimie 72:555-574. [DOI] [PubMed] [Google Scholar]
- 38.Lengauer, C., K. W. Kinzler, and B. Vogelstein. 1998. Genetic instabilities in human cancers. Nature 396:643-649. [DOI] [PubMed] [Google Scholar]
- 39.Loewith, R., M. Meijer, S. P. Lees-Miller, K. Riabowol, and D. Young. 2000. Three yeast proteins related to the human candidate tumor suppressor p33ING1 are associated with histone acetyltransferase activities. Mol. Cell. Biol. 20:3807-3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 41.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 42.Megee, P. C., B. A. Morgan, and M. M. Smith. 1995. Histone H4 and the maintenance of genome integrity. Genes Dev. 9:1716-1727. [DOI] [PubMed] [Google Scholar]
- 43.Meijsing, S. H., and A. E. Ehrenhofer-Murray. 2001. The silencing complex SAS-I links histone acetylation to the assembly of repressed chromatin by CAF-I and Asf1 in Saccharomyces cerevisiae. Genes Dev. 15:3169-3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merdes, A., R. Heald, K. Samejima, W. C. Earnshaw, and D. W. Cleveland. 2000. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149:851-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Moore, W., C. Zhang, and P. Clarke. 2002. Targeting of RCC1 to chromosomes is required for proper mitotic spindle assembly in human cells. Curr. Biol. 12:1442-1447. [DOI] [PubMed]
- 46.Munnia, A., N. Schutz, B. F. Romeike, E. Maldener, B. Glass, R. Maas, W. Nastainczyk, W. Feiden, U. Fischer, and E. Meese. 2001. Expression, cellular distribution and protein binding of the glioma amplified sequence (GAS41), a highly conserved putative transcription factor. Oncogene 20:4853-4863. [DOI] [PubMed] [Google Scholar]
- 47.Munoz-Centeno, M. C., S. McBratney, A. Monterrosa, B. Byers, C. Mann, and M. Winey. 1999. Saccharomyces cerevisiae MPS2 encodes a membrane protein localized at the spindle pole body and the nuclear envelope. Mol. Biol. Cell 10:2393-2406. [DOI] [PMC free article] [PubMed]
- 48.Musacchio, A., and K. G. Hardwick. 2002. The spindle checkpoint: structural insights into dynamic signaling. Nat. Rev. Mol. Cell. Biol. 3:731-741. [DOI] [PubMed] [Google Scholar]
- 49.Nemergut, M. E., C. A. Mizzen, T. Stukenberg, C. D. Allis, and I. G. Macara. 2001. Chromatin docking and exchange activity enhancement of RCC1 by histones H2A and H2B. Science 292:1540-1543. [DOI] [PubMed] [Google Scholar]
- 50.Nourani, A., Y. Doyon, R. T. Utley, S. Allard, W. S. Lane, and J. Cote. 2001. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol. Cell. Biol. 21:7629-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Osada, S., A. Sutton, N. Muster, C. E. Brown, J. R. Yates III, R. Sternglanz, and J. L. Workman. 2001. The yeast SAS (something about silencing) protein complex contains a MYST-type putative acetyltransferase and functions with chromatin assembly factor ASF1. Genes Dev. 15:3155-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ouspenski, I. I., U. W. Mueller, A. Matynia, S. Sazer, S. J. Elledge, and B. R. Brinkley. 1995. Ran-binding protein-1 is an essential component of the Ran/RCC1 molecular switch system in budding yeast. J. Biol. Chem. 270:1975-1978. [DOI] [PubMed] [Google Scholar]
- 53.Quimby, B. B., C. A. Wilson, and A. H. Corbett. 2000. The interaction between Ran and NTF2 is required for cell cycle progression. Mol. Biol. Cell 11:2617-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reid, J. L., V. R. Iyer, P. O. Brown, and K. Struhl. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell 6:1297-1307. [DOI] [PubMed]
- 55.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 56.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 57.Salus, S. S., J. Demeter, and S. Sazer. 2002. The Ran GTPase system in fission yeast affects microtubules and cytokinesis in cells that are competent for nucleocytoplasmic protein transport. Mol. Cell. Biol. 22:8491-8505. [DOI] [PMC free article] [PubMed]
- 58.Sanders, S. L., and P. A. Weil. 2000. Identification of two novel TAF subunits of the yeast Saccharomyces cerevisiae TFIID complex. J. Biol. Chem. 275:13895-13900. [DOI] [PubMed] [Google Scholar]
- 59.Sayed, M., S. Pelech, C. Wong, A. Marotta, and B. Salh. 2001. Protein kinase CK2 is involved in G2 arrest and apoptosis following spindle damage in epithelial cells. Oncogene 20:6994-7005. [DOI] [PubMed] [Google Scholar]
- 60.Schramm, C., S. Elliott, A. Shevchenko, and E. Schiebel. 2000. The Bbp1p-Mps2p complex connects the SPB to the nuclear envelope and is essential for SPB duplication. EMBO J. 19:421-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scolnick, D. M., and T. D. Halazonetis. 2000. Chfr defines a mitotic stress checkpoint that delays entry into metaphase. Nature 406:430-435. [DOI] [PubMed] [Google Scholar]
- 62.Sherman, F. 1991. Getting started with yeast. Methods Enzymol. 194:3-21. [DOI] [PubMed] [Google Scholar]
- 63.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Straight, A., and A. W. Murray. 1997. The spindle assembly checkpoint in budding yeast. Methods Enzymol. 283:425-440. [DOI] [PubMed] [Google Scholar]
- 65.Straight, A. F., W. F. Marshall, J. W. Sedat, and A. W. Murray. 1997. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science 277:574-578. [DOI] [PubMed] [Google Scholar]
- 66.Suka, N., K. Luo, and M. Grunstein. 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32:378-383. [DOI] [PubMed] [Google Scholar]
- 67.Taddei, A., C. Maison, D. Roche, and G. Almouzni. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3:114-120. [DOI] [PubMed] [Google Scholar]
- 68.Wang, T. H., H. S. Wang, and Y. K. Soong. 2000. Paclitaxel-induced cell death: where the cell cycle and apoptosis come together. Cancer 88:2619-2628. [DOI] [PubMed] [Google Scholar]
- 69.Wang, Y., and D. J. Burke. 1995. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 15:6838-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wigge, P. A., and J. V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]